Abstract
PPARγ (peroxisome proliferator-activated receptor-γ) is a master transcription factor involved in adipogenesis through regulating adipocyte-specific gene expression. Recently, lipin1 was found to act as a key factor for adipocyte maturation and maintenance by modulating the C/EBPα (CCAAT/enhancer-binding protein α) and PPARγ network; however, the precise mechanism by which lipin1 affects the transcriptional activity of PPARγ is largely unknown. The results of the present study show that lipin1 activates PPARγ by releasing co-repressors, NcoR1 (nuclear receptor co-repressor 1) and SMRT (silencing mediator of retinoid and thyroid hormone receptor), from PPARγ in the absence of the ligand rosiglitazone. We also identified a novel lipin1 TAD (transcriptional activation domain), between residues 217 and 399, which is critical for the activation of PPARγ, but not PPARα. Furthermore, this TAD is unique to lipin1 since this region does not show any homology with the other lipin isoforms, lipin2 and lipin3. The activity of the lipin1 TAD is enhanced by p300 and SRC-1 (steroid receptor co-activator 1), but not by PCAF (p300/CBP-associated factor) and PGC-1α (PPAR co-activator 1α). The physical interaction between lipin1 and PPARγ occurs at the lipin1 C-terminal region from residues 825 to 926, and the VXXLL motif at residue 885 is critical for binding with and the activation of PPARγ. The action of lipin1 as a co-activator of PPARγ enhanced adipocyte differentiation; the TAD and VXXLL motif played critical roles, but the catalytic activity of lipin1 was not directly involved. Collectively, these data suggest that lipin1 functions as a key regulator of PPARγ activity through its ability to release co-repressors and recruit co-activators via a mechanism other than PPARα activation.
Keywords: co-activator, co-repressor, lipin1, peroxisome proliferator-activated receptor (PPAR)
PPARγ (peroxisome proliferator-activated receptor-γ, a member of the nuclear hormone receptor family, is a master regulator of adipocyte differentiation and function via the control of gene expression [1–3]. Transcriptional regulation of nuclear hormone receptors is controlled by dynamic modulation of co-repressors and co-activators. Upon ligand binding, conformational changes induce the release of co-repressors and the recruitment of co-activators, resulting in chromatin remodeling followed by target gene transcription. However, PGC-1α (PPARγ co-activator 1α) binds to PPARγ in a ligand-independent manner with considerable PPARγ target gene selectivity [4].
Lipin1 was discovered using a positional cloning approach as a mutated gene in fld (fatty liver dystrophic) mouse characterized by a severely reduced mass of adipose tissue, resulting in insulin resistance, and hyperlipidaemia [5]. Lipin1 catalyses the conversion of PA (phosphatidic acid) into diacylglycerol for triacylglycerol synthesis [6,7]. In addition to its enzymatic function as a PAP (phosphatidate phosphatase), lipin1 also act as a co-regulator of transcription factors [8]. The importance of lipin1 's PAP activity has been evaluated by observing the elevated levels of PA after the suppression of PPARγ expression [9]. The role of triacylglycerol synthesis in adipocyte differentiation is further supported by the fact that the defect of genes involved in triacylglycerol biosynthesis blocked adipocyte differentiation [10,11]. The role of lipin1 as a transcriptional regulator has been described in in various cell types [12–15]. Although lipin1 does not have a DNA-binding motif, it has been shown to be involved in transcriptional regulation through interaction with transcription factors: a yeast homologue of lipin1 was found to be recruited to the promoters of phospholipid biosynthetic genes [16], lipin1 amplifies PPARα action by increasing its interaction with PGC-1α [12], PPARγ-dependent gene expression is regulated by lipin1 in adipocytes [14] and NFATc4 (nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4) transcriptional activity is repressed by lipin1 through protein-protein interactions [15].
Currently, the C-LIP (C-terminal conserved domain) of lipin1 is thought to be important for lipin1's transcription-regulating functions, since C-LIP is required for the association with PPARα, PPARγ and NFATc4 [12,14,15]. C-LIP contains several functional motifs that have been characterized. The DXDXT motif is required for the PAP activity and the LXXIL motif is known to be necessary for PPARα activation [12]. To repress NFATc4-mediated inflammatory gene expression lipin1 uses both the DXDXT and LXXIL motif [15]. These results suggest that lipin1 could regulate various transcription factors via utilizing its different motifs.
We reported previously that lipin1 regulates the positive feedback loop between PPARγ2 and C/EBPα (CCAAT/enhancer-binding protein α), resulting in differentiation and functional maintenance of 3T3-L1 adipocytes [14]. However, the precise mechanism by which lipin1 affects transcriptional activity of PPARγ is largely unknown. In the present study, we described how lipin1 increases the transcriptional activity of PPARγ by the following mechanisms: (i) lipin1 has the TAD (transcriptional activation domain) with a specificity for PPARγ transcriptional activation that could be modulated by p300 and SRC-1 (steroid receptor co-activator 1), but not by PCAF (p300/CBP-associated factor) and PGC-1α; (ii) the VXXLL motif located in the lipin1 C-LIP is critical for the physical interaction with PPARγ and its activation; (iii) adipocyte differentiation could be induced by the lipin1-mediated activation of PPARγ, which required the TAD and the VXXLL motif; and (iv) lipin1 disrupts the interaction between co-repressors and PPARβ. The present study provides compelling evidence that lipin1 can act as a natural regulator of PPARγ activity and reveals new insights into the mechanism of PPARγ activation.
EXPERIMENTAL
Cell culture
NIH 3T3, HEK (human embryonic kidney)-293, HeLa and PC3 (a human prostate cancer cell line) cells were obtained from the A.T.C.C. (Manassas, VA, U.S.A.). All reagents related to animal cell culture were purchased from Life Technologies. NIH 3T3, HEK-293 and HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium) and the PC3 cells were cultured in RPMI 1640 medium. All media contained 10% FBS (fetal bovine serum), 100 units/ml penicillin and 0.1 mg/ml streptomycin. The cells were cultured at 37 °C in a 5% CO2 humidified atmosphere. 3T3-L1 pre-adipocytes were propagated and maintained in DMEM supplemented with 10% FBS and were cultured at 37 °C in a 10% CO2 humidified atmosphere. The 2 mM stock solutions of the PPARγ ligand rosiglitazone and the PPARα ligand WY14643 were prepared in DMSO (Sigma–Aldrich). Ligand-treated and control cultures received the same amount of DMSO and the final DMSO concentration did not exceed 0.1 %.
Plasmid constructs
Plasmids including 3xPPRE-tk-LUC, pSG5-HA-PPARγ, pSG5-FLAG-lipin1, pSG5-FLAG-lipin1 (1-441) and pSG5-FLAG-lipin1 (425-926) were described previously [14]. The pSG5-FLAG-lipin1-(425-828) construct was generated from pSG5-FLAG-lipin1-(425-926) by removing the 3′ fragment of the lipin1 cDNA. The pSG5-FLAG-lipin1-(726-926) construct was generated from pSG5-FLAG lipin1-( 425-926) by removing the 5′ region of the lipin1 cDNA. The pSG5-lipin1ΔTAD was obtained by removing the region corresponding to the transcriptional activation domain (residues 217–399) from pSG5-FLAG-lipin1 using site-directed mutagenesis. The various mutants were created using Pfu Thrbo DNA polymerase (Stratagene). The PCR cycling conditions used in site-directed mutagenesis were 18 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 1 min and extension at 68 °C for 10 min. Amplified mixtures were treated with DpnI at 37 °C for 1 h and aliquots (10 μl) were used to transform competent Escherichia coli cells.
