Background: San1p is a nuclear ubiquitin ligase that helps degrade misfolded cytoplasmic proteins in yeast.
Results: The Hsp70, Ssa1p, facilitates an interaction between a novel misfolded substrate and San1p.
Conclusion: Chaperones play a direct role in bridging aberrant cytoplasmic proteins to ubiquitin ligases.
Significance: Understanding how chaperones select and target cytoplasmic proteins will help define how diseases associated with proteotoxic stress might be treated.
Keywords: E3 Ubiquitin Ligase, ERAD, Molecular Chaperone, Proteasome, Ubiquitin, Cdc48, Hsp40, Hsp70, Ssa1p
Abstract
Accumulation of misfolded proteins in cellular compartments can result in stress-induced cell death. In the endoplasmic reticulum (ER), ER-associated degradation clears aberrant proteins from the secretory pathway. In the cytoplasm and nucleus, this job is left to the cytoplasmic quality control (CytoQC) machinery. Both processes utilize chaperones and the ubiquitin-proteasome system to aid in protein elimination. Previous studies in yeast have drawn comparisons between these processes using data from structurally and topologically different substrates. We sought to draw a direct comparison between ERAD and CytoQC by studying the elimination of a single misfolded domain that, depending on its residence, is disposed by either of these pathways. The truncated, second nucleotide binding domain (NBD2*) from a yeast ERAD substrate, Ste6p*, resides at the cytoplasmic face of the ER. We show that a soluble form of NBD2* is cytoplasmic and unlike wild-type NBD2 is targeted for proteasome-mediated degradation. In contrast to Ste6p*, which employs the ER-localized Doa10p ubiquitin ligase, NBD2* is ubiquitinated by a nuclear E3 ligase San1p, a factor that is also required for its degradation. Although the yeast cytoplasmic Hsp70 chaperone, Ssa1p, has been thought to facilitate the nuclear import or to maintain the solubility of most CytoQC substrates, we discovered that Ssa1p facilitates the interaction between San1p and NBD2*, demonstrating that chaperones can aid in substrate recognition and San1p-dependent protein degradation. These results emphasize the diverse action of molecular chaperones during CytoQC.
Introduction
Misfolded or slowly folding proteins can aggregate, resulting in detrimental effects on cellular health. In eukaryotic cells, there are a variety of mechanisms that lead to the recognition and degradation of misfolded proteins. Ultimately, these misfolded substrates are eliminated by lysosomal/vacuolar degradation after transport through the secretory pathway, via autophagy, or by the ubiquitin-proteasome system (UPS).2 In addition to degrading misfolded cytoplasmic proteins by the cytoplasmic quality control (CytoQC) pathway, the UPS is also required for endoplasmic reticulum-associated degradation (ERAD) (1, 2). ERAD has been linked to a variety of human diseases, including cystic fibrosis and Gaucher disease, whereas the aggregation of cytoplasmic proteins can contribute to neurodegeneration, as evident in Huntington disease (3).
During ERAD, misfolded proteins are recognized by molecular chaperones, including heat shock proteins (Hsp) of ∼40, 70, and 90 kDa, ER-resident lectins, and protein-disulfide isomerases, each of which monitors folding status and can direct a terminally misfolded protein for degradation (4–7). After recognition, misfolded proteins are retrotranslocated into the cytoplasm by the action of the cytoplasmic Cdc48p complex (p97/VCP in mammals), where the substrate is polyubiquitinated by ER-localized E3 ubiquitin ligases. In yeast, the two ubiquitin ligases that function in ERAD, Hrd1p and Doa10p, are both integral membrane proteins, and in mammals there is a growing number of ubiquitin ligases that participate in ERAD and reside in both the ER and cytoplasm (8). Concomitant with continued retrotranslocation, ERAD substrates are ultimately degraded by the 26 S proteasome (9–12).
Although many of the underlying mechanistic details of the ERAD pathway have been deciphered, significantly less is known about protein quality control in the cytoplasm. Some defective cytoplasmic proteins can be sequestered in cytoplasmic inclusions (13–15), which are then destroyed via autophagy (16–20). In contrast, soluble substrates can be handled by CytoQC. Similar to ERAD, molecular chaperones play an important role during the selection and disposal of CytoQC substrates in yeast (21–23). In addition, certain components of the ERAD machinery, including Cdc48p and Doa10p, participate in the CytoQC of some substrates (2, 24, 25). Surprisingly, recent studies indicate that a nuclear E3 ligase, San1p, can contribute to CytoQC in yeast (26, 27). Moreover, Ubr1p, the classical E3 ligase of the N-end rule pathway in yeast (28), also participates in CytoQC (27, 29). These data demonstrate that aberrant cytoplasmic proteins can be ubiquitinated by a variety of E3 ligases, but the exact determinants underlying E3 specificity are largely unclear.
As noted above, cytoplasmic chaperones facilitate the turnover of some CytoQC substrates, and the cytoplasmic Hsp70, Ssa1p, is required for the degradation of several CytoQC substrates. These include Ura3-CL1 (a short artificial degron fused to the Ura3 gene), ΔssPrA (a form of the yeast vacuolar Proteinase A with its signal sequence removed), and Δ2GFP (a soluble GFP with deletion of amino acids 25–36 to disrupt folding). For these substrates, Ssa1p is critical for maintaining substrate solubility and for recruiting the ubiquitination machinery (26, 30). In contrast, for VHL (a tumor suppressor protein expressed in yeast), Ssa1p may actively target the substrate to the UPS, but in this case the E3 ligase that is employed is unknown (21). Ssa1p has also been implicated in the transport of ΔssPrA, Δ2GFP, and ΔssCPY* into the nucleus prior to ubiquitination by San1p (26, 27, 29). However, the mechanism by which Ssa1p facilitates nuclear transport is undefined.
In this study, we examined the requirements for the degradation of a novel CytoQC substrate. The substrate is a soluble version of the second nucleotide binding domain (NBD2) found in the well characterized, integral membrane yeast ERAD substrate, truncated sterile 6 protein (Ste6p*). Truncation of NBD2 results in the proteasome-dependent degradation of Ste6p* (31–33). Importantly, the selection of Ste6p* for ERAD occurs post-translationally and is mediated solely by the truncation of this otherwise well ordered domain. To explore how a soluble version of the Ste6p* NBD2, termed NBD2*, is recognized by the CytoQC machinery, we established expression systems for wild-type NBD2 and for NBD2* in yeast. Other advantages of using NBD2* as a CytoQC substrate are as follows: 1) we can directly compare the known requirements for the ERAD of Ste6p* with the requirements for degradation of NBD2*; 2) unlike ΔssPrA and ΔssCPY*, which reside in the cytoplasm due to deletion of their ER-signal sequences, NBD2* normally resides in the cytoplasm, so misfolding is independent of changes in its native subcellular environment; and 3) an examination of the misfolding of NBD2* provides a relevant system to examine the CytoQC of complex proteins and contrasts with some CytoQC substrates that are recognized based on the presence of short degrons.
Herein, we demonstrate that the cytoplasmic Hsp70, Ssa1p, along with two associated Hsp40s, are required for the efficient degradation of NBD2* and that Ssa1p is required for NBD2* to associate with San1p. This result is supported by our finding that the polyubiquitination and degradation of NBD2* do not require the action of the classical ERAD E3 ligases Doa10p or Hrd1p; rather, NBD2* degradation is primarily triggered by the action of San1p. We also uncovered a role for the Cdc48p complex in NBD2* degradation, as evident for select other CytoQC substrates. Together, these data indicate for the first time that chaperones help target misfolded cytoplasmic proteins to the San1p ubiquitin ligase. Our data also add new insights into the rules that govern the selection and degradation of CytoQC substrates.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Plasmid Construction
Yeast were grown as described previously (34), unless otherwise noted. A complete list of the Saccharomyces cerevisiae strains used in this study is presented in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference/Source |
|---|---|---|
| BY4742 | MATα, his3Δ1, leu2Δ0, lys2Δ0 ura3Δ3 | Ref. 90 |
| pdr5Δ | MATα, his3Δ1, leu2Δ0, ura3Δ3 pdr5Δ::KanMX | Ref. 90 |
| pep4Δ | MATα, his3Δ1, leu2Δ0, ura3Δ3 pep4Δ::KanMX | Ref. 91 |
| SSA1 | MATα, his3-11,15, leu2-3,112, ura3-52. trp1-Δ1, lys2, ssa2-1(LEU2), ssa3-1(TRP1), ssa4-2(LYS2) | Ref. 92 |
| ssa1-45 | MATα, his3-11,15, leu2-3,112, ura3-52. trp1-Δ1, lys2, ssa1-45, ssa2-1(LEU2), ssa3-1(TRP1), ssa4-2(LYS2) | Ref. 92 |
| KAR2 | MATa ura3-52, leu2-3,112, ade2-101 | Ref. 93 |
| kar2-1 | MATa, kar2-1, ura3-52, leu2-3,112, ade2-101 | Ref. 94 |
| HLJ1 YDJ1 | MATα, ade2, his3, leu2, ura3, trp1 | Ref. 95 |
| hlj1Δ ydj1-151 | MATα, ade2, his3, leu2, ura3, trp1, can1-100, ydj1-2::HIS3 ydj1-151::LEU2 hlj1::TRP1 | Ref. 95 |
| DOA10 HRD1 | MATa, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, gal2 | Ref. 32 |
| doa10Δ | MATa, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, gal2, doa10Δ::HIS3 | Ref. 32 |
| hrd1Δ | MATa, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, gal2, hrd1Δ::LEU2 | Ref. 32 |
| doa10Δ hrd1Δ | MATa, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, gal2 doa10Δ::HIS3, hrd1Δ::LEU2 | Ref. 32 |
| UBR1 SAN1 | MATa, his3Δ, leu2Δ0, ura3Δ0, met15Δ0 | Ref. 27 |
| ubr1Δ | MATa, his3Δ, leu2Δ0, ura3Δ0, met15Δ0, ubr1Δ::KanMX | Ref. 27 |
| san1Δ | MATa, his3Δ, leu2Δ0, ura3Δ0, met15Δ0, san1Δ::Clonat | Ref. 27 |
| ubr1Δ san1Δ | MATa, his3Δ, leu2Δ0, ura3Δ0, met15Δ0, ubr1Δ::KanMX, san1Δ::Clonat | Ref. 27 |
| UBR1 SAN1 | MATα, his3Δ1, leu2Δ1, ura3Δ0 | This study |
| ubr1Δ | MATα, his3Δ1, leu2Δ1, ura3Δ0, ubr1Δ::KanMX | This study |
| san1Δ | MATα, his3Δ1, leu2Δ1, ura3Δ0, san1Δ::NAT | This study |
| ubr1Δ san1Δ | MATα, his3Δ1, leu2Δ1, ura3Δ0, ubr1Δ::KanMX san1Δ::NAT | This study |
| SCJ1 JEM1 | MATα, ura3-52, leu2-3, 112,trp1-Δ901, his3-Δ200, lys2-801, suc2-Δ9 | Ref. 96 |
| scj1Δ jem1Δ | MATα, jem1::LEU2, scj1::TRP1, ura3–52, leu2-3,112, trp1-Δ901, his3-Δ200, lys2-801, suc2-Δ9 | Ref. 96 |
| W303 | MATα, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1 | This laboratory |
| sse1Δ | MATα, sse1::Kan, ade2-1, can1-100, his3-11, leu2-3,112, trp1-1, ura3-1 | Ref. 97 |
| cdc48-2 | Back-crossed 3X to BY4742 | Ref. 98 |
The plasmids/primers used in this study are listed in Table 2. In order to generate triple HA-tagged NBD2 and NBD2*, we first made untagged versions by PCR-amplifying the coding sequence for each protein from pSM693, a plasmid containing full-length STE6-HA, using primers OCG03, OKN52, and OKN53, thus generating pCG01 (NBD2) and pCG02 (NBD2*). Next, a two-stage PCR mutagenesis protocol (35) was used to add the sequence for the tag (that mimics the tag sequence found in pSM1911) (32) to pCG01 and pCG02, using primers OCG08 and OCG09 to generate pCG03 (3XHA-NBD2) and pCG04 (3XHA-NBD2*). DNA sequence analysis (Genewiz) was performed at each step in order to ensure the accuracy of the constructs.
