Abstract
Caspase recruitment domain-containing membrane-associated guanylate kinase protein-1 (CARMA1), a member of the membrane associated guanylate kinase (MAGUK) family of kinases, is essential for T lymphocyte activation and proliferation via T cell receptor (TCR) mediated NF-κB activation. Recent studies suggest a broader role for CARMA1 regulating other T cell functions as well as a role in non-TCR mediated signaling pathways important for lymphocyte development and functions. In addition, CARMA1 has been shown to be an important component in the pathogenesis of several human diseases. Thus, comprehensively defining its mechanisms of action and regulation could reveal novel therapeutic targets for T cell-mediated diseases and lymphoproliferative disorders.
Keywords: CARMA1, T cell, T cell receptor, NF-κB
I. INTRODUCTION
T lymphocytes are critical components of the immune response and play a major role in the defense against infections and tumors. The capacity of these cells to detect specific pathogen- or disease-specific antigens and provide immunologic memory, defines the adaptive immune response. However, inappropriate or excessive activation of T cells can cause non-infectious inflammatory diseases or lead to hematologic malignancies. Thus, an understanding of the molecular mechanisms that regulate T lymphocyte activation and functions is necessary to understand immune defense and disease, and to develop effective therapies for T cell-mediated disorders and malignancies.
T cell development and the activation of naïve T cells is dependent on T cell receptor (TCR) binding to cognate peptide-major histocompatibility complex (pMHC) complexes on antigen presenting cells (APCs). In addition, subsequent T cell proliferation and survival is dependent on signaling through the TCR, and TCR signaling strength can influence the development and function of effector T cells, memory T cells and the formation of regulatory T cells (Treg).1–6 Therefore, the intensity and nature of T cell-mediated inflammatory reactions depends on the strength of the TCR stimulus both initially and during the immune response.1–3 In addition, dysfunction in the TCR signaling pathway can contribute to lymphoproliferative disorders.4
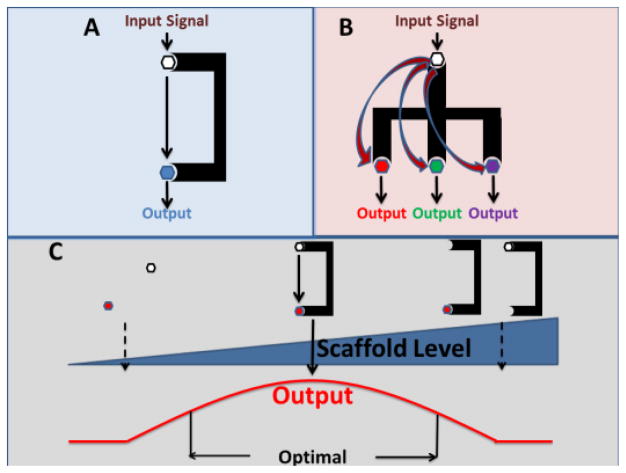
The scaffold protein caspase recruitment domain-containing membrane-associated guanylate kinase protein-1 (CARMA1), a member of the MAGUK family of kinases, has been shown to be an essential component in TCR signaling in T lymphocytes and B cell receptor (BCR) signaling in B lymphocytes. Scaffold proteins help physically assemble multiple signaling components in response to a specific stimulus, thus helping cells organize protein complexes to provide specificity in signaling pathways.5 This could involve a simple association to increase the efficiency of interactions between molecules (Figure 1A) or more complex control over multiple partners allowing several outputs from a single stimulus (Figure 1B). In addition, scaffold proteins like CARMA1 are often targets of regulation and may lead to a biphasic response with enhancement of signaling at low to moderate levels and inhibition at high levels (Figure 1C). Thus, scaffold proteins allow cells to regulate signaling for specific pathways and selectively shape output behaviors.5
Figure 1.
Scaffold proteins can mediate pathway input to a single output (A) or multiple outputs (B). C) Biphasic relationship of scaffold protein levels to output. Reduced output at high levels of scaffold mediated by sequestration of proteins away from each other.
CARMA1 is expressed selectively in T and B lymphocytes where in resting T cells it resides in a closed and inactive form. Multiple studies have now demonstrated that following engagement of the TCR, CARMA1 is activated, localizes to the TCR signalosome at the plasma membrane, and then forms multi-protein complexes that initiate NF-κB and c-jun N-terminal kinase (JNK) signaling cascades.6–8 Consistent with this, CARMA1-deficient T lymphocytes have impaired antigen-induced proliferation and activation, and have enhanced cell death with TCR stimulation.9 Recent data has suggested that CARMA1 is activated and regulated via complex interactions with numerous proteins and that CARMA1 may help mediate signaling in other non-TCR pathways in T cells.10, 11 Overall, it appears that CARMA1 is a signaling protein in T cells that has a complex role in modulating T cell activation and functions. The fact that mutations in CARMA1 can be associated with immunodeficiency, allergy, autoimmunity, and lymphoproliferative disorders demonstrates its importance for lymphocyte function,12–18 and suggests that CARMA1 is critical for T cell-mediated inflammation. Furthermore, since CARMA1 has multiple protein interactions and phosphorylation sites, it could be a prime target for therapeutic intervention. Herein, we will review the current state of knowledge regarding the role of CARMA1 in mediating T cell functions as well as its role in T cell mediated immunity and inflammation.