Transfections
NIH 3T3, PC3, HeLa and HEK-293 cells were plated in 10-cm plates or 6-well plates 1 day before transfection. When the cells reached 60–80% confluency, they were transiently transfected with the indicated plasmids using the Plus™ Reagent and Lipofectamine™ Reagent (Life Technologies) according to the supplier's protocol. Equivalent amounts of DNA were transfected by adjusting with empty vector as necessary.
Western blot analyses
Cultured cells were washed once with ice-cold PBS and harvested in whole-cell lysis buffer [0.5% Triton X-100, 20 mM Hepes (pH 7.4), 150 mM NaCl, 2 mM dithiothreitol and 1 mM PMSF]. Lysates were briefly vortex mixed and cleared by centrifugation at 10,000X g for 20 min at 4 °C. The supernatants were collected and transferred into a fresh tube. Protein concentrations were determined using the 660 nm protein assay reagent (Pierce). Equal amounts of protein extracts were subjected to SDS/PAGE (8% gel) and transferred on to nitrocellulose transfer membranes (Whatman). The membranes were blocked in Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 (Sigma–Aldrich) and 5% (w/v) non-fat Difco™ skimmed milk (BD Biosciences), followed by incubation with the primary antibodies. The following antibodies were used: anti-HA (haemagglutinin; Santa Cruz Biotechnology), anti-FLAG (Sigma–Aldrich), anti-PPARγ (Santa Cruz Biotechnology), anti-SMRT (silencing mediator of retinoid and thyroid hormone receptor; BD Biosciences) and anti-NCoR1 (nuclear receptor co-repressor 1; ATGen). The signals were developed using SuperSignal® West Pico Chemiluminescent Substrate System (Pierce) according to the manufacturer's protocol.
Luciferase assays
Luciferase activity in whole-cell lysates was measured using the Dual-Luciferase® reporter assay system (Promega). Cells are treated with 1 μM of WY14643 or rosiglitazone 24h after transfection and harvested the next day for use. The relative luciferase activity was obtained by normalizing the reporter luciferase activity with the value of Renilla luciferase using the pRL-SV40 construct. All data shown are from three independent experiments performed in triplicate.
Co-immunoprecipitation assays
PPARγ and lipin1 C-terrninal fragments were overexpressed in HEK-293 cells. The transfected cells were incubated for 2 days, washed twice with PBS, scraped and ntrifuged at 5,000X g for 2 min at 4 °C. Pellets were lysed in * [50 mM Tris (pH 7.4), 150 mM NaCl, 0.5% Triton X-100 and 0.5% Nonidet P40] and centrifuged at ?? g for 10 min at 4 °C. Supernatants were transferred to a new tube and 1-ml aliquots were rotated for 10 min at 4 °C with 30 μl of Protein A/G plus–agarose (Santa Cruz Biotechnology), followed by centrifugation at 10,000X g for 3 min at 4 °C. Supernatants were transferred into a new tube and incubated with rotation for 12 h at 4 °C with the anti-HA antibody (Santa Cruz Biotechnology). The lysates were added to 30 μl of Protein A/G plus–agarose and rotated for 90 min at 4 °C. After centrifugation at 10,000X g for 3 min at 4 °C, the pellets were resuspended in wash buffer (*/PBS, 1:2) and incubated for 10 min at 4 °C. This wash step was repeated three times. The immunoprecipitates were suspend in 30 μl of 2× SDS sample buffer and heated at 95 °C for 5 min. After a brief centrifugation, the supernatants were subjected to Western blot analyses.
Yeast transformation
Yeast vectors encoding fusion proteins between the GAL4-DBD (galactose metabolism 4 DNA-binding domain) and lipin1 fragment were generated by cloning lipin1 cDNAs into the pGBKT7 vector (Clontech). Vectors were introduced into AH109 yeast cells (Clontech) using the yeast transformation system (Clontech) according to the manufacturer's instructions. Transformed yeast cells were plated on SD (synthetic defined dropout) agar plates, in which appropriate nutrients, such as tryptophan, histidine and/or adenine, were depleted. Plates were incubated for 4 days at 30°C.
Transfection and in vitro differentiation of 3T3-L1 cells
The 3T3-L1 cells were trypsinized, washed with 1× PBS and finally resuspended in 10 μl of 1% SDS, 60 mM Tris-HCl, pH 6.8 and 0.5 μg of plasmid at a concentration of 2×105 cells per reaction. The cells were then microporated utilizing OneDrop MicroPorator MP-100 (Digital Bio) at 1300 V, 20-ms pulse width and two pulses. Following microporation, 1 × 105 cells were seeded in 35-mm cell culture dishes and placed at 37 °C in a 10% CO2-humidified atmosphere. The next day the differentiation of transfected 3T3-L1 cells were induced by the addition of a 0.5 mM IBMX (isobutylmethylxanthine), 1 μM dexamethasone and 1 μg/ml insulin to DMEM containing 10% FBS. At 2 days later the medium was replaced by DMEM containing 1 μg/ml insulin and 10% FBS. Culture media were renewed every 2 days thereafter with DMEM supplemented with 10% FBS.
Oil Red O staining
Cells were washed with PBS then fixed with 3.7% formaldehyde for 2 min at room temperature (23 °C). The cells were then washed with water, stained with 0.5% Oil Red O solution (isopropanol/water, 3:2) for 1 h at room temperature, washed again with water and then observed.
Reverse transcription and real-time PCR
Total RNA was purified from cultured cells with TRizol® reagent according to the manufacturer's instructions (Invitrogen). First-strand cDNA synthesis was performed using oligo(dT) primers with SuperScriptTM IT reverse transcriptase (Invitrogen) following the supplier's protocol. Quantitative real-time PCR analyses were performed on the 7300 Real Time PCR System (Applied Biosystems) using SYBR® Green PCR Master Mix (Applied Biosystems). Each assay was performed in triplicate and included four serial dilution points of a control cDNA, a no-template control and each sample cDNA. The relative concentrations of the genes of interest, and the endogenous control (36B4) were calculated by plotting the threshold cycle (Ct) against the log of the serial dilution points. The relative expression of the gene of interest was determined after normalizing to the endogenous control. The sequences of primers used for real-time PCR was described previously [14].
PAP activity measurement
To assay the enzymatic activities of wild-type lipin1 and its mutants, the cell lysates were prepared from HEK-293 cells transfected with plasmids expressing wild-type lipin1 or its mutants in 10-cm dish and then cultured for 2 days. Cell were detached by rubber policeman, washed once with PBS, and then disrupted using a Dounce homogenizer at 4 °C in 50 mM Tris/HCl (pH 7 .5) buffer containing 0.25 M sucrose, 1 mM EDTA, 10 mM 2-mercaptoethanol, 1 mM benzamidine, 0.5 mM PMSF and 5 μg/ml of aprotinin, leupeptin and pepstatin. The lysed cells were centrifuged at 1000 g for 10 min at 4 °C and the supernatant was used as a cell extract. Total PAP activity (Mg2+ dependent and Mg2+ independent) was measured at 37 °C for 20 min in the reaction mixture (total volume of 100 μl) containing 50mM Tris/HCl (pH 7.5), 1 mM MgCl2, 10 mM 2-mercaptoethanol, 0.2 mM [32P]PA (5000 c.p.m./nmol), 2 mM Triton X-100 and enzyme protein. The radioactive [32P]PA was synthesized enzymatically from diacylglycerol and [γ-32P]ATP with E. coli diacylglycerol kinase. The Mg2+ independent PAP activity, which was measured in the same reaction mixture except 2 mM EDTA was used instead of 1 mM MgCl2, which was low to be negligible. A unit of PAP activity was defined as the amount of enzyme that catalysed the formation of 1 nmol of product/min. Specific activity was expressed as units/mg protein. The average S.D. of the assays was ±5%. The reactions were linear with time and protein concentration.