TABLE 2.
Primers and plasmids used in this study
| Primer/Plasmid | Sequence/Genotype | Reference/Source |
|---|---|---|
| Primer | ||
| OKN51 | 5′-GCGCCCGGGATGTACCCATACGATGTTCCAGATT ACGCTATACCCGATATAAGTAGA-3′ | This study |
| OKN52 | 5′-GCGCCGCGGTTAACTGCTTTGGTTGGA-3′ | This study |
| OKN53 | 5′-GCGCCGCGGTTATTCACTATGCGTTAT-3′ | This study |
| OCG03 | 5′-GCGCCCGGGATGATACCCGATATAAGTAGAGGC-3′ | This study |
| OCG08 | 5′-GAATTCCTGCAGCCCGGGATGTACCCATACGATGTT CCTGACTATGCGGGCTATCCCTATGACGTCCCGGACTA TGCAGGATCCTATCCATATGACGTTCCAGATTACGCTA TACCCGATATAAGTAGAGGC-3′ | This study |
| OCG09 | 5′-GCCTCTACTTATATCGGGTATAGCGTAATCTGGAACG TCATATGGATAGGATCCTGCATAGTCCGGGACGTCATAGGGATAGCCCGCATAGTCAGGAACATCGTATGGGTACATCCCGGGCTGCAGGAATTC-3′ | This study |
| OCG71 | 5′-AAGGATATCGACGGTGAATACATTGAGATT-3′ | This study |
| OCG72 | 5′-AAGGAATTCGTAAAAGATTTTATGGTAAGA-3′ | This study |
| Plasmid | ||
| pSM693 | 2μ URA3 STE6:HA | Ref. 99 |
| pSM1911 | 2μ URA3 P-PGK ste6–166-3HA | Ref. 32 |
| pSM1912 | 2μ URA3 P-PGK ste6–166-HA-GFP | Ref. 32 |
| pCG01 | 2μ URA3 STE6NBD2 | This study |
| pCG02 | 2μ URA3 ste6–166NBD2 | This study |
| pCG03 | 2μ URA3 STE6NBD2–3HA | This study |
| pCG04 | 2μ URA3 ste6–166NBD2–3HA | This study |
| pKN31 | 2μ HIS3 Pcup1-mycUb-Tcyc1 | Ref. 33 |
| pRG472 | Int, TRP 3HSV-SAN1 | Ref. 54 |
Standard PCR cloning procedures were used to transfer 3XHSV-SAN1 from pRG472 (a kind gift from Richard Gardner (University of Washington, Seattle, WA)) into pRS423 (36). Briefly, the San1p promoter, coding sequence, and terminator were amplified using primers OCG71 and OCG72. The resulting PCR product was digested with EcoRV and EcoRI and ligated into the same sites in pRS423. The sequence of the entire gene was confirmed (Genewiz).
Assays to Monitor Protein Degradation
To measure NBD2 and NBD2* stability, a cycloheximide chase assay was used, essentially as described (37). Briefly, yeast expressing HA-tagged NBD2 or NBD2* were grown in SC (synthetic complete) medium lacking uracil (−ura) and containing glucose to log phase (A600 0.5–1.5), and a 1-ml aliquot was removed for the zero time point. Cycloheximide was then added to the culture to a final concentration of 187.5 μg/ml. The culture was incubated in a shaking water bath at 26, 30, or 37 °C (as indicated) at ∼200 rpm. At each time point, 1-ml aliquots were removed and mixed with 35 μl of a 0.5 m NaN3 solution (final concentration, 17.5 mm), and the cells were lysed as described (37). The pelleted protein samples were then disrupted with a mechanical pestle in trichloroacetic acid (TCA) sample buffer (80 mm Tris, pH 8, 8 mm EDTA, 3.5% SDS, 15% glycerol, 0.08% Tris base, 0.01% bromphenol blue) supplemented with freshly prepared β-mercaptoethanol (final concentration, 5%), and the solution was incubated at 37 °C for 30 min. Cellular debris was cleared by centrifugation at 14,000 rpm in a microcentrifuge at room temperature, samples were run on denaturing 10% polyacrylamide gels, and the proteins were transferred onto nitrocellulose (BioTrace NT, Pall Corp.) using the Fast Semi-Dry Blotter system (Thermo Scientific).
Antibodies used in this study were mouse monoclonal anti-HA (HA-11, Covance) used at a 1:5000 dilution; rat monoclonal anti-HA-horseradish peroxidase (HRP) high affinity (3F10, Roche Applied Science) used at 1:2500; rabbit polyclonal anti-Myc (sc987, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) used at 1:5000; rabbit anti-ubiquitin (FL-76, Santa Cruz Biotechnology, Inc.) used at 1:250; rabbit anti-glucose-6-phosphate dehydrogenase (G6PD) (A9521, Sigma-Aldrich) used at 1:5000; polyclonal rabbit anti-Sec61p used at 1:5000, as described previously (38); rabbit anti-Anp1p, a gift from Sean Munro (Cambridge University, Cambridge, UK), used at 1:4000; polyclonal rabbit anti-Pma1p (a gift from Amy Chang (University of Michigan, Ann Arbor, MI)) used at 1:3000; mouse anti-Pdi1p (a gift from Vlad Denic (Harvard University, Cambridge, MA)) used at 1:1000; and mouse anti-HSV (EMD Millipore) used at 1:10,000. After an overnight incubation of the immunoblots with primary antibody at 4 °C, they were decorated with either HRP-conjugated goat anti-mouse or anti-rabbit secondary antibody (Jackson ImmunoResearch) at 1:5000 for 2 h at room temperature. Proteins were then visualized with SuperSignal Chemiluminescence (Thermo Scientific). Images of the blots were taken on a Kodak Image Station 440CF (Eastman Kodak Co.) and quantified using ImageJ version 1.47m software (National Institutes of Health).
Sucrose Gradient Analysis for NBD2* Residence
Sucrose gradients were performed essentially as described previously (39). Approximately 60 A600 equivalents of log phase BY4742 cells expressing HA-tagged NBD2* were grown in SC−ura/glucose medium. Cells were pelleted for 3 min at 3000 rpm in a clinical centrifuge at room temperature and washed with 30 ml of ice-cold water, and the pellets were flash-frozen in liquid nitrogen and stored at −80 °C. A 70% sucrose solution was prepared in 10 mm Tris, pH 7.6, 10 mm EDTA and subsequently diluted with 10 mm Tris, pH 7.6, 10 mm EDTA to create a 10–70% layered sucrose gradient (see below). The cells were thawed, resuspended in 400 μl of 10% sucrose solution plus protease inhibitors (3 mm PMSF, 3 μg/ml leupeptin, and 1.5 μg/ml pepstatin A with 1 mm DTT), and lysed by agitation using glass beads on a Vortex mixer three times for 1 min with a 1-min rest on ice between each round. The homogenate was removed to a new tube and combined with another 400 μl of 10% sucrose solution supplemented with protease inhibitors. Unbroken cells were removed via three rounds of centrifugation for 2 min at 2000 rpm in a tabletop microcentrifuge at 4 °C, and the supernatants from each round were combined.
A non-continuous sucrose gradient was created in an SW41 high speed 14 × 89-mm Polyallomer ultracentrifuge tube (Beckman Instruments, Inc.). A total of 2 ml of each percentage was layered on top of one another, with 70% on the bottom to 20% at the top, although only 1.5 ml each of the 30 and 20% sucrose solutions was used. Finally, 450 μl of the homogenate was loaded onto the gradient, and 4.5 μl (1%) of the homogenate was retained at −20 °C as the load fraction. The gradient was ultracentrifuged in an SW41 rotor for ∼18 h at 28,500 rpm (∼140,000 × g) at 4 °C. After the spin, 500-μl fractions were carefully removed from the top of the gradient until the entire gradient was aliquoted. Each fraction was mixed, and a 15-μl aliquot was removed and combined with 3.5 μl of 5× sample buffer (0.325 m Tris, pH 6.8, 10% SDS, 50% glycerol, 25 mg/ml bromphenol blue, 5% β-mercaptoethanol), heated at 37 °C for 30 min, and loaded onto a 10% denaturing polyacrylamide gel. The resolved proteins were transferred to nitrocellulose, and anti-HA-HRP was used to visualize NBD2* (see below). Antibodies for the following organelle markers were used: anti-Anp1p for the Golgi, anti-Pma1p for the plasma membrane, and anti-Sec61p for the ER (see above).
Assays to Monitor Protein Ubiquitination
To measure the amount of protein ubiquitin in yeast, a 30-ml culture of the indicated yeast strains expressing HA-tagged NBD2* was grown to an A600 of ∼1.0 in SC−ura/glucose medium. For the experiments examining proteasome-dependent degradation, the cultures were split, MG132 (final concentration, 50 μm) or DMSO was added, and the cells were incubated at 30 °C for 1 h. The yeast were then harvested by centrifugation for 3 min at 3000 rpm in a clinical centrifuge at room temperature and washed with ice-cold water, and the cell pellets were frozen at −80 °C. The cells were thawed on ice; resuspended in 1 ml of a detergent solution containing 50 mm Tris, pH 8, 100 mm EDTA, pH 8, 0.4% sodium deoxycholate, 1% Nonidet P-40, protease inhibitors (1 mm PMSF, 1 μg/ml leupeptin, and 0.5 μg/ml pepstatin A), N-ethylmaleimide (final concentration, 10 mm); and transferred to 13 × 100-mm glass culture tubes. The yeast were disrupted with glass beads by agitation on a Vortex mixer (as described above). The supernatant was removed and combined with a 500-μl wash of the beads with the detergent solution described above. Unbroken cells and cell membrane fractions were removed using a tabletop centrifuge by centrifugation for 3 min at 13,000 rpm at 4 °C. An A280 of a 200-fold dilution of each sample in 2% SDS was used to obtain the relative protein concentrations. Equal amounts of lysate were then immunoprecipitated overnight at 4 °C using 30 μl of anti-HA-conjugated agarose beads (Roche Applied Science). Next, the beads were pelleted via tabletop centrifugation for 1 min at 13,000 rpm at 4 °C and washed four times with the detergent solution. Samples were processed for immunoblotting as described above, except prior to blocking, the nitrocellulose membrane was incubated in a boiling water bath for 1 h in order to expose the antibody epitopes on the polyubiquitin chains. Polyubiquitin was visualized using anti-ubiquitin (FL76) or anti-Myc for cells expressing Myc-tagged ubiquitin.