II. STRUCTURE AND FUNCTION OF CARMA1
A. Structural Features of CARMA1
CARMA1 is a scaffolding protein comprised of five domains connected by intervening stretches of linker regions. The domains are: 1) an N-terminal caspase activation and recruitment (CARD) domain; 2) a coiled-coil (CC) domain; 3) a PDZ homology domain (Postsynaptic Density Protein [PSD95], Drosophila Disc Large Tumor Suppressor [DLG1] and Zona Occludans-1 protein [ZO1]); 4) a SRC homology 3 (SH3) domain; and 5) a guanylate kinase (GUK) domain. Among these, the PDZ, SH3 and GUK domains together constitute the membrane associated guanylate kinase (MAGUK) domain, a catalytically inactive domain that is evolutionarily conserved in proteins. This domain plays a major role in cellular adhesion, formation of multi-protein complexes, and signal transduction.9, 19, 20 It appears that this region may be critical for localization of CARMA1 to the plasma membrane and for CARMA1 multimerization, two processes thought to be important for CARMA1 function.8, 9, 21–24 Among the CARMA1 domains, only the PDZ domain has been shown to be dispensable for CARMA1 driven NF-κB activation.21, 24–27
CARD domains are protein motifs that facilitate multi-protein binding via CARD-CARD interactions. These motifs have long been associated with a multitude of outcomes such as caspase activation, apoptosis and inflammasome assembly in innate immunity. Accumulating evidence over the last decade strongly suggests that CARD domain containing proteins such as CARMA1 also play a crucial role in the adaptive immune system by the regulation of NF-κB and JNK transcription factor pathways.12, 14, 22, 28–31 Importantly, the CARD domain in CARMA1 interacts with a CARD motif in B-cell CLL-lymphoma 10 (Bcl10), which helps mediate NF-κB activation following TCR engagement.32, 33
The coiled-coil motif is a common domain found in many types of proteins. Its primary role is to facilitate oligomerization of proteins,34, 35 and it has been postulated that this region is needed for CARMA1 oligomerization following activation and may help mediate binding to mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1), another essential protein for NF-κB activation following TCR engagement.22, 23, 36 The domain appears to be critical for CARMA1 function based on studies in a mouse strain with a mutation in this domain of CARMA1.25
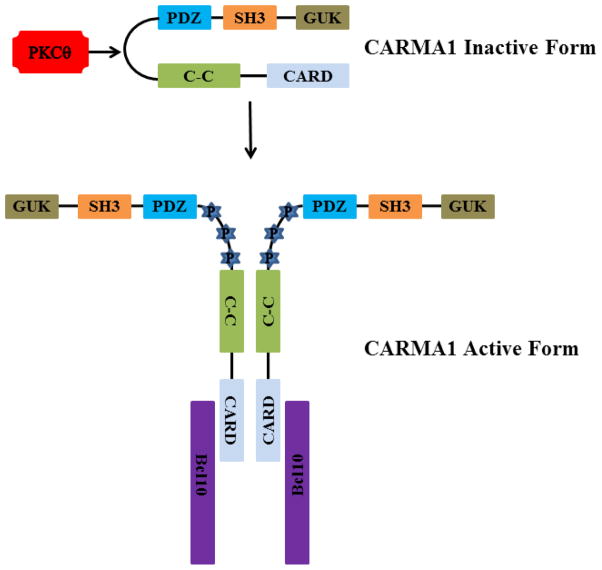
The region linking the coiled-coil domain to the MAGUK domain (linker region) appears to stabilize CARMA1 in an inactive or “closed” form (Figure 2).30 This is suggested by studies which demonstrated that deletion of this region leads to constitutive activation of NF-κB and high-level CARMA1 activation.27 Phosphorylation of serine residues in this region by PKCβ in B cells or PKCθ in T cells leads to activation of the protein presumably by “opening” up its conformation and allowing it to interact with other proteins as well as form multimers with itself.27, 37
Figure 2.
Proposed model of TCR-mediated activation of CARMA1 via PKCθ in T cells. Phosphorylation (P) of the linker region between the C–C and PDZ domains results in opening up of the CARMA1 protein allowing Bcl10 to bind to the CARD domain. This then initiates further downstream signaling.
T cell activation typically involves the interaction of the TCR with pMHC complexes and also the interaction of T cell and APC co-stimulatory molecules at the immunological synapse. Formation of the immunological synapse during the interaction of T cells and APCs is the first step in shaping of the T cell driven adaptive immune response. The CARD-CC-MAGUK domains in CARMA1 ideally suits its involvement in plasma membrane tethering, oligomerization and formation of multi-protein complexes at the immune synapse.38
B. Binding Partners of CARMA1
In resting T cells, CARMA1 is constitutively expressed and primarily localized under the cytoplasmic membrane in an inactive conformation.8, 30 TCR stimulation leads to the activation of PKCθ, which phosphorylates and activates both CARMA1 and TGFβ associated kinase 1 (TAK1).39, 40 After activation CARMA1 acquires an active conformation (Figure 2) and is recruited to the immunological synapse where it self-associates in multimers and interacts with multiple other proteins to initiate signaling cascades.8, 27, 37 Overexpression and mutation studies of CARMA1 and its binding partners suggest that oligomerization of the proteins is both necessary and sufficient for activation of downstream signaling pathways such as NF-κB.22, 40 Many of the proteins that interact with the CARMA1 signalosome are kinases and ubiquitylating enzymes which probably help regulate CARMA1 activity (Table 1).