Statistical analyses
All results are expressed as means ± S.E.M. Statistical comparisons of groups were performed using an unpaired Student's t test.
RESULTS
Lipin1 enhances basal and ligand-mediated PPARγ activity
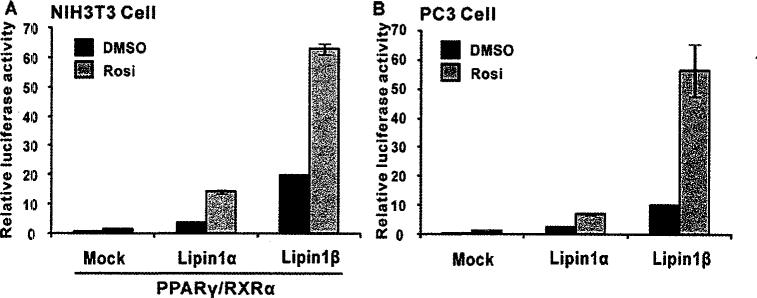
To elucidate the effect of lipin1 on PPARγ transcriptional activity, promoter-reporter assays were performed in the presence or absence of the PPARγ ligand rosiglitazone after co-expression of lipin1 with PPARγ/RXRα (retinoid X receptor). We tested lipin1α and lipin1β, two known lipin1 isoforms resulting from alternative splicing of the lipin1 gene [17]. Noticeably, overexpression of either lipin1α or lipin1β significantly stimulated PPARγ activity even in the absence of ligand (Figure 1A). Moreover, this effect was further enhanced by the PPARγ ligand rosiglitazone (1 μM), whereas overexpression of PPARγ/RXRα alone only caused a slight increase in the PPRE (peroxisome proliferator response element)-tk reporter activity at a given concentration of rosiglitazone. The enhancing effect of lipin1 on PPARγ activity was recapitulated in PC3 cells, prostate cancer line which express high endogenous PPARγ (Figure 1B). It is also noteworthy that lipin1β was more potent than lipin1α with regard to activating PPARγ. These results indicate that lipin1 is a crucial regulator in ligand-dependent and -independent activity of PPARγ.
Figure 1. Lipin1 enhances PPARγ transcriptional activity.
(A) NIH 3T3 cells were cotransfected with 3× PPRE-tk luciferase constructs, PPARγ and RXR with a combination of pSG5-KH2M1 (Mock), pSG5-lipin1α or lipin1β. (B) PC3 cells were transfected with 3× PPRE-tk luciferase constructs along with empty vector, pSG5-lipin1α or lipin1β. Rosiglitazone (Rosi; 1 μM) was added 1 day after transfection. Relative luciferase activity was calculated as described in Experimental section.
Lipin1 C-terminus plays a critical role in the interaction with PPARγ
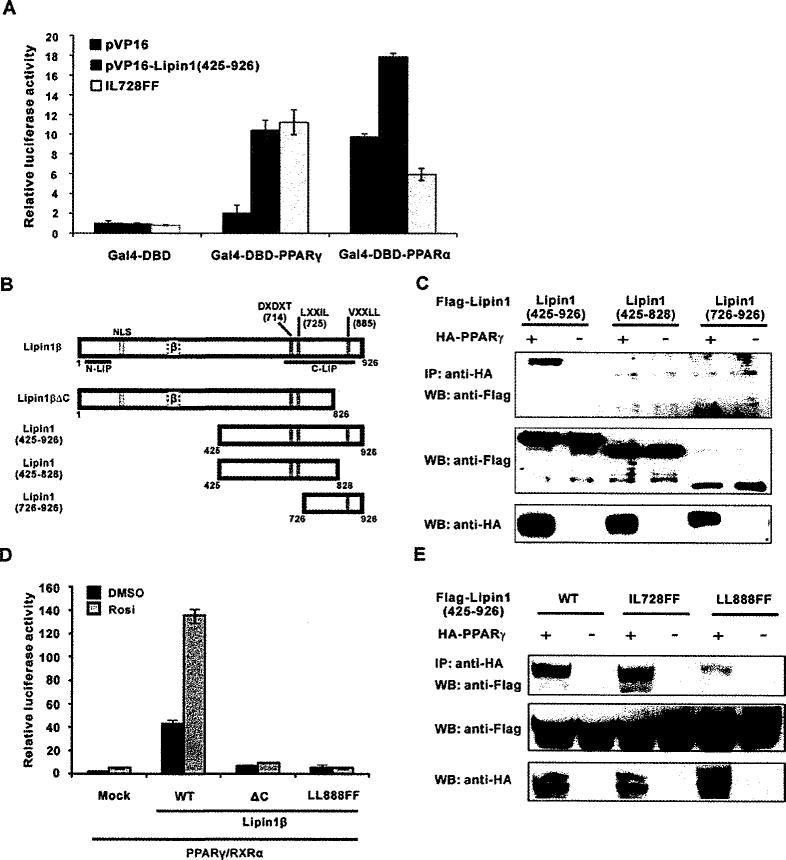
We reported previously that the C-terminal region of lipin1 (residues 425–926) interacts with PPARγ, and there are reports showing lipin1 C-terminus interacts with transcriptional regulators, such as PGC-1α, PPARα and NFATc4 [12,14,15]. Since the lipin1 C-terminal region contains an LXXIL motif (residues 725–729), which has been shown to be important for PPARα activation [12], it was intriguing to see whether it is also required for PPARγ activation. First, to confirm that PPARγ or PPARα interacts with the lipin1 C-terminus, a mammalian two-hybrid assay was performed in NIH 3T3 cells by overexpressing PPARs and lipin1 (residues 425–926) fused with GAL4-DBD and the VP16 activation domain respectively. We measured the luciferase activity of pFR-Luc reporter that has the UAS (upstream activation sequence) as the binding element for GAL4-DBD (Figure 2A). The wild-type lipin1 (residues 425–926) fusion protein significantly enhanced the activities of both GAL4-DBD–PPARγ and the GAL4-DBD–PPARα fusion proteins, but not GAL4-DBD itself. Consistent with the data from a previous study [12], mutation of the LXXIL motif to LXXFF (IL728FF) of lipin1 severely attenuated its interaction with PPARα. However, the interaction of lipin1 with PPARγ was not affected by this mutation. Next, we confirmed this result by carrying out co-immunoprecipitation. We constructed several C-terminal fragments of lipin1 to find the region of lipin1 to which PPARγ binds (Figure 2B). As shown in Figure 2(C), lipin1 (residues 725–926) C-terminal small fragment still strongly binds to PPARγ, whereas lipin1 (residues 425–828), even though the LXXIL motif is intact, was not co-immunoprecipitated with PPARγ. This result indicates that PPARγ interacts with most C-terminal region of lipin1 (residues 725–926) and LXXIL motif is not critical to this interaction.
Figure 2. Lipin1 interacts with PPARγ via its C-terminal region.