To measure the level of substrate ubiquitination in vitro, we developed an assay based on previously described methods (27, 33). Briefly, NBD2*-expressing wild-type yeast cells were grown to an A600 of ∼1.0, and for each reaction, material from 30 A600 of yeast cells was used. The cells were washed twice with cold water and once with buffer 88 (20 mm HEPES, pH 6.8, 150 mm KOAc, 250 mm sorbitol, 5 mm MgOAc). After the last pelleting step, the cells were resuspended in 1 ml of buffer 88 plus protease inhibitors (1 mm PMSF, 1 μg/ml leupeptin, and 0.5 μg/ml pepstatin A) per 60 A600 of cells and lysed by agitation using glass beads on a Vortex mixer five times for 1 min with a 1-min rest on ice between each round. The lysate was transferred to a new tube, and another 1 ml of buffer 88 plus protease inhibitors per 30 A600 of cells was added to the glass beads. The lysates were combined, and unbroken cells were pelleted at 5000 rpm for 5 min. The resulting supernatant was transferred to a new tube. In a total volume of 1.5 ml, NBD2* was immunoprecipitated after a 2-h incubation at 4 °C using 35 μl of anti-HA-conjugated agarose beads (Roche Applied Science). The beads were then pelleted in a clinical centrifuge for 3 min at 3000 rpm, combined into a 15-ml conical tube, and washed four times with 1 ml per 30 A600 of cells with 50 mm NaCl, 10 mm Tris, pH 7.5, plus protease inhibitors. After the last wash, the beads were resuspended in 40 μl of buffer 88 per 30 A600 of cells. Next, the beads were gently mixed, and 40 μl of the mixture was aliquoted into microcentrifuge tubes. Each reaction contained an ATP-regenerating system (1); 3 μl of 125I-labeled ubiquitin, as described previously (33); and cytosol at a final concentration of 1 mg/ml. Buffer 88 was then added to make the final reaction volume 50 μl. All components except for the iodinated ubiquitin were added, and the reaction was prewarmed at room temperature for 10 min. After the addition of the 125I-ubiquitin, the reaction was allowed to proceed at room temperature for the indicated time. To quench the reaction, 1 ml of 15 mm NaPO4, pH 7.2, 150 mm NaCl, 10 mm EDTA, 2% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate (27) plus protease inhibitors (1 mm PMSF, 1 μg/ml leupeptin, and 0.5 μg/ml pepstatin A) and N-ethylmaleimide (final concentration, 10 mm) was added, and the solution was mixed and centrifuged at 13,000 rpm for 1 min at 4 °C to pellet the beads. The beads were then washed three times with 1 ml of 50 mm Tris, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.2% SDS plus protease inhibitors and N-ethylmaleimide. After the last wash, the beads were aspirated to dryness, and the immunoprecipitated NBD2* was eluted from the beads using 35 μl of TCA sample buffer supplemented with 5% β-mercaptoethanol and incubated at 37 °C for 30 min. NBD2* was resolved using SDS-PAGE, and the polyacrylamide gels were dried at 80 °C with a vacuumed gel drier for 1.5 h. The signal corresponding to iodinated ubiquitin was detected by phosphorimaging. For normalization, equal loading of NBD2* was determined by Western blot analysis, as described above.
Isolation of Yeast Cytosol
Cytosol was isolated from yeast cells of specific genetic backgrounds using a method similar to that described previously (27). Yeast cells were grown to an A600 of ∼2.0, and a total of 800–1000 A600 cells were pelleted for 3 min at 3000 rpm in a clinical centrifuge. For temperature-sensitive strains, a 45-min incubation at the non-permissive temperature was performed prior to harvesting. Each pellet was washed twice with cold water and once with cold buffer 88. The pellet was resuspended in 100 μl of buffer 88, and this mixture was slowly pipetted into liquid nitrogen. The frozen pellets were then ground for 5 min using a mortar and pestle that were precooled with liquid nitrogen. Liquid nitrogen was continuously added to the ground cells as needed to keep them frozen. The resulting powder was transferred into a microcentrifuge tube and allowed to thaw, and DTT was added to a final concentration of 1 mm. Next, the cytosol was clarified with three centrifugations at 4 °C: 5000 × g for 15 min, 17,000 × g for 15 min, and 100,000 × g for 1 h. The protein concentration of the cytosol (i.e. the final supernatant) was measured using Bradford reagent (Bio-Rad), and aliquots were flash-frozen and stored at −80 °C until use.
Indirect Immunofluorescence Microscopy
Immunofluorescence was performed essentially as described previously with slight modifications (40). In brief, cells were grown in SC−ura/glucose medium to A600 ∼0.5 and fixed for 1 h in 4% formaldehyde. The cells were spheroplasted by incubation with zymolyase 20T (U.S. Biological) at 37 °C and spotted onto polylysine-treated slides and permeabilized using a methanol/acetone treatment. The slides were blocked with 0.5% BSA, 0.5% ovalbumin, 0.6% fish skin gelatin, and stained with mouse anti-HA (HA-11, Covance) at 1:250 and rabbit anti-Kar2p (41) at 1:250. The primary antibodies were decorated with Alexa Fluor 488 goat anti-mouse and Alexa Fluor 568 goat anti-rabbit, both at 1:500, and nuclei were stained with DAPI. The slides were mounted with Prolong Antifade Gold (Invitrogen) and imaged with an Olympus FV1000, ×100 UPlanSApo oil immersion objective, numerical aperture 1.40.
Membrane Association Assay
BY4742 yeast transformed with pCG03 (NBD2) or pCG04 (NBD2*) was grown in SC−ura/glucose medium to A600 of ∼1. Approximately 30 A600 of cells were harvested by centrifugation for 3 min at 3000 rpm at room temperature and washed once with ice-cold water. The cells were lysed by agitation using glass beads on a Vortex mixer four times for 30 s with a 1-min rest on ice between each round in a 50-ml conical tube. Unbroken cells were cleared two times by centrifugation for 3 min at 3000 rpm. The lysate was spun at 50,000 rpm at 4 °C in a Sorvall RCM120EX (S100AT5 rotor) for 15 min, and the resulting supernatant was retained as “S1.” The pellet was washed with lysis buffer (0.1 m sorbitol, 50 mm KOAc, 2 mm EDTA, 20 mm HEPES, pH 7.4) and recollected by centrifugation. The pellet was resuspended in lysis buffer and split into three portions. P1 was retained, whereas P2 was mock-treated with lysis buffer and P3 was treated with lysis buffer supplemented with 6 m urea for 15 min at 4 °C. After this treatment, the P2 and P3 fractions were collected as described above and separated from S2 and S3. Proteins were precipitated using TCA, and pellets were resuspended into equal volumes of TCA sample buffer for analysis by SDS-PAGE and Western blotting.
Coimmunoprecipitation Assay
Mutant ssa1-45 yeast were transformed with either empty vector (pRS423) or pCG57 (pRS423–3XHSV-San1p) and pCG04 (NBD2*). Cells were grown in filter-sterilized SC−ura−his/glucose medium to A600 of ∼1. Cultures were split and regrown to A600 of ∼1, at which point half of the culture was harvested, and the remainder was shifted to 37 °C for 45 min prior to harvesting. The pellets were flash-frozen and stored at −80 °C. The pellets were thawed on ice and resuspended in 1 ml of co-IP buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100 plus protease inhibitors). Cells were lysed with glass beads, and the lysate was removed, and the beads were washed with 500 μl of co-IP buffer. The lysate and wash fractions were combined and centrifuged at 18,000 × g to clear unbroken cells, and the resulting supernatant was precleared with 70 μl of Pansorbin cells (Calbiochem) for 30 min with rotation at 4 °C. NBD2* was then precipitated from the cleared lysate for 2 h at 4 °C with 35 μl of anti-HA-conjugated agarose beads (Roche Applied Science) per 30 A600 of cells. The beads were washed four times with co-IP buffer lacking Triton X-100 but supplemented with protease inhibitors. The beads were aspirated to dryness, and the bound protein was eluted with 35 μl of TCA sample buffer supplemented with 5% 2-mercaptoethanol and incubated at 37 °C for 30 min. The protein eluate was resolved using SDS-PAGE, and NBD2*, San1p, and G6PD were detected by Western blot as described above.
RESULTS
NBD2* Is an Unstable Cytoplasmic Protein
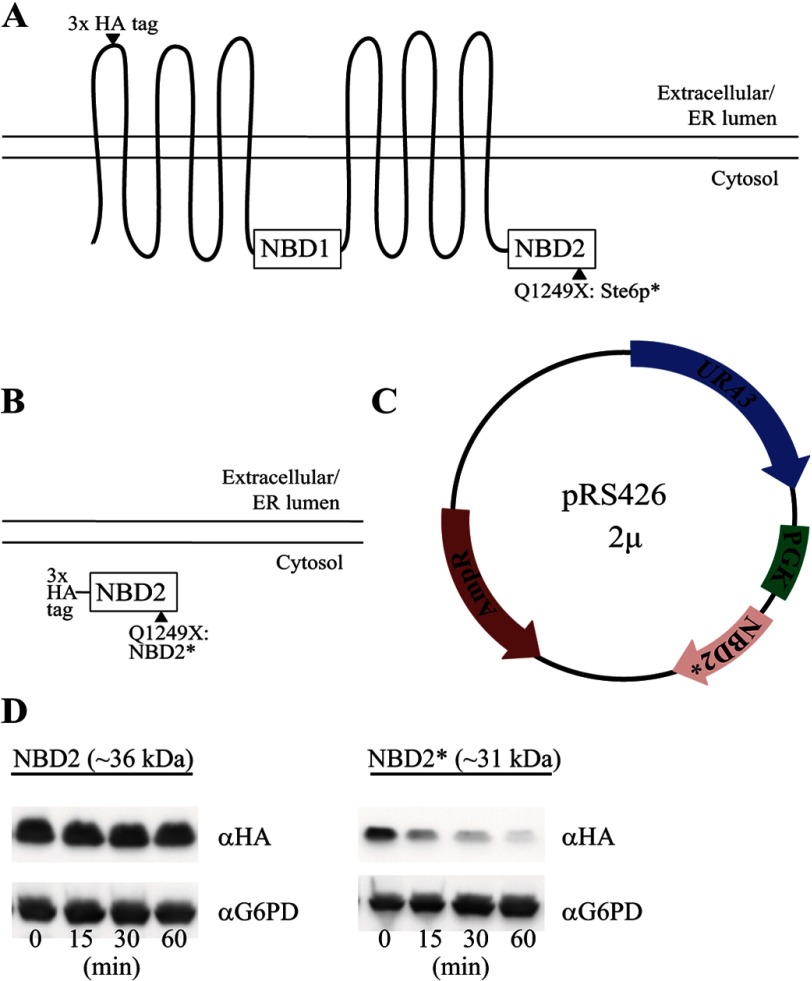
Our goal was to compare and contrast the degradation requirements for an ERAD and CytoQC substrate containing an identical, well defined misfolded protein motif. The motif employed is a truncated version of the second nucleotide binding domain, termed NBD2*, derived from the well characterized yeast ERAD substrate, Ste6p*. The wild-type protein, Ste6, is the yeast a mating factor transporter (42, 43), but a premature stop codon in NBD2 (Q1249X; Fig. 1A), results in targeting of Ste6p* for ERAD. One advantage of using NBD2* as a quality control module is that the degradation decision will be made post-translationally, because full-length Ste6 is not targeted for ERAD (31). To validate the use of NBD2* as a quality control substrate, we developed expression systems for NBD2 (the full-length second nucleotide binding domain) and NBD2* (Fig. 1B). Given that these proteins lack signal sequences or hydrophobic domains sufficient to support membrane integration, we predicted that NBD2* would reside in the cytosol and help define the requirements for CytoQC. A comparison of the requirements for the degradation of NBD2* to the known requirements for the degradation of Ste6p* would then allow us to define the similarities and differences between ERAD and CytoQC for a single misfolded domain in its native cellular compartment, in this case the cytoplasm. For this comparison, the NBD2 and NBD2* expression systems mimicked that used in previous studies of Ste6p* (Fig. 1C) (32), and in order to facilitate the detection of NBD2 and NBD2*, the proteins contained a triple HA tag at their N termini (see “Experimental Procedures” and Fig. 1B).