Table 1.
Proteins that Interact with the CARMA1 Signalsome
| Kinaseses | Ubiquitin Ligases | Other |
|---|---|---|
| PKC θ/β | UBC13-UEV1A | Bcl10 |
| IKK Complex | TRAF6 | MALT1 |
| PDK1 | TRAF2 | ADAP |
| CaMKII | cIAP2 | Caspase 8 |
| HPK1 | NEDD4 | Net1162 |
| CK1α | ITCH | |
| Akt | CBL-b | |
| TAK1 | COP9 | |
| RIP2 | STUB1 | |
| MKK7 (JNK activation) | CYLD (de-ubiquitylating enzyme) | |
| Calcineurin (phosphatase) PP2A (phosphatase) | A20 (de-ubiquitylating enzyme) |
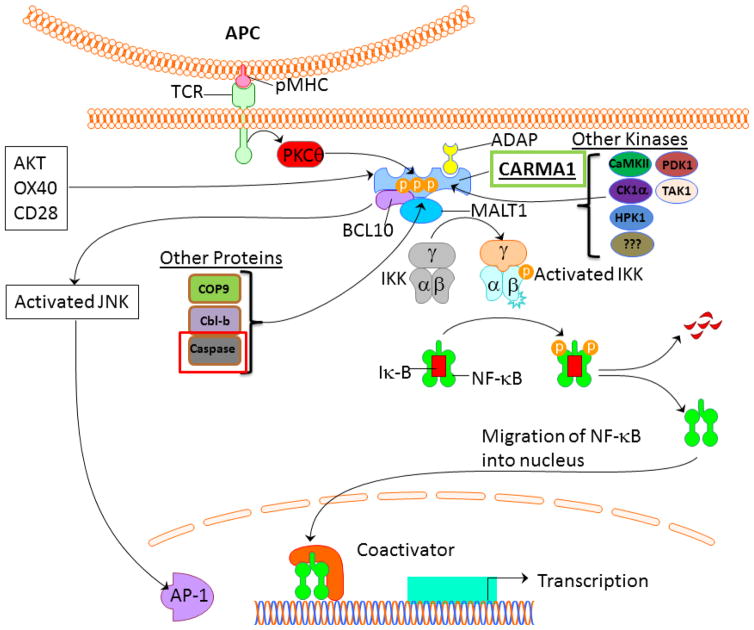
Once activated, CARMA1 binds to Bcl10 via CARD-CARD interactions, and then this complex binds the immunoglobulin-like domains in MALT1 via the distal end of the Bcl10 CARD domain to complete the assembly of the CARMA1-Bcl10-MALT1 (CBM) complex at the TCR signalosome. There may also be direct association between CARMA1 and MALT1 through the C–C domain.36 The CBM complex is essential for activation of transcription factors such as NF-κB and JNK (see signaling sections), and is critical for the recruitment and activation of the IκB kinase (IKK) complex to the TCR signalosome (Figure 3). Other proteins also interact with CARMA1 and may have a role in mediating its activity. The adhesion and degranulation-promoting adapter protein (ADAP) has been shown to interact with CARMA1 and may help translocate CARMA1 to lipid rafts at the TCR signalosome.41 Of note, ADAP-deficient T cells have impaired TCR-induced NF-κB activation. Another study has demonstrated that phosphoinositide-dependent kinase 1 (PDK1) binds to CARMA1 and may help recruit it to lipid rafts.42 However, it is not entirely clear whether PDK1 is necessary for TCR-induced NF-κB activation as experiments have given contradictory results.42, 43 It also seems likely that CARMA1 binds to a different membrane-associated protein to localize to the cytoplasmic membrane.30 Other kinases in addition to PKCθ and PDK1 have been shown to associate with CARMA1 and/or phosphorylate it including AKT, TAK1, RIP, PKCδ, hematopoetic progenitor kinase-1 (HPK1), IKKβ, casein kinase 1α (CK1α) and calmodulin-dependent protein kinase II (CaMKII). However, it is unclear how critical all of these are for optimal CARMA1 function.11, 37, 42–50 The CBM also associates with the ubiquitin ligase TNF receptor-associated factor 6 (TRAF6) which ubiquitinates Bcl10 and MALT1 which may facilitate IKK complex recruitment to the CBM via binding to the IKKγ subunit.40 TRAF6 may also directly activate the IKK complex by ubiquination of IKKγ.51 Other ubiquitin ligases such as TRAF2, Cbl-b, and STUB1 have also been shown to bind to CARMA1 and may regulate its function.11, 47, 52 A complete list of proteins found to interact with CARMA1 is shown in Table 1.
Figure 3.