(A and B) NIH 3T3 cells were transfected with the reporter plasmid pFR-Luc along with the vectors encoding GAL4-DBD–PPARα or –PPARγ. VP16–lipin1 (residues 425–927) or the IL728FF mutant construct were also co-transfected. Ligands for PPARα (WY14643) or PPARγ (rosiglitazone) was treated the next day and cells were incubated for another 24 h before the assay. (C) Lipin1 constructs used in the experiments. NLS, nuclear localization signal; DXDXT, PAP enzymatic motif; LXXIL, nuclear receptor-interaction motif; VXXLL, PPARγ-interacting motif. (D) NIH 3T3 cells were co-transfected with the 3× PPRE-tk luciferase construct and the PPARγ2 and RXR vectors, along with pSG5-KH2M1 (Mock), pSG5-lipin1β, pSG5-lipin1ΔC or pSG5-lipin1 LLBBBFF as indicated. The next day, cells were treated with 1 μM rosiglitazone (Rosi.) and incubated for another 24 h. Relative luciferase activity was calculated as described in Experimental section. (E) FLAG–triacylglycerolged lipin1 fragments or its mutants were overexpressed with or without HA–PPARγ in HE-K293 cells. Co-immunoprecipitation performed as described in Experimental section. IP, immunoprecipitation; WB, Western blot.
We further investigate the role of this C-terminal region in terms of regulating the activity of PPARγ. As shown in Figure 2(D), a mutant lipin1β (ΔC), which was constructed by deletion of residues 827–926, drastically attenuated the activation of PPARγ, indicating that lipin1-mediated activation of PPARγ requires the C-terminal region from residues 827 to 926 of lipin1. Having shown the LXXIL motif in lipin1 is not necessary for its binding with PPARγ and that the lipin1 C-terminal residues 827–926 are involved in interacting with PPARγ, we speculated that residues 827–926 have another motif that is necessary for the binding with PPARγ. We found VFPLL motif (residues 885–889) as a candidate sequence similar to the LXXLL motif. We mutated two lysine residues of VFPLL to phenylalanines (LL888FF) and its abilit to activate PPARγ activity was assessed. As shown in Figure 2(D), the LL888FF mutant totally lost the ability to activate PPARγ. Furthermore, the LL888FF mutant did not interact with PPARγ, whereas the IL728FF mutant could (Figure 2E). Collectively, these data indicate that lipin1 physically interacts with PPARγ via its C-terminal VXXLL motif and this interaction is critical for facilitating the activity of PPARγ.
The N-terminal region of lipin1 contains the TAD
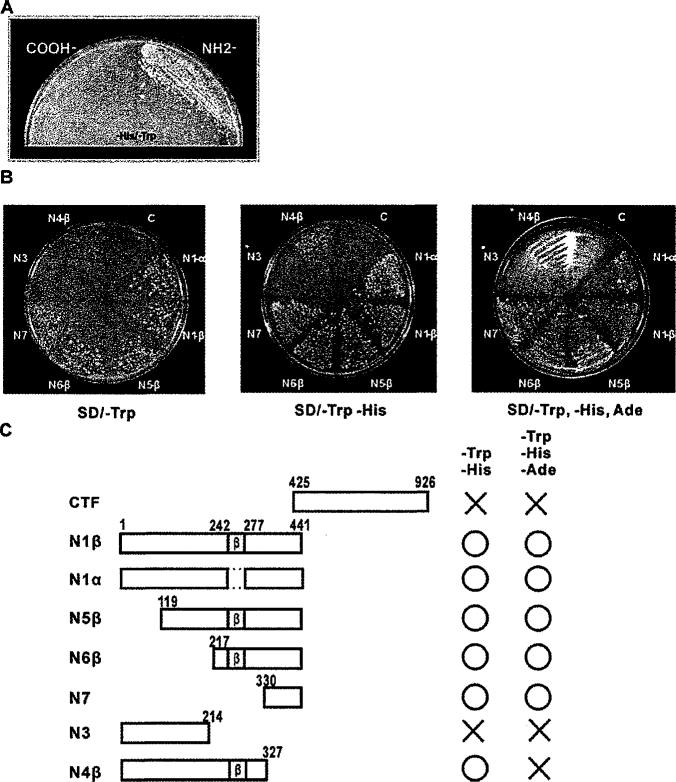
To examine whether lipin1 possesses transcriptional activation function, we constructed yeast vectors encoding the fusion proteins of GAL4-DBD with lipin1 fragments. We evaluated their transcriptional activity by the ability of yeast to grow in media lacking marker nutrients, such as histidine and adenine. Because lipin1 interacts with transcription factors and co-activators through its C-terminal region [12], we expected the C-terminal region to contain the TAD. In contrast to this expectation, the N -terminal fragment of lipin1 (residues 1–441), but not the C-terminal fragment, permitted yeast growth in histidine-deficient medium (Figure 3A), indicating that the N-terminal region contains the TAD. To further delineate the location of TAD in the N-terminal region, yeast vectors encoding various N-terminal lipin1 fragments were constructed and the ability of transformed yeast to grow in selection media was evaluated (Figures 3B and 3C). The N1β (residues 1–441) and N1α fragments, which lack the β isoform-specific region (residues 243–276), showed similar activity. Deletion of the N-terminal portion of the N1β fragment downstream to residue 216 (N5β and N6β fragments) did not significantly affect the transcriptional activity of the N1β fragment. The N-terminal region to residue 214 (N3 fragment) had no activation function, but the addition of residues 215–327 (N4β fragment) induced a weak transcriptional activity. The minimal fragment from residue 330 to 441 (N7 fragment) had a strong activity.
Figure 3. Identification of the lipin1 TAD using a yeast two-hybrid system.
(A) AH109 yeast cell were transformed with the pGBKT7-lipin1 C-terminus or the pGBKT7-lipin1 N-terminus, and plated on SD/ – His/ – Trp agar plates followed by incubation for 4 days at 30°C. (B) The pGBKT7 constructs, containing various fragments of lipin1, were introduced into AH109 yeast cells, and cultured on SD/ – Trp, SD/ – His-Trp or SD/ – His – Trp – Ade agar plates for 4 days at 30°C. (C) Schematic representation showing the ability of the lipin1 fragments to rescue yeast cells from nutritional dependency. CTF, C-terminal fragment.
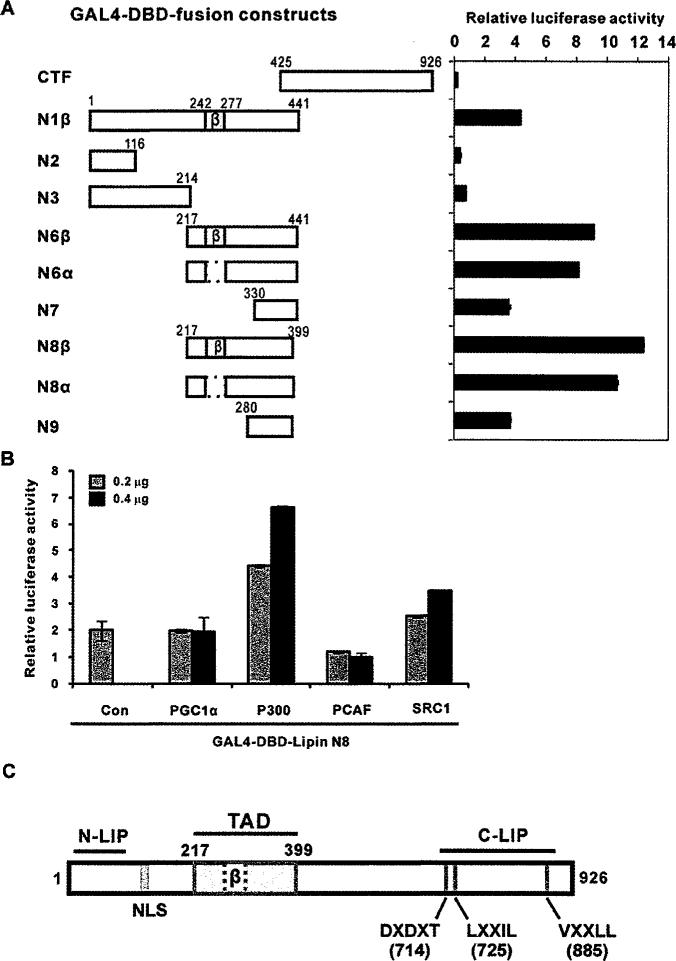
To further evaluate the transcriptional activity of the TAD in animal cells, we co-transfected vectors encoding fusion proteins of the GAL4-DBD flanked with various lipin1 fragments and pFR-Luc reporter vector followed by luciferase activity was measured in HEK-293 cells (Figure 4A). Consistent with the results from the yeast experiments, lipin1 fragments expressing residues 217–399 showed the strongest transactivation activity. Thus we defined this region as the TAD of lipin1, which is not conserved in lipin2 and lipin3 (Figure 4C, TAD). The TAD includes the region corresponding to the alternative exon 7 of the β isoform. The removal of residues 242–277, which are present in the β isoform, slightly decreased the transcriptional activation function (N6β and N8β compared with the N6α and N8α fragments).