FIGURE 1.
NBD2* is an unstable protein. A, Ste6p is a yeast ATP-binding cassette transporter containing 12 transmembrane domains and two cytoplasmic nucleotide binding domains. A mutation, Q1249X, in the second nucleotide binding domain (NBD2) of Ste6p (termed Ste6p*) results in its recognition as an ERAD substrate. B, soluble NBD2 constructs were designed with an N-terminal triple HA tag as either full-length NBD2 or NBD2 containing the identical Q1249X mutation found in Ste6p* (NBD2*). C, all constructs were cloned into a multicopy (2μ) plasmid (pRS426) and were placed under the control of the yeast phosphoglycerate kinase (PGK1) promoter. D, a cycloheximide chase assay (as described under “Experimental Procedures”) was used to measure protein stability over time, and yeast lysates were blotted with anti-HA-HRP to detect NBD2 and NBD2*. G6PD was probed as a loading control.
A study examining the CytoQC of a C-terminally tagged version of NBD2* was published during the preparation of this manuscript (44). Among other differences (see “Discussion”), a full-length wild type version of NBD2* (i.e. NBD2) was not examined in that study. Therefore, it is unclear whether instability is caused by the NBD2 truncation, the addition of the C-terminal HA tag (which is the site of the truncation), or another inherent feature of the protein. Therefore, we first compared the stabilities of NBD2 and NBD2* in a wild-type yeast strain using a cycloheximide chase assay. Parallel to what was observed for Ste6p (31), NBD2 is a stable protein, whereas NBD2* is highly unstable and is degraded nearly to completion during a 1-h time course (Fig. 1D). These results indicate that a truncation of NBD2 is sufficient to destabilize this domain, regardless of whether the domain is part of the full-length Ste6 protein.
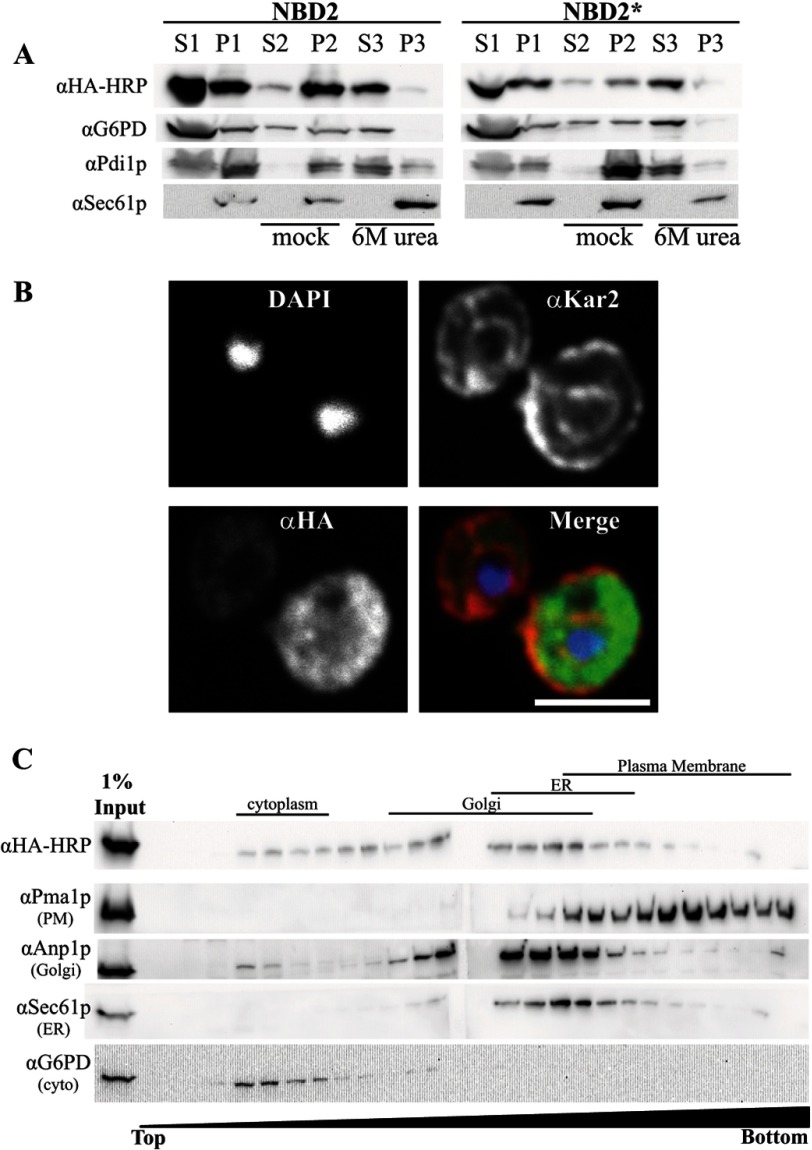
To begin to characterize the cellular properties of NBD2*, we prepared cell lysates and subjected them to various treatments and centrifugation in order to assess protein solubility. A significant NBD2 and NBD2* pool (∼40%) sedimented after cell lysis. Upon treatment with 6 m urea, both proteins were stripped from the membrane fraction, whereas the integral membrane ER protein, Sec61p, remained membrane-associated (Fig. 2A). This treatment also liberated a majority of Pdi1p, which is soluble and resides in the ER lumen (45). Because NBD2 and NBD2* do not differ significantly in their membrane association or aggregation propensity, this result suggests that there are no gross NBD2* folding defects and that the preferred selection of NBD2* for degradation is triggered by a more subtle conformational change.
FIGURE 2.
NBD2* is a cytoplasmic membrane-associated protein. A, yeast expressing NBD2 or NBD2* were grown to log phase and disrupted by glass bead lysis (as described under “Experimental Procedures”) to generate a membrane fraction (pellet 1 (P1)) and cytosolic fraction (supernatant 1 (S1)). Equal portions of the pellet fraction were either mock-treated or treated with 6 m urea to remove peripheral proteins (S2/P2 and S3/P3, respectively). Immunoblots were performed with anti-HA-HRP for NBD2, anti-G6PD (cytoplasmic marker), anti-Sec61 (integral membrane marker), and anti-Pdi1p (ER lumenal/peripheral membrane marker). B, NBD2* localization was determined by indirect immunofluorescence confocal microscopy, using mouse anti-HA (NBD2*), rabbit anti-Kar2p (ER lumen), and DAPI to label nuclei. Primary antibodies were decorated with Alexa goat anti-mouse 488 and goat anti-rabbit 568, respectively. Scale bar, ∼5 μm. C, yeast lysates containing NBD2* were analyzed by sucrose gradient centrifugation. Fractions were taken from the top, and the density of sucrose in each fraction increases throughout the gradient. An aliquot of each fraction was analyzed by SDS-PAGE, and after transfer, the nitrocellulose was immunoblotted for Kar2p (ER marker), Anp1 (Golgi marker), Pma1 (plasma membrane marker), and NBD2* (anti-HA-HRP). A lane containing 1% of the total protein loaded on the gradient was also included (1% Input).
Next, to examine the subcellular localization of NBD2*, we performed indirect immunofluorescence confocal microscopy. NBD2* staining appeared in both a diffuse pattern in the yeast cytoplasm and in more discrete punctae that do not colocalize with the ER marker Kar2p (Fig. 2B). As in the sedimentation assay, NBD2 showed no marked difference in localization pattern (data not shown). These punctae may represent the association of NBD2* with internal membranous structures or cytoplasmic inclusions. Therefore, we performed sucrose gradient sedimentation of lysates prepared from a wild-type yeast strain expressing NBD2*. Under these conditions, the sucrose gradient resolves ER, Golgi, and the plasma membrane fractions. As expected for a cytoplasmic protein, a large portion of NBD2* resided in the lowest density sucrose fractions, which are populated by cytoplasmic proteins (Fig. 2C, compare with G6PD). However, NBD2* also sedimented in heavier fractions associated with the ER (Sec61p) and Golgi (Anp1). Of note, NBD2* does not appear to co-migrate with fractions containing the plasma membrane marker, Pma1, consistent with the immunofluorescence data. Taken together, these results suggest that NBD2* is primarily a soluble, cytoplasmic protein but may possess characteristics that mediate its interaction with the cytoplasmic face of select organelles and/or that the protein is recruited to these sites by virtue of its affinity for other proteins.
NBD2* Degradation Is Ubiquitin- and Proteasome-dependent
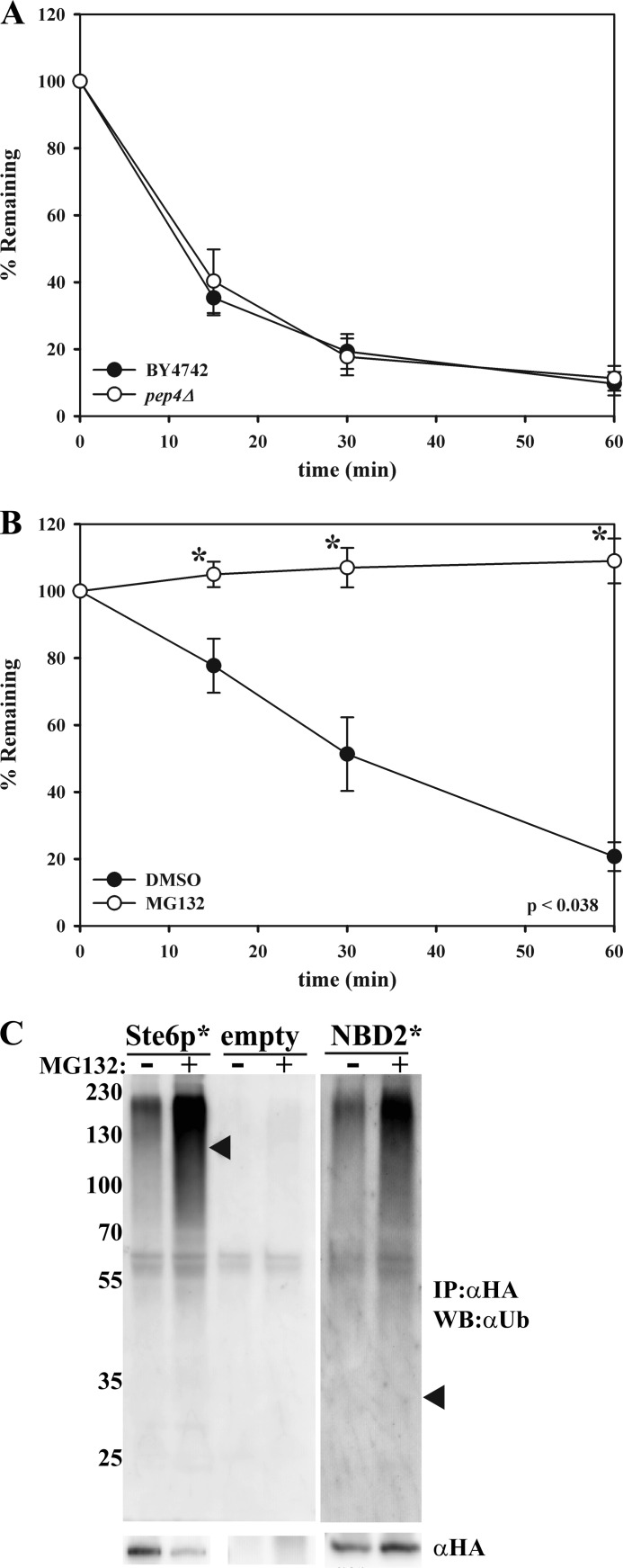
Like other ERAD substrates, Ste6p* is destroyed by the UPS (32, 33, 46). We hypothesized that NBD2* would also be degraded by the UPS, although it was possible that NBD2* might form inclusions and would then be subject to autophagic degradation in the vacuole (16–19, 47). To test this possibility, we assessed the degradation of NBD2* in a pep4Δ strain, which lacks >90% of functional vacuolar protease activity (48). No defect in NBD2* degradation in the pep4Δ strain was evident (Fig. 3A). In contrast, when NBD2* degradation was assessed in a multidrug pump-deficient yeast strain (pdr5Δ) that was treated with either DMSO or the proteasomal inhibitor MG-132 (49, 50), NBD2* was completely stabilized in the presence of MG-132 (Fig. 3B). To confirm these results, NBD2* was immunoprecipitated from cells treated with DMSO or MG-132, and the precipitated protein was then immunoblotted to detect the levels of conjugated ubiquitin. As shown in Fig. 3C, an elongated “smear” of high molecular weight products, typical of polyubiquitination, was present in the samples expressing NBD2*. The ubiquitin signal was significantly stronger in cells treated with MG-132, as expected for a proteasome-targeted, ubiquitinated protein. These collective data establish NBD2* as a new CytoQC substrate.