Schematic of TCR signaling pathway leading to NF-κB and JNK activation.
C. CARMA1-Mediated Signaling Pathways
1. NF-κB Signaling
Activation of NF-κB is critical for multiple facets of T cell driven adaptive immune responses, including T cell development, activation, polarization, proliferation and survival.53, 54 Engagement of the TCR in T cells is one of the major triggers for activation of NF-κB, and this signaling pathway is dependent on CARMA1.12, 14 Exactly how CARMA1 helps mediate NF-κB activation is not fully understood, however, many of the key steps and mediators are now known.
Five individual transcription factors (RelA or p65, RelB, c-Rel, p50 and p52) constitute the NF-κB family of proteins. Among these, RelA and c-Rel form heterodimers with p50, and RelB heterodimerizes with p52 to form functional units of NF-κB. These complexes, when allowed to translocate to the nucleus, will then induce transcriptional activation of NF-κB target genes.55 NF- κB can be activated via the classical (or canonical) pathway which depends on p50, RelA, or c-Rel, or by the alternative pathway that involves RelB and p52. TCR-induced NF-κB activation is through p50, RelA, and c-Rel, so for this review we will focus on the classical pathway.7, 14
In resting T cells NF-κB is sequestered in the cytoplasm by association with the inhibitor of κB (IκB). This protein prevents nuclear translocation of the transcription factor by masking its nuclear localization motif. NF-κB is activated when the IκB complex is degraded, which allows the transcription factor to travel to the nucleus and initiate gene transcription. Stimulation of cells with a wide range of stimuli ultimately results in activation of the IKK complex, which is made up of two catalytic subunits (IKKα and IKKβ) and a regulatory subunit (IKKγ or NEMO). Once activated, IKK phosphorylates IκB which then triggers ubiquitination and degradation of the protein, allowing NF-κB to translocate into the nucleus. All stimuli that activate the classical NF-κB pathway utilize IKK, however, specificity is obtained upstream of IKK by signaling intermediates such as CARMA1, which tend to be both cell- and pathway-specific mediators of NF-κB activation. In TCR-induced NF-κB activation, it is the formation of the CBM complex that ultimately activates IKK and initiates NF-κB activation.
The exact mechanism(s) by which the CBM complex leads to NF-κB activation has not been fully delineated. Some studies have suggested that the CBM complex facilitates the phosphorylation of IKKβ via the TAK1 kinase.40, 44 Whereas a more recent study suggests that the phosphorylation of IKKα and IKKβ occurs via TAK1 independently of CARMA1, suggesting that the major mechanisms by which the CBM complex initiates IKK activation is via recruitment of IKKγ to the TCR signalosome leading to its polyubiquitination.39 Consistent with this, studies have shown that MALT1 functions as an ubiquitin ligase that induces polyubiquitination of IKKγ leading to activation of IKK,56 while other studies have suggested that TRAF6 associates with the CBM and is the primary ligase involved in the polyubiquitination of IKKγ.40 A different study has suggested that TRAF6 directly ubiquinates MALT1 which then leads to association of MALT1 and IKK.51 It is also possible that other E3 ligases are involved in the process.
Another potential mechanism by which the CBM mediates NF-κB activation involves the paracaspase activity of MALT1.36 Among the members of the CBM complex, MALT1 is the only enzymatically active component with a caspase like functional domain whose catalytic activity is enabled upon its assembly in the CBM complex. Following TCR stimulation, MALT1 in the CBM signalosome can cleave Bcl10, the NF-κB inhibitory protein A20 (a deubiquitinating enzyme), and the NF-κB subunit protein RelB at the same consensus cleavage site.57–59 The protease activity of CBM-associated MALT1 has been linked to several functional outcomes in T-cells following TCR stimulation. For example, cleavage of Bcl10 by CBM-associated MALT1 in activated T-cells was not essential for NF-κB activation or IL-2 production, but enhanced T cell adhesion to integrin ligands such as fibronectin.58 In this context, it has been proposed that MALT1 in the CBM complex may have a dual function – the first being driven by MALT1 in the CBM complex that degrades IkB proteins to allow nuclear translocation of NF-κB, and the second being driven by MALT1 protease activity that degrades RelB to prolong the DNA binding capacity of RelA- and c-Rel- containing NF-κB complexes through a yet unclear mechanism.59 Caspase-8 has also been shown to be associated with the CBM complex following TCR stimulation,60 and it was later shown that MALT1 directly induces Caspase-8 activation to induce NF-κB stimulation and IL-2 production.61
2. JNK Signaling