Figure 4. GAL4-DBD fusion proteins containing the lipin1 TAD activate UAS-driven transcription.
(A) Left-hand panel, pFA-CMV-lipin1 constructs encoding GAL4-DBD fusion proteins with various fragments of lipin1 and the reporter vector pFR-luciferase were introduced into HEK-293 cells. Right-hand panel, luciferase activity was measured 48 h after transfection. (B) NIH 3T3 cells were co-transfected with pFR-Luc and GAL4-DBD-lipin1 TAD fusion constructs (GAL4-DBD–lipin1 N8) along with co-activator plasmids (PGC-1α, P300, PCAF and SRC-1). Luciferase activity was normalized to Renilla luciferase and the data shown are representative of three independent experiments performed in triplicate. (C) Schematic diagram depicting the domains and motifs of the human lipin1. N-LIP and C-LIP indicate evolutionarily conserved N-terminal and C-terminal lipin domains. TAD indicates the transcriptional activation domain delineated in the present study. The nuclear localization signal (NLS), PAP enzymatic motif (DXDXT) and nuclear receptor interaction motifs (LXXIL and VXXLL) are also shown.
Next, various co-activators were tested for their co-activation effects on the chimaeric transcription factor, which was constructed by fusion of GAL4-DBD and the lipin1 TAD (residues 217–399 N8 (3 fragment) (Figure 4B and S1). Among the co-factors we tested, p300 showed the strongest activation effects, SRC-1 also showed good level of activation in a dose-dependent manner, but PGC-1α and PCAF did not.
Taken together, the results indicate that lipin1 has a TAD (residues 217–399) the activity of which is enhanced by p300 and SRC-1.
The mechanism for lipin1-mediated PPARγ activation differs from that of PPARα activation
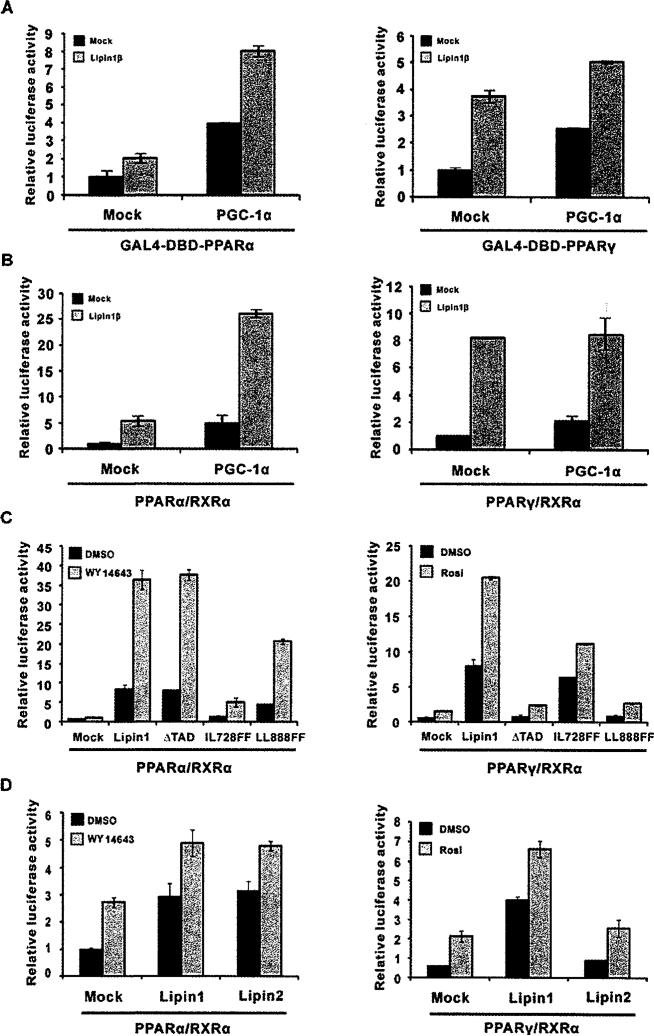
In the liver, where PPARα is highly expressed and controls fatty acid oxidation in response to dietary status, lipin1 plays an important role during a fasting state as a co-activator of PPARα, which, with PGC-1α, results in enhanced fatty acid oxidation [12]. Therefore we tested whether PGC-1α also modulates the lipin1-mediated activation of PPARγ. Consistent with the previous study, the chimaeric transcription factor, GAL4-DBD–PPARα was activated by PGC-1α and showed a higher level of activation when both PGC-1α and lipin1 were present (Figure 5A, left-hand panel). Although, although lipin1 and PGC-1α increased the transcriptional activity of the GAL4-DBD–PPARγ chimaera they did not show a synergistic action (Figure 5A, right-hand panel). Using the same assay, the synergistic action of lipin1 and PGC-1α was confirmed by measuring PPARα-mediated PPRE-tk reporter activity (Figure 5B, left-hand panel). However, no cooperative action between PGC-1a and lipin1 in PPARγ-mediated transcription was observed (Figure 5B, right-hand panel), indicating that PGC-1α is not involved in lipin1-mediated PPARγ activation.
Figure 5. The mechanism of lipin1-mediated PPARγ activation differs from PPARα activation.
(A) Expression vectors for GAL4-DBD–PPARα (left-hand panel) or the GAL4-DBD–PPARγ (right-hand panel) fusion proteins and pFR-Luc were transfected into NIH 3T3 cells along with pcDNA-PGC-1α and/or pSG5-lipin1, as indicated. (B–D) PPRE-tk-luciferase and expression vectors for PPARα (left-hand panels), PPARγ (right-hand panels), RXRα, PGC1α, lipin1, lipin1 mutants (ΔTAD, LL728FF and LL888FF) or lipin2 were transfected into NIH 3T3 cells as indicated. Rosi., rosiglitazone.
Next, we evaluated whether the various motifs and TAD of lipin1 have distinctive functional features for the transcriptional co-activation of PPARα and PPARγ (Figure 5C). When the TAD was deleted (ΔTAD), lipin1-mediated PPARγ activation was inhibited to the basal level in the absence and presence of rosiglitazone. In contrast, ΔTAD did not affect the activation of PPARα. These results indicate that the TAD of lipin1 is required for the specific activation of PPARγ. Given the fact that the LXXIL motif of lipin1 is not required for its interaction with PPARγ (Figures 2A and 2E), we used the IL728FF mutant to evaluate its ability for the activation of PPARγ (Figure 5C, right-hand panel). The IL728FF mutant showed little or no effect in the absence of rosiglitazone, whereas a marked decrease was observed in the presence of ligand, indicating that the LXXIL motif might be important only when the ligand is present. Furthermore, it is noteworthy that the LL888FF mutation of the VXXLL motif is not able to interact with and co-activate PPARγ (Figurs 2D, 2E and 5C, right-hand panel), whereas this mutant had significant co-activating function of PPARα (Figure 5C, left-hand panel), indicating the importance of the VXXIL motif for the specific activation of PPARγ.