FIGURE 3.
NBD2* is a degraded by the ubiquitin proteasome system. Cycloheximide chase analyses were performed as described under “Experimental Procedures” in wild-type (BY4742) or pep4Δ yeast (A) and a pdr5Δ yeast strain that was either treated with DMSO (closed circles) or 100 μm MG-132 (open circles) for 20 min prior to the assay (B). Lysates were probed with anti-HA-HRP, and data for each graph represent the means ± S.E. (error bars) for three independent experiments, each performed in triplicate; *, p < 0.038. C, a pdr5Δ yeast strain containing vectors engineered for the expression of Ste6p* or NBD2* or that contained an empty vector was treated with DMSO (−) or 50 μm MG-132 (+) for 1 h prior to lysate preparation. An immunoprecipitation using anti-HA-agarose beads was then performed with each lysate. Proteins were detected with either anti-HA-HRP (αHA) or anti-ubiquitin (Ub). Arrowheads represent the location on the ubiquitin blot where full-length Ste6p* and NBD2* would migrate, and molecular weight markers (× 103) are shown to the left.
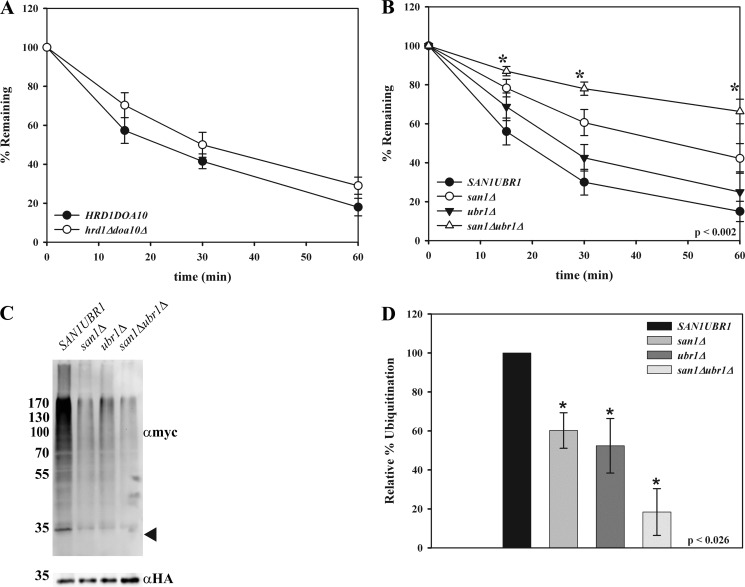
NBD2* Polyubiquitination Requires San1p and Ubr1
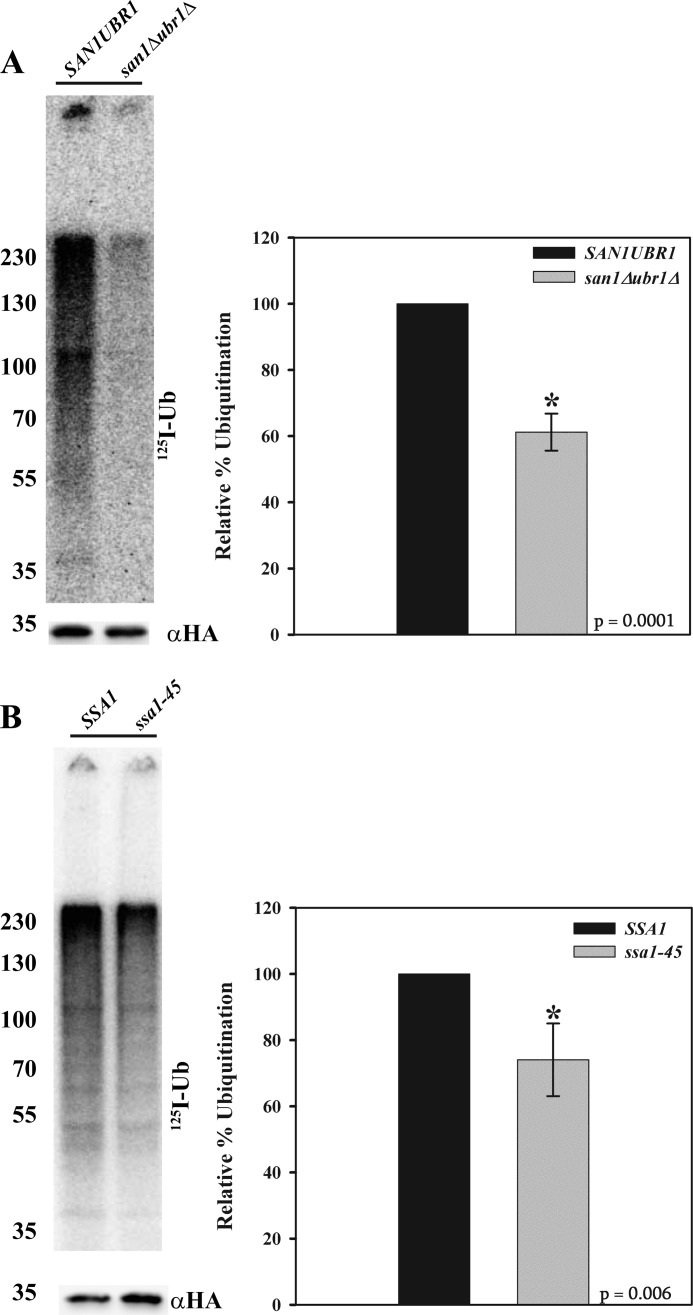
ERAD substrates have been classified as those that contain lesions in the ER lumen (ERAD-L), membrane (ERAD-M), or cytoplasm (ERAD-C) (51). In yeast, ERAD-L and ERAD-M substrates are ubiquitinated primarily by the Hrd1p ubiquitin ligase, whereas ERAD-C substrates are modified by Doa10p, another E3 ligase that resides in the ER membrane. Recent data also indicate that substrates that slowly translocate into the ER and/or that remain associated with the translocon are ubiquitinated by Hrd1p (52). Because the misfolded lesion in Ste6p* resides in the cytoplasm and is largely ubiquitinated by Doa10p, it is classified as an ERAD-C substrate (32). Interestingly, Doa10p contributes to the degradation of some previously studied CytoQC substrates (2, 24, 25). However, we found that the ER-localized E3 ligases, Doa10p and Hrd1p, were dispensable for the degradation of NBD2* (Fig. 4A). We therefore investigated the roles of other E3 ligases on the fate of NBD2* and expressed the protein in yeast strains lacking Ubr1p and/or San1p, which have also been implicated in CytoQC (26, 27, 29). As evident in Fig. 4B, NBD2* degradation was highly dependent on San1p, and Ubr1p contributed much less to the rate and extent of NBD2* turnover. Because NBD2* was further stabilized in a san1Δubr1Δ strain, it is likely that Ubr1p partially compensates for the loss of San1p. Consistent with these data, NBD2* polyubiquitination was decreased in a san1Δ mutant, and this effect was slightly more pronounced in a san1Δubr1Δ strain (Fig. 4, C and D).
FIGURE 4.
NBD2* degradation relies on both cytoplasmic and nuclear E3 ligases. Cycloheximide chases were performed as described under “Experimental Procedures” in HRD1DOA10 and hrd1Δdoa10Δ yeast (data represent the means ± S.E., n = 4) (A) or SAN1UBR1, san1Δ, ubr1Δ, and san1Δubr1Δ yeast (B) (data represent the means ± S.E. (error bars), n = 4–5); *, p ≤ 0.002. C, NBD2* was immunoprecipitated from the strains listed in B using anti-HA-agarose beads and analyzed by immunoblotting with either anti-HA-HRP to detect NBD2* or anti-Myc antibody to detect exogenously expressed Myc-tagged ubiquitin. The arrowhead on the αmyc blot denotes where NBD2* migrates, and molecular weight markers (× 103) are shown to the left. D, quantification of the ubiquitin signal from three independent experiments. Error bars, means ± S.E. *, p ≤ 0.026.
San1p is a nuclear-localized E3 ligase that was originally thought to contribute only to protein quality control in the nucleus, but this enzyme has since been shown to polyubiquitinate select, misfolded substrates that reside in the cytoplasm (26, 27, 53, 54). Because NBD2* degradation and ubiquitination depend mostly on San1p, NBD2* may have been transported into the nucleus, as proposed for other San1p-dependent CytoQC substrates (26, 27). Indeed, some other San1p-dependent CytoQC substrates appear to accumulate in the nucleus in a san1Δ mutant (26, 27). To explore whether NBD2* might also accumulate in the nucleus in this genetic background, confocal microscopy was performed on wild-type and san1Δ yeast expressing NBD2*. NBD2* did not appear to accumulate in the nucleus in san1Δ cells (data not shown). This observation is not altogether surprising, given the lack of a defined nuclear localization signal in NBD2* and the strong cytoplasmic residence of NBD2* in wild-type cells (Fig. 2B). In support of our findings, a recent study demonstrated that a short-lived, modified form of GFP is a San1p-dependent substrate that also does not accumulate in the nucleus of a san1Δ strain (55).