The JNK family is comprised of the ubiquitously expressed kinases JNK1 and JNK2 and the tissue restricted kinase JNK3. Activation of the TCR triggers the activation of JNK, which helps mediate T-cell proliferation and differentiation.62–65 Unlike constitutively expressed transcription factors such as NF-κB, TCR stimulation induces the expression of JNK, while CD28 co-stimulation is necessary for phosphorylation and activation of JNK.66 Following TCR activation, CARMA1 and Bcl10 form a molecular scaffold on which the kinases TAK1, MKK7 and JNK2 are assembled, leading to JNK2 phosphorylation.12, 30, 67 Phosphorylated JNK2, together with the independently phosphorylated JNK1, induce the activation of the transcription factor AP-1, which regulates T cell proliferation and survival.68 AP-1 is a heterodimeric protein formed by the members of the Jun family (c-Jun, JunB and JunD) with Fos or ATF/CREB families of transcription proteins.69 Engagement of the TCR leads to transcriptional upregulation of c-Jun, which is then capable of upregulating its own expression through a feedback amplification loop.70, 71 T cells from CARMA1-deficient mice fail to activate JNK following TCR stimulation suggesting that CARMA1 mediates JNK activation.67, 72 Consistent with this, TCR-induced accumulation of c-Jun is severely impaired in a CARMA1-deficient T cell line,67 and it appears that deficiency of CARMA1 leads to enhanced ubiquitination and the subsequent degradation of c-Jun.73 Despite these functional relationships between CARMA1 and JNK, deficiency of CARMA1 does not lead to defective AP-1 activity in T cell lines or primary T cells following TCR stimulation.21, 26, 67, 72 The uncoupling of CARMA1 dependent JNK phosphorylation from AP-1 activity therefore suggest that signaling through JNK1 and other Jun family members can still drive AP-1 activity in the absence of CARMA1.12, 30 Since JNK phosphorylation leads to the activation of a plethora of transcription factors, it remains to be determined if CBM-complex driven JNK signaling in T cells participates in the activation of transcription factors other than AP-1, and how important CBM-mediated JNK activation is for overall T cell functions independent of NF-κB activation. It is also not clear whether CBM-mediated JNK activation depends on unique binding partners and regulatory pathways different from those needed for NF-κB activation.
D. Regulation of CARMA1
1. Phosphorylation of CARMA1
While the conserved domains in CARMA1 drive its function, the activation of CARMA1 in T cells is largely determined by the phosphorylation status of key residues throughout the protein. These phosphorylation events presumably change the conformation of CARMA1 allowing it to interact with downstream binding partners. The dominant activation signal seems to be phosphorylation of serine residues S552 and S645 (S564 and S657 for murine CARMA1) in the linker region that connects the coiled-coil and PDZ domains by PKCθ in T-cells and PKCβ in B-cells. This phosphorylation event is essential for the assembly of the CBM complex and the subsequent activation of NF-κB and JNK.27, 37 Multiple other kinases have been shown to phosphorylate and regulate CARMA1 function (Table 2). These include, CaMKII, which localizes to the immune synapse following TCR activation and phosphorylates CARMA1 at residue S109 (S116 in mice) to enhance NF-κB activation.45 HPK1 also phosphorylates CARMA1 at residues S549, S551, and S552, of which the residues S549 and S551 are obligatory for TCR-induced NF-κB activation and IL-2 production.48 In addition, IKKβ has been shown to phosphorylate CARMA1 at S555 and is important for NF-κB and JNK activation as well as CBM complex formation.43 CARMA1 T110 has been shown to be phosphorylated with antigen receptor activation, and mutation of the residue impairs CBM formation.43 However, the kinase that acts on T110 has not been identified. On the other hand, mutation of two other residues in murine CARMA1, S620 (targeted by CK1a) and S649 (targeted by PKCθ), resulted in enhanced activation of NF-κB upon TCR stimulation, suggesting that phosphorylation of these residues may serve to downregulate CARMA1 driven NF-κB activation.49, 74 In a recent report, PKCδ was co-purified with the CARMA1 signalosome upon TCR activation, and overexpression of PKCδ inhibited TCR driven NF-κB activation in T cells by interfering with the physical interactions between MALT1 and TRAF6.50 However, the kinase activity of PKCδ was dispensable for this activity and does not seem to play a role in limiting CBM complex driven T cell responses.
Table 2.
CARMA1 Phosphorylation Sites
| Human CARMA1 | S109 | T110 | S551 | S552 | S555 | S565 | S608 | S637 | S645 |
| Mouse CARMA1 | S116 | T117 | S563 | S564 | S567 | S577 | S620 | S649 | S657 |
| Kinase | CaMKII | PKCθ | HPK1 | PKCθ | IKKβ | ? | CK1α | PKCθ | PKCθ |
| Effect of Mutation | |||||||||
| NF-κB | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↑ | Normal | ↓ |
| JNK | ? | ↓ | ? | ↓ | ↓ | ? | Normal | ? | ↓ |
Adapted from14.