Lipin2, another member of the lipin family, has high sequence homology with lipin1 at both N-LIP and C-LIP (Figure 4C). Lipin2 is enriched in the liver and is able to activate PPARα similarly to lipin1 [18]. In agreements with this, lipin2 enhanced both ligand-dependent and -independent activities of PPARα in a similar manner to lipin1 (Figure 5D, left-hand panel). In contrast with PPARα, PPARγ activity was increased by lipin1, but not by lipin2. These data suggest that lipin1 has unique feature to co-activate PPARγ through a different mechanism from that of its co-activation of PPARα.
Lipin1 mediates adipocyte differentiation through its PPARγ-co-activating function
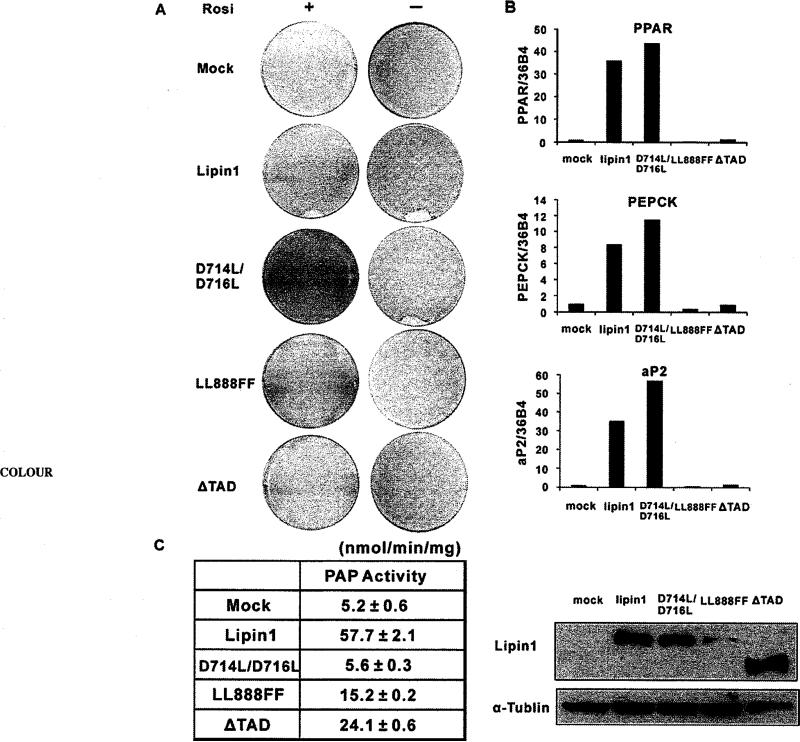
To assess the biological function of lipin1 as a co-activator of PPARγ in adipocyte differentiation, we overexpressed lipin1 and three mutants (D714L/D716L, LL888FF and ΔTAD) in 3T3-L1 pre-adipocytes. Transfected 3T3-L1 cells were induced to differentiate, as described in Experimental section, to 70% confluency. Using this method, the efficiency of adipocyte differentiation is very low even in the presence of rosiglitazone (Figure 6A, Mock). Lipin1 expression enhanced adipocyte differentiation and efficiently induced lipid accumulation both in the absence and presence of rosiglitazone. Interestingly, the mutant which has no catalytic activity for PAP (D714L/D716L), induced adipocyte differentiation as efficiently as the wild-type (Figures 6A and 6C). However, the mutants which lack PPARγ activating function (LL888FF and ΔTAD; Figure 5C), but still conserve the activity of PAP (Figure 6C), did not induce adipocyte differentiation and expression of adipocyte-specific genes, such as PPARγ2, PEPCK (phosphoenolpyruvate carboxykinase) and aP2 [also known as FABP4 (fatty acid-binding protein 4)] (Figures 6A and 6B). These results indicate that PPARγ-co-activating activity of lipin1 plays a significant role in 3T3-L1 preadipocyte differentiation.
Figure 6. Lipin1 mediates differentiation of 3T3-L1 pre-adipocytes through its PPARγ-co-activating activity.
3T3-L1 cells were transfected with pSG5-KF2M1 (Mock), pSG5-lipin1 and lipin1 mutants (D714L/D716L, LL888FF and ΔTAD) using electroporation and induced to differentiate when cells reached below 70% confluncy. (A) Oil Red O staining of transfected cells on day 7 after induction of differentiation with (+) or without (–) 1 μM rosiglitazone. Forced expression of lipin1 and mutants were detected at day 2 after induction of differentiation. (B) The expressions of adipocyte-specific genes were analysed by real-time PCR at day 7 after differentiation in the presence of 1 μM rosiglitazone. Relative mRNA levels of each gene were normalized to the levels of 36B4 mRNA. The data shown are representative of three independent experiments performed in triplicate. (C) PAP1 activity was measured with the extracts from HEK-293 cells which overexpress wild-type or mutant lipin1 (left). The data shown are representative of three independent experiments performed in triplicate. Western Blots of wild-type or mutant lipin1 are shown (right).
Lipin1 releases co-repressors from PPARγ-containing complexes
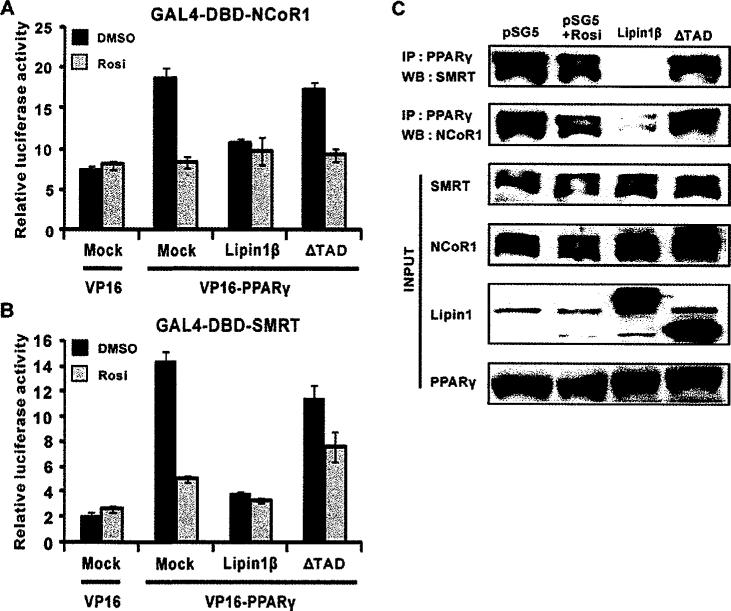
It is generally accepted that transcriptional activation of nuclear hormone receptors are mediated by sequential and/or concomitant dynamic interactions of co-repressors and co-activators. It has been reported that pioglitazone treatment releases co-repressors from PPARγ which results in its target gene expression during adipogenesis probably through recruiting co-activator complexes [19]. Thus we hypothesized that lipin1 would activate PPARγ through dissociating two major co-repressors of nuclear receptors, NCoR1 and SMRT, from PPARγ. We performed mammalian two-hybrid system and co-immunoprecipitation assays in order to address this hypothesis. Overexpression of VP16–PPARγ increased luciferase activity by 4- and 7-fold in cells co-transfected with GAL4-DBD–NCoR1 and GAL4-DBD–SMRT respectively, implying that PPARγ interacts with NCoR1 and SMRT (Figures 7A, 7B and S2). This interaction was completely lost in the presence of rosiglitazone, consistent with the data from the previous study [19]. Given the fact that lipin1 expression itself facilitated PPARγ activity (Figure 1), we raised the possibility of lipin1 would be able to release co-repressors from PPARγ-containing complexes. Strikingly, lipin1 expression drastically inhibite VP16–PPARγ-mediated luciferase activity with no further decrease in activity upon the treatment with the ligand suggesting that interaction of PPARγ with co-repressors is completely diminished by lipin1. Next, we asked whether the TAD of lipin1 is required for the clearing of co-represssors from PPARγ-containing complexes. The ΔTAD mutant did not release co-repressors, indicating that the interaction of PPARγ and co-repressors is dependent on the TAD of lipin1. These results were evaluated further by co-immunoprecipitation assays using HeLa cell extracts. Cells were treated with rosiglitazone or transfected with the wild-type lipin1 or ΔTAD constructs (Figure 7C). Whole-cell extracts were immunoprecipitated using an anti-PPARγ antibody and analysed by Western blotting. Consistent with the data from the mammalian two-hydrid assay, lipin1 strongly inhibited the interaction of PPARγ with NCoR1 or SMRT, although rosiglitazone treatment at the given concentration led to partial reduction in this interaction. The mutant lipin1 (ΔTAD) did not show any effects on the PPARγ–co-repressor interactions. Collectively, these results demonstrate that lipin1 itself acts like a ligand for dissociating co-repressors, NCoR1 and SMRT, from PPARγ-containing complexes.