NBD2* Degradation Requires Cdc48p and Molecular Chaperone Function
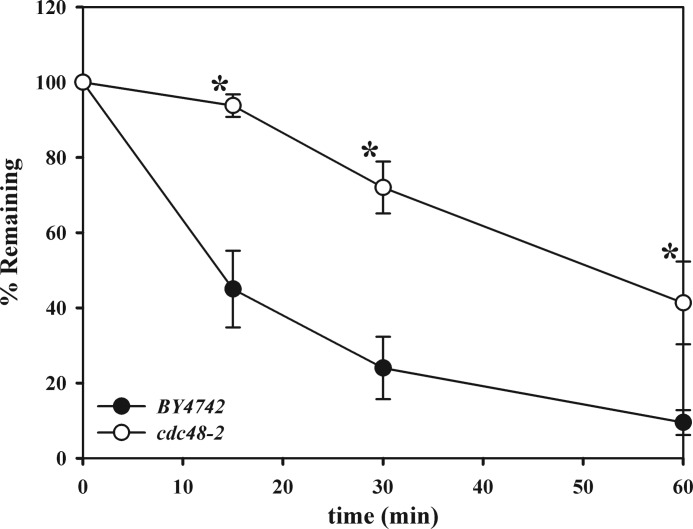
In order to be processed by the cytoplasmic 26 S proteasome, ERAD substrates undergo retrotranslocation, whereby the proteins are dislocated from the ER lumen or membrane coincident with or following polyubiquitination (10, 11). For many ERAD substrates, including Ste6p* (32, 33), this process depends on the AAA+ ATPase Cdc48p (p97 or VCP in mammals), which is thought to provide the mechanical force required for extraction (56–61). Some previously studied CytoQC substrates also require Cdc48p function for degradation (2, 62, 63), whereas other groups have found that Cdc48p is dispensable for the degradation of these substrates (25, 64, 65). To examine whether the degradation of NBD2* was Cdc48-dependent or -independent, cycloheximide chases were performed in a strain containing a temperature-sensitive mutation in Cdc48p (cdc48-2). Following a 2-h shift to the non-permissive temperature of 37 °C, we found that NBD2* degradation was significantly delayed in the mutant strain (Fig. 5). These data suggest that Cdc48p may prevent NBD2* aggregation, similar to its proposed role in preventing the formation of toxic poly(Q)-containing protein aggregates (reviewed in Ref. 11), or through its action as a disaggregase (66). Alternatively, Cdc48p may help shuttle cytoplasmic substrates to the proteasome or liberate the membrane-associated pool of NBD2* prior to degradation.
FIGURE 5.
NBD2* degradation utilizes Cdc48p. Cycloheximide chases were performed as described under “Experimental Procedures” in BY4742 and cdc48-2 yeast following a 2-h temperature shift to 37 °C. Data represent the means ± S.E. (error bars) for four independent experiments, each performed in triplicate; *, p ≤ 0.036.
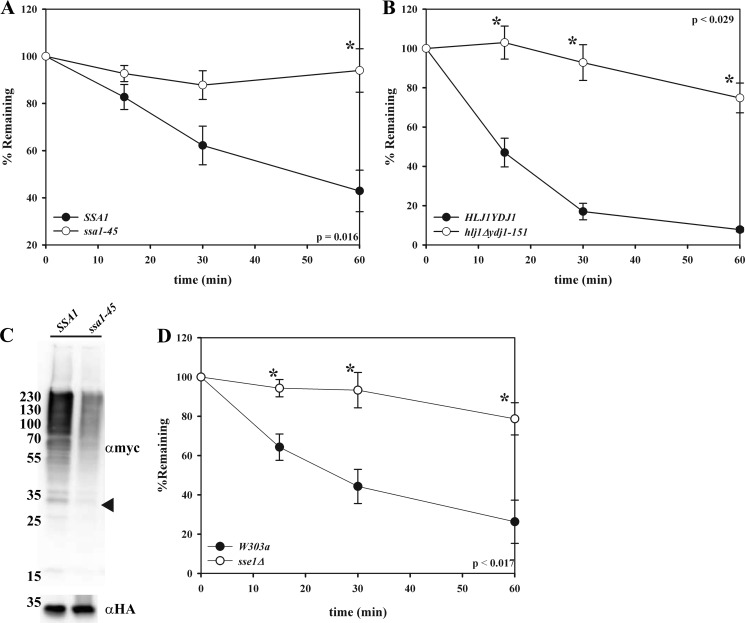
ERAD substrate recognition is mediated by molecular chaperones and chaperone-like proteins (4–7, 10). Perhaps the most prominent and well studied proteins in this family are the Hsp70s (67). In yeast, the ER lumenal Hsp70 is Kar2p (BiP), and the major cytoplasmic Hsp70 is Ssa1p. Ste6p* degradation relies on Ssa1p but not Kar2p, which is not surprising, given that the folding lesion in Ste6p* is cytoplasmic (32). To examine whether the chaperone dependence of NBD2* degradation mirrored the requirements for the degradation of Ste6p*, we utilized strains containing temperature-sensitive mutations in the genes encoding either Kar2p or Ssa1p. NBD2* degradation was highly dependent on Ssa1p, as evidenced by the nearly complete stabilization of the protein in the ssa1-45 strain at the non-permissive temperature (Fig. 6A). NBD2* degradation also required the cytoplasmic Hsp40 co-chaperones, Hlj1p and Ydj1p (Fig. 6B), which function with Ssa1p to support the degradation and ubiquitination of Ste6p* (32, 33). As predicted, the ER-lumenal Hsp70, Kar2p, and Hsp40s, Scj1p and Jem1p, were dispensable when NBD2* degradation was examined in yeast containing mutations in the genes encoding these proteins (data not shown).
FIGURE 6.
NBD2* degradation and polyubiquitination are Hsp70- and Hsp40-dependent. Cycloheximide chases, as described in the preceding figure legends, were performed in SSA1 and ssa1-45 (A) and HLJYDJ1 and hlj1Δydj1-151 (B) yeast at 37 °C, and W303a and sse1Δ yeast (D) at 26 °C. Data represent the means ± S.E. (error bars) from either three (SSA1), four (YDJ1HLJ1), or three (W303a) independent experiments, each performed in triplicate (*, p < 0.016 (SSA1); *, p < 0.029 (YDJ1HLJ1); *, p < 0.017 (W303a)). C, SSA1 and ssa1-45 yeast expressing NBD2* were shifted to 37 °C for 12 min prior to lysis, NBD2* was precipitated using anti-HA-agarose beads, and the precipitate was analyzed by immunoblotting with either anti-HA-HRP to detect NBD2* or anti-Myc to detect exogenously expressed Myc-ubiquitin. The arrowhead on the αmyc blot denotes where NBD2* migrates, and molecular weight markers (× 103) are shown to the left.
Hsp70 Facilitates NBD2* Polyubiquitination
During the ERAD of Ste6p*, Ssa1p acts as a bridging factor to mediate the interaction between Ste6p* and Doa10p, allowing for efficient polyubiquitination (33). For CytoQC substrates, Ssa1p plays many roles, including substrate recognition, maintaining protein solubility, and facilitating nuclear transport; in one case, this chaperone was also thought to bridge a substrate to the cytoplasmic ubiquitination machinery, but in this example, the E3 ligase was not defined (2, 26, 27, 29, 68). In order to explore how Ssa1p impacts NBD2* degradation, we examined NBD2* polyubiquitination in a wild-type strain (SSA1) and in the ssa1-45 mutant strain. We found that NBD2* polyubiquitination decreased by ∼2-fold in the ssa1-45 mutant strain (Fig. 6C), suggesting that Hsp70 function is required prior to NBD2* ubiquitination.
Another chaperone that impacts the degradation of CytoQC substrates is Sse1p (27, 69, 70), which is the constitutively expressed Hsp110 homolog in yeast. Hsp110 chaperones are potent “holdases,” which prevent substrate aggregation, and they function as Hsp70 nucleotide exchange factors (71). As shown in Fig. 6D, NBD2* degradation was blocked in yeast lacking Sse1p. Ste6p* degradation was also slowed in sse1Δ yeast (72), suggesting that this chaperone participates in the degradation pathway of the NBD2* module, regardless of whether it is integrated into the ER membrane or resides in the cytoplasm.
Ssa1p Facilitates NBD2* Polyubiquitination via Its Interaction with San1
Based on our in vivo ubiquitination data (Fig. 3C), we hypothesized that Ssa1p augments NBD2* degradation prior to the ubiquitination step. As discussed above, it is possible that Ssa1p aids in the transport of NBD2* into the nucleus for ubiquitination by San1p and/or that Ssa1p facilitates an interaction between NBD2* and San1p. To our knowledge, an Hsp70-facilitated association between a CytoQC substrate and San1p has not been reported. The former possibility seems unlikely, given that NBD2* fails to accumulate in the nucleus in san1Δ cells, a phenomenon that reportedly relies on Ssa1p (26).
In order to distinguish between these possibilities, we developed an in vitro ubiquitination assay similar to that published for both ERAD and CytoQC substrates (27, 33). The advantages of the assay include 1) enhanced and quantitative ubiquitination signal by using 125I-lableled ubiquitin and 2) the ability to alter single components by generating cytosol from mutant yeast strains. To validate the use of this assay, we examined the ubiquitination of NBD2* incubated with either wild-type yeast cytosol or cytosol prepared from san1Δubr1Δ yeast cells. We measured an ∼40% decrease in NBD2* ubiquitination in the absence of San1p and Ubr1p (Fig. 7A), indicating that this assay recapitulates data using the in vivo assay shown in Fig. 3C.
FIGURE 7.
Hsp70 function contributes to NBD2* polyubiquitination. In vitro ubiquitination assays were performed using cytosol prepared from SAN1UBR1 or san1Δubr1Δ yeast (A) or cytosol prepared from SSA1 or ssa1-45 yeast (B) following a 45-min shift to 37 °C. Graphs represent the means ± S.E. (error bars) from five independent experiments performed with replicates utilizing cytosol made three separate times (*, p ≤ 0.006 (SSA1) and p ≤ 0.0001 (SAN1UBR1)). Molecular weight markers (× 10−3) are shown to the left.
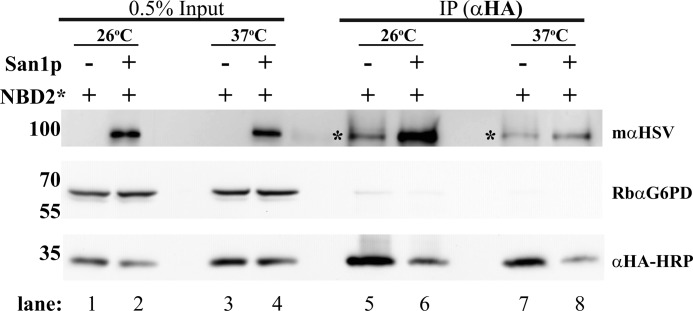
In this in vitro assay, we immunoprecipitated NBD2* from mechanically lysed cells, followed by incubation with wild-type or mutant cytosol. Therefore, the role of any factor that impacts the association between San1p and NBD2* can be measured directly, regardless of whether the factor is required for nuclear transport. If the only role of Ssa1p is to facilitate NBD2* nuclear transport, then performing the in vitro ubiquitination assay in the presence of SSA1 or ssa1-45 cytosol should be without consequence. However, if ssa1-45 cytosol results in a decrease in ubiquitination, this would suggest that Ssa1p plays a more direct role in ubiquitination. When this assay was performed, we measured an ∼25% reduction in NBD2* ubiquitination when exposed to ssa1-45 cytosol (Fig. 7B). We posit that the more robust effect on NBD2* turnover than the effect observed in this experiment using ssa1-45-derived reagents stems from the possibility that the Ssa1p chaperone plays other roles during NBD2* CytoQC and/or that different functionally redundant chaperones compensate in vitro. Regardless, to examine more directly whether Ssa1p aids in the formation of an NBD2*-San1p complex, we immunoprecipitated NBD2* from ssa1-45 yeast grown at either the permissive or non-permissive temperature that were co-transformed with either an empty vector or an HSV-tagged form of San1p. Although a robust interaction between NBD2* and San1p was evident from cells incubated at the permissive temperature (Fig. 8, lane 6), when NBD2* was isolated from cells following a 45-min shift to the non-permissive temperature, the interaction decreased by ∼3-fold (Fig. 8, compare lanes 6 and 8). These data support a previously unreported role for Ssa1p in facilitating substrate interaction with San1p.