While phosphorylation by kinases generally leads to the assembly of the CBM complex and T cell activation, counter-regulatory mechanisms such as dephosphorylation by phosphatases may also be present to downregulate T cell activation. There is now accumulating evidence that there are specific phosphatases that target members of the CBM complex to limit NF-κB activation in T cells. A subunit of the serine/threonine protein phosphatase PP2A (PPP2R1A) was constitutively associated with CARMA1 in resting and activated cells.75 Upon T cell activation, PP2A removed PKCθ dependent phosphorylation of CARMA1 at residue S645, leading to the attenuation of NF-κB responses. Similary, the calcium dependent phosphatase calcineurin dephosphorylated Bcl10 to disrupt the formation of CBM complexes and NF-κB activation.76, 77 Most recently, another protein phosphatase (PP4R1) subunit was found to be associated with the CBM complex following TCR stimulation.78 PP4R1 is a component of the phosphatase holoenzyme PP4, which dephosphorylated IKKα and IKKβ to suppress NF-κB activation in T cells, a phenomenon that reduced CARMA1-mediated NF-κB activity.78
In summary, CARMA1 is phosphorylated at multiple sites by a diverse set of kinases including PKCθ/β, IKKβ, CaMKII, HPK1, and CK1α. Whether other kinases, such as PDK1, TAK1, and Akt, which are known to associate with CARMA1, also phosphorylate CARMA1 remains to be determined. Phosphorylation of S608 and S637, or dephosphorylation of other sites mediates negative regulation of CARMA1 function. The fact that CARMA1 has such a complex array of phosphorylation sites suggests that the activity of this protein must require tight regulation for effective T cell function. Exactly how all of these events modulate CARMA1 binding activity as well as NF-κB and JNK activation remains to be fully determined.
2. Ubiquitination of CARMA1
Not surprisingly, post-translational modifications that target CARMA1 for degradation also downregulate TCR-mediated NF-κB activation. For example, ubiquitination of lysine residues in the SH3 and GUK domains of CARMA1, are induced following TCR activation and lead to proteasomal degradation of CARMA1.79 In addition, elimination of these ubiquitination sites in CARMA1 resulted in elevated NF-κB and JNK activation and increased persistence of CARMA1 protein in activated lymphocytes. In natural killer T cells (NKT cells), repeated doses of the agonistic ligand α-galactosylceramide leads to the ubiquitination and degradation of CARMA1 by the E3 ligase Cbl-b, inducing NKT cell anergy.80 Similar processes also regulate Bcl10 as demonstrated by a recent study which showed that TCR activation in primary T cells was associated with ubiquitination of Bcl10 and degradation by the autophagy dependent proteolysis machinery.81 Inhibition of Bcl10 autophagy resulted in enhanced activation of NF-κB upon TCR stimulation. Multiple ubiquitinating and deubiquitinating enzymes have been shown to target the CARMA1 signalosome (Table 1), however, more work is needed to fully understand which enzymes regulate CARMA1 activity and how this effects T cell functions.
III.Role of CARMA1 in TCR signaling
A. TCR signaling pathway
TCR induced T-cell activation is a critical event in adaptive immune responses. The ability of a T cell to recognize an antigen is dictated by the interaction of the TCR and peptides presented in the groove of the MHC molecule on APCs. This TCR-pMHC interaction initiates a complex process that transforms a naïve T cell into a cell capable of executing effector functions.82 The TCR is composed of two ligand binding chains (TCRα and TCRβ) which form a complex with a cluster of four different CD3 signaling invariant chains organized into three dimers (CD3γε, CD3δε, and CD3ζζ). These chains contain immunoreceptor tyrosine-based activation motifs (ITAMs) in their cytoplasmic tails.83, 84 The TCR-CD3 complex is accompanied by a CD4 molecule in T helper cells or a CD8 molecule in cytotoxic cells.
It is thought that engagement of the TCR complex with the appropriate pMHC triggers the TCR signaling pathway via TCR aggregation and conformational changes in the complex that induces changes in the CD3 subunits.85 These conformational changes allow phosphorylation of the ITAMs by the protein tyrosine kinases Lck and Fyn and the subsequent recruitment of the Syk kinase ζ-associated protein of 70 kDa (ZAP-70) to the complex. This is followed by the recruitment and phosphorylation of linker for activation of T cells (LAT),86–88 which serves as a platform for several signaling proteins. The recruitment of multiple proteins to the TCR as well as lipid rafts creates the TCR signaling complex at the point of contact with the APC forming the immune synapse. Sequential amplification of downstream signals leads to the recruitment of an array of molecules to the immunological synapse and activation of a network of signaling pathways (reviewed in Smith-Garvin et al85). These dynamic and highly regulated signaling complexes induce PLCγ1-dependent calcium- and diacylglycerol (DAG)-mediated responses. TCR-mediated calcium and DAG release is instrumental in activation of PKCθ which then activates CARMA1 and starts the cascade that stimulates NF-κB and AP-1 activity as outlined in previous sections.85 Co-stimulatory signals from proteins such as CD28 augment these signaling pathways.
The signaling pathways activated in response to TCR engagement include NF-κB, AP-1 (via JNK activation), and NFAT.85 Once activated, these transcription factors mediate the expression of numerous genes involved in T cell activation. Furthermore, the profile and degree of activation of these pathways will ultimately determine aspects of the T cell response including effector functions. The magnitude of activity of these pathways is mediated in part by the strength of the TCR signal. Consistent with this, TCR signal strength has been shown to determine short-term T cell activity as well as long-term T cell functions and inflammatory events.89 These include effects on T cell proliferation, cytokine production, and cytotoxicity.89–93 These data suggest that alterations in the activity of downstream signaling proteins in the TCR cascade could be utilized to influence T cell-mediated inflammatory disorders.3 We propose that in T cells CARMA1 has as its primary function the amplification of TCR signaling, providing a mechanism to enhance signals from pMHC complexes. It is unclear if the output generated by CARMA1 is digital (switch-like) or analog (the greater the CARMA1 activity the stronger the TCR signal), but prior work has suggested that TCR-mediated NF-κB activation is digital in nature and that the digitization occurs upstream of activation the IKK complex.94 This suggests that CARMA1 may amplify the TCR signal above a threshold that leads to NF-κB activation. However, CARMA1 could alter the duration of NF-κB signaling, modulate JNK signaling in an analog fashion, or modulate NF-κB signaling via interactions with co-stimulatory pathways. Overall, we feel that CARMA1 provides a mechanism to fine-tune TCR-mediated signaling in response to an antigen.