Figure 7. Lipin1 releases co-repressors from the PPARγ-containing complex.
pGAL4-NCoR1 (A) or pGAL4-SMRT (B) and pUAS-Luc reporter plasmids were transfected into NIH 3T3 cells, along with pVP16-PPARγ, pSG5-FLAG-lipin1 or pSG5-FLAG -lipin1ΔTAD. (C) pSG5-KH2M1 (Mock), pSG5-FLAG-lipin1 (Lipin1) or pSG5-FLAG-lipin1ΔTAD (ΔTAD) were transfected into Hela cells. In the ligand-treated group, 1 μM rosiglitazone (Rosi) was added to the media 1 day after transfection. At 2 days after transfection, cell extracts were prepared and expression of SMRT, NCoRI, lipin1 and PPARγ were confirmed in whole-cell lysates by Western blot analyses. The anti-PPARγ antibody was used for immunoprecipitation (IP) and SMRT and NCoR1 were detected by Western blot (WB) analyses.
DISCUSSION
Since PPARγ had been discovered as the biological receptor for the TZD (thiazolidinedione) class insulin-sensitizing anti-diabetic drugs with high-affinity binding, there have been intense efforts to synthesize new agonist ligands for therapeutic purposes and identify naturally occurring ligands to elucidate its physiological function. Although there are putative endogenous ligands for PPARγ, including non-esterified fatty acids and prostanoids, its physiological relevance is still unclear since they have relatively low-binding affinity to PPARγ [5] and their levels are too low to be a biological active during adipogenesis [20]. Even though PPARγ is shown to be activated by oxidized fatty acids, which are present at significant concentrations in atherosclerotic lesions [21], it is not clear that they act as a ligand in other tissues. Thus it is probable that different ligands could mediate the activity of PPARγ depending on different cell contexts.
Our previous report suggested that lipin1 has important roles in the differentiation and functional maintenance of 3T3-L1 adipocytes by modulating a positive feedback loop between PPARγ2 and C/EBPα [14]. In the present study, we showed that lipin1 acts as a natural regulator of PPARγ transcriptional activity and identifed a novel VXXLL motif and the TAD domain of lipin1. Furthermore, the biological relevance of the PPARγ-activating function of lipin1 was assessed by its ability to differentiate 3T3-L1 cells into adipocytes.
We showed that PPARγ transcriptional activity was facilitated by lipin1α and lipin1β (Figure 1). Interestingly, lipin1β showed more potent PPARγ activating capacity than lipin1α. Consistent with this result, the TAD of lipin1β (N8β fragment) showed higher transactivation effects than the TAD of lipin1α (N8α fragment) (Figures 4A and 4C). Although the functional difference between lipin1α and lipin1β was reported by observing different cellular localization of lipin1 isoforms and triacylglycerol contents in lipin1α- or lipin1β-overexpressing 3T3-L1 cells and fld MEFs (mouse embryonic fibroblasts) [17], further investigations are necessary to explicate the different effects of lipin1α or lipin1β on the PPARγ-activating capacity (Figure 1).
We first described that the VXXLL motif is required for binding with PPARγ (Figure 2D). Since the VXXLL motif in C-LIP region is also found in other lipin isoforms, such as lipin2 and lipin3, and, furthermore, lipin2 does not have PPARγ-transactivating capacity (Figure 5D, right-hand panel), the VXXLL motif by itself does not seem to be sufficient to make lipin1 as an activator of PPARγ. Thus the identification of the TAD of lipin1 is important in the elucidation of the mechanism of lipin1-mediated PPARγ activation. This notion is supported by several observations. First, deletion of the TAD disrupted lipin1-mediated PPARγ activation (Figure 5C, right-hand panel). Secondly, the TAD has critical role in dissociating co-repressors from PPARγ (Figure 7). Thirdly, the activity of the TAD could be regulated by various co-activators, such as p300 and SRC-1 (Figure 4B). Finally, lipin2 and lipin3 do not have any sequence homology to the TAD and accordingly lipin2 did not affect PPARγ activity. This suggests that lipin1 has unique function to activate PPARγ transcriptional activity.
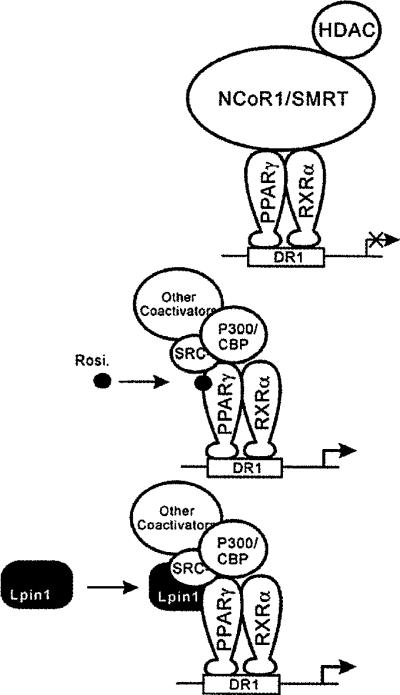
As shown in Figures 5(A) and 5(B), PGC-1α is not involved in lipin1-mediated PPARγ activation in contrast with lipin1-mediated PPARα activation which was synergistically increased by PGC-1α. These results indicate that PGC-1α is not a critical factor in lipin1-mediated PPARγ activation, reflecting the fact that PGC-1α is scarcely expressed in adipocytes where PPARγ performs its major actions. Collectively, these results imply that lipin1-mediated PPARγ transcriptional regulation could be dependent on diverse cellular context with the ability of releasing co-repressors and utilizing its TAD domain and motifs as proposed in Figure 8.
Figure 8. A schematic representation of the proposed function of lipin1 in PPARγ regulation.