FIGURE 8.
Interaction of NBD2* with San1p is Hsp70-dependent. Mutant ssa1-45 yeast expressing NBD2* or both a tagged form of San1p and NBD2* were harvested at 26 °C or following a 45-min shift to 37 °C. Lysates from each strain were prepared, and NBD2* was immunoprecipitated. After SDS-PAGE, the indicated antibodies were used to detect San1p (mαHSV), G6PD (RbαG6PD, as a loading control), and NBD2* (αHA-HRP). 0.5% of the input was also analyzed to demonstrate equal loading. *, a background band that cross-reacted with the αHSV antibody and migrated close to the San1p band (note its presence in the San1p (−)-lanes). A comparison of lanes 6 and 8 corresponding to the mαHSV signal shows a loss of interaction between NBD2* and San1p following the temperature shift. Molecular weight markers (× 103) are shown to the left.
DISCUSSION
During protein biosynthesis, folding errors can result in the targeting of a nascent polypeptide for degradation by the UPS. Key features of a misfolded substrate, which designate the factors utilized for destruction, are the location of the misfolded lesion and the protein's residence. In this study, we report on the generation of a soluble version of the misfolded domain in Ste6p*, NBD2*, in order to allow for a direct comparison between the requirements for the ERAD and CytoQC of a single domain. Understanding the factors that mediate protein quality control in different cellular compartments is essential for modulating CytoQC pathways that are deficient in specific disease states. Herein we report that NBD2* can be removed from Ste6p* and is a substrate for the cytoplasmic UPS (Fig. 3B). In contrast, full-length NBD2 is stable (Fig. 1D), suggesting that the isolated domain can attain proper structure and that the CytoQC machinery exhibits a high degree of substrate selectivity.
To date, the rules underlying the E3 ubiquitin ligase requirements for different CytoQC substrates are not defined. There are multiple E3 ligases that participate in CytoQC, but most substrates are primarily handled by Doa10p, Ubr1p, and/or San1p. Some of the previously studied CytoQC substrates are normally translocated into the ER but were engineered to instead reside in the cytoplasm by ablating the ER-targeting, hydrophobic signal sequence (26, 27, 73). These substrates might display multiple misfolded lesions because they may lack proper post-translational modifications due to their aberrant localization and because they are excluded from the folding machinery and environment that they normally encounter. Because NBD2* resides in the cytoplasm (whether in the context of Ste6p* or as used in this study), this issue is circumvented. Nevertheless, NBD2* does not retain the Doa10p dependence observed for Ste6p* and instead utilizes San1p and, to a lesser extent, Ubr1p for degradation and ubiquitination (Fig. 4). These data indicate that a substrate removed from the ER “platform” must, by definition, utilize another ligase. It is also notable that NBD2* is not completely stabilized in a san1Δubr1Δ strain, suggesting that other E3 ligases, such as Hul5p, Rsp5p, or Rkr1p/Ltn1p (74–80), might facilitate NBD2* degradation. It is worth reiterating that some CytoQC substrates in yeast, including a mammalian kinase expressed in this organism (81), preferentially use the ER platform/Doa10 in order to become modified with ubiquitin (2, 24, 25).
Because a lack of membrane anchoring releases NBD2* from Doa10p-dependent modification, it is tempting to ask which feature(s) of NBD2* direct it toward the San1p-dependent degradation pathway. A recent report from the Gardner laboratory demonstrates that San1p can recognize substrates via continuous stretches of at least five hydrophobic amino acids (82, 83). However, NBD2* does not appear to contain any obvious San1p recognition motifs (data not shown). Two previously studied CytoQC substrates, ΔssCPY* and ΔssPrA, show a strong dependence on San1p (26, 27), and because both are mislocalized substrates, they probably expose otherwise buried hydrophobic motifs, which are recognized by San1p. To this end, San1p contains natively unfolded regions at its N and C termini (82, 83) that may be sufficient to mediate its association with unfolded protein substrates. Thus, we were surprised to discover a strong Ssa1p/Hsp70 dependence on the interaction between San1p and NBD2* (Fig. 8). However, nearly every CytoQC substrate relies on Ssa1p for efficient degradation (Table 3), but the contribution of Ssa1p to the degradation of each substrate is different. For example, Ssa1p can maintain solubility (2), promote substrate ubiquitination (2, 27, 29), and facilitate substrate transport into the nucleus (26, 27). Using our in vitro ubiquitination assay, we show that there is a nuclear transport-independent role for Ssa1p in NBD2* degradation (Fig. 7). Our data indicate for the first time that chaperones can directly assist San1p-mediated recognition of CytoQC substrates.
TABLE 3.
Survey of the degradation requirements for select CytoQC substrates
The table demonstrates the factors that have been tested for their involvement in the CytoQC of the indicated substrates, and a brief description of the substrates is also provided. The assays used to examine the contribution of each factor represent a mixture of both pulse-chase and cycloheximide chase analyses. +, a requirement; (+), a slight requirement; −, no requirement; ND, not determined.
| Substrate | Description | Hsp70 (Ssa1p) | Hsp110 (Sse1p) | Hsp40 (Ydj1p) | E3 (Doa10p) | E3 (San1p) | E3 (Ubr1p) | Cdc48p Complex | Reference/Source |
|---|---|---|---|---|---|---|---|---|---|
| HA-NBD2* | C-terminal nucleotide binding domain of Ste6p* | + | + | + | − | + | (+) | + | This Study |
| ΔssCPY* | CPY* (ERAD substrate) lacking ER signal sequence | + | − | + | − | ND | ND | − | Ref. 68 |
| ΔssCPY* | + | + | + | ND | + | (+) | ND | Ref. 27 | |
| Ura3p-CL1 | CL1 degron fused to Ura3p | + | ND | + | + | ND | ND | (+) | Ref. 2 |
| Matα2 | Short-lived transcription factor | ND | ND | ND | + | ND | ND | − | Ref. 24 and 25 |
| ΔssPrA | Vacuolar proteinase A lacking signal sequence | + | + | + | ND | + | (+) | ND | Ref. 26 |
| Δ2GFP | GFP with a disrupted β-barrel | + | + | + | ND | + | (+) | ND | Ref. 26 |
| tGnd1 | Truncated phosphogluconate dehydrogenase | + | + | ND | ND | (+) | + | ND | Ref. 27 |
| VHL | Tumor suppressor protein with point mutation | + | + | − | ND | ND | ND | ND | Ref. 21 |
To date, there is no consensus on whether the Cdc48p/p97 complex is required for CytoQC (Table 3), and this may reflect distinct, undefined features embedded in each substrate. Cdc48p may prevent and/or reverse aggregation or perhaps shuttle substrates to the proteasome by virtue of its association with the proteasome (84). We found that NBD2* degradation depends on Cdc48p function (Fig. 5), and similarly, the Cdc48p complex is required for the degradation of Ura3p-CL1 (2) and for fusions of the Matα2 degron (Deg1) to GFP (Deg1-GFP) (62, 63). In contrast, others have examined Deg1-GFP fusions and found no role for the Cdc48 complex (25, 85). These differences are puzzling and point toward a need to further explore the role of Cdc48p in CytoQC. One possible explanation for the inconsistent results could stem from different cdc48 alleles that were used, because among the studies that have looked at Cdc48 complex dependence, at least seven different mutant alleles have been used (2, 25, 62, 63, 73, 85). In addition, the tested alleles are temperature-sensitive to varying extents, which raises the possibility that some studies may have been completed at a non-optimal temperature to elicit a phenotype or that eliciting a phenotype for CytoQC requires conditions different from those previously used. Based on recent publications describing the role of Cdc48p in the degradation of nascent proteins at the ribosome (78, 86), it is also possible that Cdc48p function during CytoQC may occur co-translationally in some cases.
During the preparation of this manuscript, a preliminary report was published that examined the quality control of a C-terminally tagged version of NBD2* (44). This work uncovered some of the same co-factors required for substrate degradation but did not explore the underlying mechanism of quality control and lacked several controls to validate the use of the substrate. Moreover, there was a critical difference between the substrate used in their study and NBD2*, as reported here. Specifically, we explored the CytoQC of an N-terminally tagged form of the domain and compared the stability of our N-terminally tagged version with that of wild-type NBD2 (Fig. 1D), which was lacking from the other report. The use of a C-terminal tag might significantly alter how NBD2* is selected for CytoQC. In Ste6p*, the NBD2* truncation is necessary and sufficient to direct this protein for ERAD because the full-length protein is significantly more stable (31), but by appending a tag at this position and thus extending the open reading frame, the “decision” to degrade the protein might be altered. For example, the C-terminally tagged form of NBD2* exhibited a more pronounced Ubr1p dependence (44) than seen in the current study (Fig. 4B). The underlying reason for this phenomenon may be an artifact of the C-terminally appended tag, or it might arise from recognition of the protein by the N-end rule pathway, which recognizes N-terminal destabilizing amino acids (87, 88). However, the free N terminus of NBD2* lacks any of the characterized destabilizing residues or known susceptibility to destabilizing modifications (89). Importantly, the addition of the HA tag to the N terminus of NBD2* also does not add any of the N-end rule recognition signals. Further evidence of the negative effects of adding a C-terminal tag to NBD2* is that there was no Cdc48 dependence of this substrate (44). In contrast to our exploration of the mechanism of Hsp70 dependence, the other study also did not report whether ssa mutants affected the solubility or ubiquitination of the substrate, which is essential to properly define the role of Hsp70. For example, we found that Ssa1 facilitates ubiquitination and that NBD2 and NBD2* have a propensity to aggregate (Fig. 2A), which may differ for C-terminal tagged versions of NBD2* and have important implications for the role for Hsp70 on CytoQC.
In summary, our work demonstrates that the same misfolded domain/degron recognized for ERAD in a chaperone- and Doa10-dependent fashion can also be recognized by the San1p-dependent CytoQC pathway. Moreover, we find that the degron has retained its requirement for chaperone-mediated selection. Given the many conformations misfolded proteins may adopt, future work will be needed to define new factors and mechanisms that work together to aid in the clearance of misfolded cytoplasmic proteins.
Acknowledgments
We thank Davis Ng, Richard Gardner, Susan Michaelis, and Randy Hampton for reagents and Kunio Nakatsukasa for cloning early NBD2* constructs that were not used in this study.
This work was supported, in whole or in part, by National Institutes of Health National Research Service Award 1F32GM090364 (to C. J. G.) and Grants GM75061 and DK79307 (University of Pittsburgh George O'Brien Kidney Research Center) (to J. L. B.). This work was also supported by American Heart Association Grant 09POST2140058 (to C. J. G.) and a Beckman Scholars award (to K. F. W.).
- UPS
- ubiquitin-proteasome system
- ER
- endoplasmic reticulum
- CytoQC
- cytoplasmic quality control
- ERAD
- endoplasmic reticulum-associated degradation
- Hsp
- heat shock protein
- Ste6p
- sterile 6 protein
- NBD2
- full-length second nucleotide binding domain from Ste6p
- NBD2*
- truncated second nucleotide binding domain from Ste6p*.