B. Role of Carma1 in naïve T cells
1. T cell development
Naïve T cells emerge from the thymus after undergoing a complex selection process that produces cells that react to a highly diverse set of foreign-pMHC complexes without reacting with self-antigens for the most part.95, 96. Although the development of T cells in the thymus requires TCR signaling, studies in CARMA1-deficient mice have revealed normal cell numbers in the thymus and normal T cell numbers and ratios in lymph nodes.97 However, there is some variation in the numbers of thymocyte subsets within the double-negative compartment (CD4−CD8−). This may be due to premature maturation of these cells as well as an accelerated rate of apoptosis. Not surprisingly, this thymic phenotype is very similar to that described in Bcl10-deficient mice.98 The impact of these thymocyte subset abnormalities does not seem to be significant. Overall it appears the development of naïve T cells is largely unaffected by CARMA1-deficiency despite the dependence of the selection process on TCR signaling. However, whether alterations in CARMA1 activity will affect the TCR repertoire of naive T cells has not been determined.
2. T cell activation
Naïve T cells congregate in secondary lymphoid tissues and migrate continuously from one lymphoid organ to another via the blood and lymph. In the lymph node, T cells enter from the blood thru the HEV into the T cell zone in a process mediated by homing receptors CD62L and CCR7. Naïve T cells in the periphery constitute a stable, diverse and functional repertoire of cells, maintained by homeostatic mechanisms.99 The activation of a naïve T cell depends on strong TCR binding to cognate pMHC complexes on APCs.85, 100 As discussed in the previous sections, CARMA1 is required for optimal activation of T cells, and deletion of CARMA1 abolishes TCR-induced activation of JNK and NF-κB. Interestingly, it appears that activation of naïve CD8+ T cells is not as impaired as it is in CD4+ T cells, suggesting that these cells are less dependent on CARMA1 and that an alternate pathway may be active in CD8+ T cell activation (unpublished observations).
3. T cell proliferation
An effective adaptive immune response relies on the capability of activated cells to undergo rapid expansion. A productive engagement of the TCR signal guides the fate of the cells towards a proliferative state. The early activation responses that induce T cell proliferation and signal amplification, is characterized by the secretion of pro-inflammatory cytokines (i.e. IL-2) and upregulation of the alpha chain of the IL-2 receptor (CD25). T cell proliferation and the expression of these cytokines are highly dependent on NF-κB as well as AP-1 activation.85 As discussed in the previous section, TCR-mediated NF-κB and AP-1 activation is dependent on CARMA1, thus induction of gene expression of these cytokines and their effects on proliferation is also dependent on CARMA1. This has been demonstrated in studies where deficiency in CARMA1 correlates with impaired proliferation signals.72, 97 In addition, CARMA1-deficient CD4+ and CD8+ T cells show defective proliferation and display markedly abrogated cytokine production. These data demonstrate that T cell proliferation is intrinsically dependent on the formation of the CBM complex.31, 101
4. T cell survival
An effective T cell-mediated immune response against infection requires expansion of antigen-specific T cells against the invading pathogen, and then contraction of T cell numbers after the pathogen is cleared. The downregulation of the immune response is important for limiting organ damage from the inflammatory response, but memory T cells specific for the pathogen must be maintained to preserve cellular immunity. Thus, signals that control T cell survival after activation are important components of adaptive immunity. TCR-mediated NF-κB activation provides an important survival signal to activated T cells. In addition, protection of cells against activation induced cell death during the immune response is mediated by an increase of co-stimulatory signals,102 and cytokines such as IL-2, IL-4, IL-7 and IL-15.103, 104 The survival of activated T cells is not fully dependent on CARMA1,72, 97 however, recruitment of PKCθ to the immunological synapse and its association with CARMA1 upregulates the anti-apoptotic molecule Bcl-x, suggesting an indirect requirement of CARMA1 in T cell survival.105 This is also demonstrated by studies in which CARMA1-deficient T cells have a high rate of apoptosis following activation, and in human studies of lymphomas which have demonstrated that mutations in CARMA1 provide high-level proliferative and survival signals in malignant cells.15, 16, 97, 106, 107
C. Role of CARMA1 in effector and memory T cells
TCR induced activation of naïve CD4+ T cells lead to proliferation and differentiation into effector T cells that fight infection by producing cytokines that promote a number of activities that help eliminate invading pathogens.108 As stated above the success of an effector T cells is attained upon initial T cell activation, a process largely dependent on CARMA1. In addition, effector T cells likely continue to encounter antigen and receive TCR signals which provide survival signals and enhance their effector function.109 Following the clearance of infection and the reduction in TCR signaling, many of the effector cells are eliminated via apoptosis. However, some of the effector cells appear to differentiate into memory T cells that provide long-term cellular immunity to the specific pathogen. These cells can then be reactivated in recall responses to recurrent infections where they lead to a rapid and robust immune reaction.110–113
While the role of CARMA1 and CBM complex has been extensively investigated in T cell activation, proliferation and survival responses, there is a paucity of studies that describe the role of CARMA1 and the CBM complex in effector and memory T cells. The signals requirements for the re-activation of memory T cells are not the same as those in naïve and effector T cells,114–116 and Bcl10 seems to be dispensable for activation of memory T cells,117 suggesting that CARMA1 may not be critical for TCR signaling in effector and memory T cell responses. However, we have recently demonstrated that CARMA1 is necessary for optimal effector T cell and memory T cell function.109 Deletion of CARMA1 from T cells after activation lead to impaired re-activation of memory T cells and reduced allergic inflammation in a model of memory T cell recall responses. In addition, CARMA1-deficient effector T cells produced less allergic inflammation in an adoptive transfer model of allergic airway inflammation compared to wild-type effectors. Interestingly, there was no survival defect in the CARMA1-deficient memory cells. These data suggest that TCR signaling in effector and memory T cells is also dependent on CARMA1 activity. Whether the signaling outputs from CARMA1 differ in these cell types compared to naïve T cells is unclear at this time.