In the absence of exogenous PPARγ ligands, PPARγ recruits the corepressors NCoR1/SMRT. The mechanism of action of lipin1 involves triggering a conformational change in PPARγ causing co-repressor release and co-activator recruitment.
In our previous report, lipin1 overexpression accelerated 3T3-L1 differentiation and knockdown of lipin1 impaired adipogenesis through regulating PPARγ-transactivating function [14]. Therefore we assessed the biological function of wild-type lipin1 and PPARγ-co-activating function-deficient mutants (LL888FF and ΔTAD) in 3T3-L1 adipocyte differentiation. Our data show that 3T3-L1 cells expressing wild-type lipin1 or the D714L/D716L mutants, which have PPARγ-activating capacity, differentiated into adipocytes with no substantial difference (Figure 6A). In contrast, cells expressing PPARγ-co-activating function-deficient mutants (LL888FF and ΔTAD) failed to differentiate into adipocytes even though these mutants retain PAP activity (Figure 6C), suggesting that the PPARγ-co-activating activity oflipin1 is necessary for 3T3-L1 preadipocyte differentiation.
However, the results of the present study appear contradictory to a previous report suggesting that PAP activity of lipin1, but not its co-activator activity, is required during adipogenesis through the induction of PPARγ [9]. The explanation for the discrepancy is not clear, however, the previous results are from using fld MEF cells, whereas we used 3T3-L1 pre-adipocytes which express normal lipin1 during adipogenesis. A possible explanation underlying the lipid droplet accumulation caused by the D714L/D716L mutant expression is that endogenous lipin1 is sufficient to produce diacylglycerol from PA in our 3T3-L1 differentiating method.
Previous studies have suggested that the function of lipin1 would not be reserved only to PAP in adipocytes. Catalytically active or inactive lipin1 directly interacts with NFATc4 and suppresses NFATc4 transcriptional activity through recruitment of histone deacetylases [15]. Also, our previous report suggested that the role of lipin1 as a regulator of PPARγ in the differentiation and functional maintenance of 3T3-L1 adipocytes is via being recruited to the promoter region of PEPCK, one of the target genes of PPARγ, [14].
Collectively, we propose that the regulation of PPARγ transcriptional activity by lipin1 is a novel mechanism required for adipogenesis, although we cannot rule out the role of lipin1 as a PAP in adipogenesis.
As mentioned above, the identity of endogenous PPARγ ligands is still vague. The present study provides compelling evidence that lipin1 could act as a natural regulator of PPARγ activity and provides new understanding of the mechanisms of PPARγ activation. Future work is needed to determine whether lipin1 can increase the sensitivity of PPARγ in response to natural ligands at physiological concentrations.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. 2011-0016206 and 2011-0030705) and partly by National Institutes of Health of USA (GM-28140).
Abbreviations used
- C/EBPα
CCAAT/enhancer-binding protein α
- C-LIP
C-terminal conserved domain
- DMEM
Dulbecco's modified Eagle's medium
- fld
fatty liver dystrophic
- GAL4-DBD
galactose metabolism 4 DNA-binding domain
- HA
haemagglutinin
- HEK
human embryonic kidney
- MEF
mouse embryonic fibroblast
- NcoR1
nuclear receptor co-repressor 1
- NFATc4
nuclear factor of activated T-cells
- cytoplasmic
calcineurin-dependent 4
- N-LIP
N-terminal conserved domain
- PA
phosphatidic acid
- PAP
phosphatidate phosphatase
- PCAF
p300/CBP-associated factor
- PEPCK
phosphoenolpyruvate carboxykinase
- PPAR
peroxisome proliferator-activated receptor
- PPRE
peroxisome proliferator response element
- PGC-1α
PPARγ co-activator 1α
- RXR
retinoid X receptor
- SD
synthetic defined dropout
- SMRT
silencing mediator of retinoid and thyroid hormone receptor
- SRC-1
steroid receptor co-activator 1
- UAS
upstream activation sequence
Footnotes
AUTHOR CONTRIBUTION HeeEun Kim, Eunjin Koh and Kyung-Sup Kim designed the experiments. Min-Ji Kim and Won-ji Jin performed the yeast experiments, HeeEun Kim, Eunju Bae, Deok-yoon Jeong and Eunjin Koh performed the animal cell experiments. Gil-Soo Han and George Carman performed PAP assay. HeeEun Kim, Eunjin Koh, Shang-wook Park and Kyung-Sup Kim analysed the data. HeeEun Kim, Eunjin Koh and Kyung-Sup Kim wrote the paper. Eunjin Koh and Kyung-Sup Kim managed and advised the project.
REFERENCES
- 1.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR)γ: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 2.Hu E, Tontonoz P, Spiegelman BM. Transdifferentiation of myoblasts by the adipogenic transcription factors PPARγ and C/EBPα. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1 alpha (PGC-1γ): transcriptional coactivator and metabolic regulator. Endocr. reviews. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 5.Rosen ED, Spiegelman BM. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 6.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scavello GS, Jr, Paluru PC, Zhou J, White PS, Rappaport EF, Young TL. Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes. Mol. Vis. 2005;11:97–110. [PubMed] [Google Scholar]
- 8.Reue K, Zhang P. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 2008;582:90–96. doi: 10.1016/j.febslet.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, Takeuchi K, Csaki LS, Reue K. Lipin-1 phosphatidic phosphatase activity modulates phosphatidate levels to promote peroxisome proliferator-activated receptor gamma (PPARγ) gene expression during adipogenesis. J. Biol. Chem. 2012;287:3485–3494. doi: 10.1074/jbc.M111.296681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale SE, Frolov A, Han X, Bickel PE, Cao L, Bowcock A, Schaffer JE, Ory DS. A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 2006;281:11082–11089. doi: 10.1074/jbc.M509612200. [DOI] [PubMed] [Google Scholar]
- 11.Shan D, Li JL, Wu L, Li D, Hurov J, Tobin JF, Gimeno RE, Cao J. GPAT3 and GPAT4 are regulated by insulin-stimulated phosphorylation and play distinct roles in adipogenesis. J. Lipid Res. 2010;51:1971–1981. doi: 10.1194/jlr.M006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Liu GH, Gerace L. Sumoylation regulates nuclear localization of lipin-1α in neuronal cells. PLoS ONE. 2009;4:e7031. doi: 10.1371/journal.pone.0007031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh YK, Lee MY, Kim JW, Kim M, Moon JS, Lee YJ, Ahn YH, Kim KS. Lipin1 is a key factor for the maturation and maintenance of adipocytes in the regulatory network with CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma 2. J. Biol. Chem. 2008;283:34896–34906. doi: 10.1074/jbc.M804007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HB, Kumar A, Wang L, Liu GH, Keller SR, Lawrence JC, Jr, Finck BN, Harris TE. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol. Cell. Biol. 2010;30:3126–3139. doi: 10.1128/MCB.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterfy M, Phan J, Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 2005;280:32883–32889. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 18.Donkor J, Zhang P, Wong S, O'Loughlin L, Dewald J, Kok BP, Brindley DN, Reue K. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 2009;284:29968–29978. doi: 10.1074/jbc.M109.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor γ transcriptional activity and repress 3T3-L1 adipogenesis. J. Biol. Chem. 2005;280:13600–13605. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- 20.Tzameli I, Fang H, Ollero M, Shi H, Hamm JK, Kievit P, Hollenberg AN, Flier JS. Regulated production of a peroxisome proliferator-activated receptor-γ ligand during an early phase of adipocyte differentiation in 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:36093–36102. doi: 10.1074/jbc.M405346200. [DOI] [PubMed] [Google Scholar]
- 21.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.