REFERENCES
- 1. McCracken A. A., Brodsky J. L. (1996) Assembly of ER-associated protein degradation in vitro. Dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Metzger M. B., Maurer M. J., Dancy B. M., Michaelis S. (2008) Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 283, 32302–32316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guerriero C. J., Brodsky J. L. (2012) The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 92, 537–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vembar S. S., Brodsky J. L. (2008) One step at a time. Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feige M. J., Hendershot L. M. (2011) Disulfide bonds in ER protein folding and homeostasis. Curr. Opin. Cell Biol. 23, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pearse B. R., Hebert D. N. (2010) Biochim. Biophys. Acta Mol. Cell Res. 1803, 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) N-Glycan structures. Recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 8. Claessen J. H., Kundrat L., Ploegh H. L. (2012) Protein quality control in the ER. Balancing the ubiquitin checkbook. Trends Cell Biol. 22, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagiwara M., Nagata K. (2012) Redox-dependent protein quality control in the endoplasmic reticulum. Folding to degradation. Antioxid. Redox Signal. 16, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 10. Wolf D. H., Stolz A. (2012) Biochim. Biophys. Acta Mol. Cell Res. 1823, 117–124 [DOI] [PubMed] [Google Scholar]
- 11. Meyer H., Bug M., Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 12. Hampton R. Y., Sommer T. (2012) Finding the will and the way of ERAD substrate retrotranslocation. Curr. Opin. Cell Biol. 24, 460–466 [DOI] [PubMed] [Google Scholar]
- 13. Kopito R. R., Sitia R. (2000) Aggresomes and Russell bodies. Symptoms of cellular indigestion? EMBO Rep. 1, 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnston J. A., Ward C. L., Kopito R. R. (1998) Aggresomes. A cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaganovich D., Kopito R., Frydman J. (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarkar S., Perlstein E. O., Imarisio S., Pineau S., Cordenier A., Maglathlin R. L., Webster J. A., Lewis T. A., O'Kane C. J., Schreiber S. L., Rubinsztein D. C. (2007) Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 3, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carmichael J., Chatellier J., Woolfson A., Milstein C., Fersht A. R., Rubinsztein D. C. (2000) Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease. Proc. Natl. Acad. Sci. U.S.A. 97, 9701–9705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamamoto A., Cremona M. L., Rothman J. E. (2006) Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J. Cell Biol. 172, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Filimonenko M., Isakson P., Finley K. D., Anderson M., Jeong H., Melia T. J., Bartlett B. J., Myers K. M., Birkeland H. C., Lamark T., Krainc D., Brech A., Stenmark H., Simonsen A., Yamamoto A. (2010) The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 38, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. (2005) Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 102, 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McClellan A. J., Scott M. D., Frydman J. (2005) Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 [DOI] [PubMed] [Google Scholar]
- 22. Lee do H., Goldberg A. L. (2010) Hsp104 is essential for the selective degradation in yeast of polyglutamine expanded ataxin-1 but not most misfolded proteins generally. Biochem. Biophys. Res. Commun. 391, 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee D. H., Sherman M. Y., Goldberg A. L. (1996) Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 4773–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swanson R., Locher M., Hochstrasser M. (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15, 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ravid T., Kreft S. G., Hochstrasser M. (2006) Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prasad R., Kawaguchi S., Ng D. T. (2010) A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell 21, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heck J. W., Cheung S. K., Hampton R. Y. (2010) Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U.S.A. 107, 1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Varshavsky A. (2006) The early history of the ubiquitin field. Protein Sci. 15, 647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nillegoda N. B., Theodoraki M. A., Mandal A. K., Mayo K. J., Ren H. Y., Sultana R., Wu K., Johnson J., Cyr D. M., Caplan A. J. (2010) Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell 21, 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Metzger M. B., Michaelis S. (2009) Analysis of quality control substrates in distinct cellular compartments reveals a unique role for Rpn4p in tolerating misfolded membrane proteins. Mol. Biol. Cell 20, 1006–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loayza D., Tam A., Schmidt W. K., Michaelis S. (1998) Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 33. Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (1997) Media and stock preservation. in Methods in Yeast Genetics, pp. 145–160, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Wang W., Malcolm B. A. (1999) Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 36. Mumberg D., Müller R., Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y., Michaelis S., Brodsky J. L. (2002) CFTR expression and ER-associated degradation in yeast. Methods Mol. Med. 70, 257–265 [DOI] [PubMed] [Google Scholar]
- 38. Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. (1992) Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 3, 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sullivan M. L., Youker R. T., Watkins S. C., Brodsky J. L. (2003) Localization of the BiP molecular chaperone with respect to endoplasmic reticulum foci containing the cystic fibrosis transmembrane conductance regulator in yeast. J. Histochem. Cytochem. 51, 545–548 [DOI] [PubMed] [Google Scholar]
- 40. Amberg D. C., Burke D., Strathern J. N., and Cold Spring Harbor Laboratory (2005) Yeast immunofluorescence. in Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, pp. 149–152, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Brodsky J. L., Schekman R. (1993) A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berkower C., Michaelis S. (1991) Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 10, 3777–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuchler K., Sterne R. E., Thorner J. (1989) Saccharomyces cerevisiae STE6 gene product. A novel pathway for protein export in eukaryotic cells. EMBO J. 8, 3973–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prasad R., Kawaguchi S., Ng D. T. (2012) Biosynthetic mode can determine the mechanism of protein quality control. Biochem. Biophys. Res. Commun. 425, 689–695 [DOI] [PubMed] [Google Scholar]
- 45. Mizunaga T., Katakura Y., Miura T., Maruyama Y. (1990) Purification and characterization of yeast protein disulfide isomerase. J. Biochem. 108, 846–851 [DOI] [PubMed] [Google Scholar]
- 46. Loayza D., Michaelis S. (1998) Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 48. Jones E. W. (1984) The synthesis and function of proteases in Saccharomyces. Genetic approaches. Annu. Rev. Genet. 18, 233–270 [DOI] [PubMed] [Google Scholar]
- 49. Gaczynska M., Osmulski P. A. (2005) Small-molecule inhibitors of proteasome activity. Methods Mol. Biol. 301, 3–22 [DOI] [PubMed] [Google Scholar]
- 50. Lee D. H., Goldberg A. L. (1996) Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271, 27280–27284 [DOI] [PubMed] [Google Scholar]
- 51. Vashist S., Ng D. T. (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rubenstein E. M., Kreft S. G., Greenblatt W., Swanson R., Hochstrasser M. (2012) Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J. Cell Biol. 197, 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dasgupta A., Ramsey K. L., Smith J. S., Auble D. T. (2004) Sir antagonist 1 (San1) is a ubiquitin ligase. J. Biol. Chem. 279, 26830–26838 [DOI] [PubMed] [Google Scholar]
- 54. Gardner R. G., Nelson Z. W., Gottschling D. E. (2005) Degradation-mediated protein quality control in the nucleus. Cell 120, 803–815 [DOI] [PubMed] [Google Scholar]
- 55. Summers D. W., Wolfe K. J., Ren H. Y., Cyr D. M. (2013) The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One 8, e52099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ye Y., Meyer H. H., Rapoport T. A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 57. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bays N. W., Hampton R. Y. (2002) Cdc48-Ufd1-Npl4. Stuck in the middle with Ub. Curr. Biol. 12, R366–R371 [DOI] [PubMed] [Google Scholar]
- 59. Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139 [DOI] [PubMed] [Google Scholar]
- 61. Hitchcock A. L., Krebber H., Frietze S., Lin A., Latterich M., Silver P. A. (2001) The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12, 3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 63. Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005) Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 64. Lipson C., Alalouf G., Bajorek M., Rabinovich E., Atir-Lande A., Glickman M., Bar-Nun S. (2008) A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J. Biol. Chem. 283, 7166–7175 [DOI] [PubMed] [Google Scholar]
- 65. Park S.-Y., Ye H., Steiner D. F., Bell G. I. (2010) Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted. Biochem. Biophys. Res. Commun. 391, 1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 67. Kriegenburg F., Ellgaard L., Hartmann-Petersen R. (2012) Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 279, 532–542 [DOI] [PubMed] [Google Scholar]
- 68. Park S. H., Bolender N., Eisele F., Kostova Z., Takeuchi J., Coffino P., Wolf D. H. (2007) The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 18, 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mandal A. K., Gibney P. A., Nillegoda N. B., Theodoraki M. A., Caplan A. J., Morano K. A. (2010) Hsp110 chaperones control client fate determination in the Hsp70-Hsp90 chaperone system. Mol. Biol. Cell 21, 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. (2002) Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1–151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell 13, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shaner L., Morano K. A. (2007) All in the family. Atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hrizo S. L., Gusarova V., Habiel D. M., Goeckeler J. L., Fisher E. A., Brodsky J. L. (2007) The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic reticulum-associated degradation (ERAD). J. Biol. Chem. 282, 32665–32675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Medicherla B., Kostova Z., Schaefer A., Wolf D. H. (2004) A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fang N. N., Ng A. H., Measday V., Mayor T. (2011) Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat. Cell Biol. 13, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 76. Tofaris G. K., Kim H. T., Hourez R., Jung J. W., Kim K. P., Goldberg A. L. (2011) Ubiquitin ligase Nedd4 promotes α-synuclein degradation by the endosomal-lysosomal pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang S., Thibault G., Ng D. T. (2011) Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity. J. Biol. Chem. 286, 29376–29387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Verma R., Oania R. S., Kolawa N. J., Deshaies R. J. (2013) Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2, e00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Theodoraki M. A., Nillegoda N. B., Saini J., Caplan A. J. (2012) A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J. Biol. Chem. 287, 23911–23922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bengtson M. H., Joazeiro C. A. (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Arteaga M. F., Wang L., Ravid T., Hochstrasser M., Canessa C. M. (2006) An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc. Natl. Acad. Sci. U.S.A. 103, 11178–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fredrickson E. K., Rosenbaum J. C., Locke M. N., Milac T. I., Gardner R. G. (2011) Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol. Biol. Cell 22, 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fredrickson E. K., Gallagher P. S., Clowes Candadai S. V., Gardner R. G. (2013) Substrate recognition in nuclear protein quality control degradation is governed by exposed hydrophobicity that correlates with aggregation and insolubility. J. Biol. Chem. 288, 6130–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Proteasomal proteomics. Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3, 24–29 [DOI] [PubMed] [Google Scholar]
- 86. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C., Li G. W., Zhou S., King D., Shen P. S., Weibezahn J., Dunn J. G., Rouskin S., Inada T., Frost A., Weissman J. S. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bachmair A., Finley D., Varshavsky A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 88. Varshavsky A. (1991) Naming a targeting signal. Cell 64, 13–15 [DOI] [PubMed] [Google Scholar]
- 89. Tasaki T., Sriram S. M., Park K. S., Kwon Y. T. (2012) The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 91. Latterich M., Fröhlich K. U., Schekman R. (1995) Membrane fusion and the cell cycle. Cdc48p participates in the fusion of ER membranes. Cell 82, 885–893 [DOI] [PubMed] [Google Scholar]
- 92. Becker J., Walter W., Yan W., Craig E. A. (1996) Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16, 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sanders S. L., Whitfield K. M., Vogel J. P., Rose M. D., Schekman R. W. (1992) Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69, 353–365 [DOI] [PubMed] [Google Scholar]
- 94. Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. (1999) The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274, 3453–3460 [DOI] [PubMed] [Google Scholar]
- 95. Youker R. T., Walsh P., Beilharz T., Lithgow T., Brodsky J. L. (2004) Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15, 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shaner L., Trott A., Goeckeler J. L., Brodsky J. L., Morano K. A. (2004) The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 279, 21992–22001 [DOI] [PubMed] [Google Scholar]
- 98. Moir D., Stewart S. E., Osmond B. C., Botstein D. (1982) Cold-sensitive cell-division-cycle mutants of yeast. Isolation, properties, and pseudoreversion studies. Genetics 100, 547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Berkower C., Loayza D., Michaelis S. (1994) Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol. Biol. Cell 5, 1185–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]