D. Role of CARMA1 in regulatory T cell development
The maintenance of peripheral tolerance and establishment of controlled T-cell responses is dependent of the generation of regulatory CD4+ T cells (Tregs). Tregs can be generated from a bone marrow derived progenitor cell in the thymus (thymic Tregs) or from a naïve T cells during an inflammatory response in the periphery (peripheral Tregs).118 Tregs have been characterized by their ability to suppress immune responses and regulate peripheral tolerance.119 CARMA1-deficiency has been shown to alter the frequency of Tregs demonstrating its requirement for Treg cell lineage commitment.120–122 The role of CARMA1 in each type of Tregs is discussed below.
1. Thymic Tregs
Thymic Tregs are characterized by their involvement in the regulation of autoimmune interactions via active suppression of self-reactive T cells.123 These cells evolve from bone marrow progenitor cells that acquire linage commitment and mature in the thymus. Tregs develop from moderately self-reactive T cells that have escaped negative selection in a process that requires TCR triggering, CD28 costimulation, and stimulation by IL-2.124–127 After selection in the thymus, these cells move to the periphery where they constitute a small percentage of the total peripheral CD4+ T cell population.128 However, the majority of Treg cells present in the periphery are of thymic origin as peripheral Treg cell generation has specific prerequisites.129
Although TCR signaling is known to be an essential requirement for thymic Treg development, the signaling molecules that regulate Treg development are still a work in progress.129, 130 Prior work has shown that CARMA1 is necessary for Treg development,121, 122, 131, 132 as is PKCθ, Bcl10, and IKK.122, 133–135 Interestingly, the defect in Treg development with PKCθ-deficiency is not as severe as the defect with CARMA1-deficiency. This may be due to a role for CARMA1 in IL-2 signaling via CD25, which is necessary for optimal Treg development.132 Overall these data suggest that TCR induced NF-κB signaling via CARMA1 is necessary for thymic Treg development. Recent data suggests that CARMA1-mediated activation of NF-κB provides a survival signal that opposes the clonal deletion of self-reactive T cells allowing the development of organ-specific Tregs.121, 136
2. Peripheral Tregs
In contrast to thymic Tregs, peripheral Tregs develop from naïve T cells that acquire their suppressive activity as a consequence of activation in the setting of a unique set of stimulatory conditions.137–139 Peripheral Tregs seem tailored to respond to foreign antigens and neoantigens (such as tumor antigens), but it is likely that they are also generated in response to self-antigens and synergize with thymic Tregs in the control of autoimmunity.140 Studies suggest that the mechanisms for differentiation of peripheral Tregs differ from thymic Tregs.141, 142 However, CARMA1 is also important for optimal peripheral Treg development,122 although it is possible to induce peripheral Tregs in CARMA1-deficient T cells in vitro and in vivo with very strong TCR stimulation.122, 143, 144 It has been suggested that the different requirements for CARMA1 for generation of thymic derived Tregs versus peripheral Tregs might be a consequence TCR signaling strength, but further supportive evidence remains to be generated.129
E. Differential role of CARMA1 in different T cell subtypes
Following activation, CD4+ and CD8+ T cells differentiate into effector cells. Depending on the conditions during activation the effector cells can have different properties as defined by their functions and cytokine production. CD4+ T cells can develop into multiple different effector subtypes such as Th1, Th2, and Th17 cells. CD8+ T cell subtypes are not as well defined but may also develop different functional properties based on the conditions during activation. The specific role of CARMA1 in effector differentiation has not been fully delineated, but it does appear that CARMA1 has a differential role in the development of various T cell subsets.
1. T helper subtypes
CD4+ effector T cells are polarized into specific subsets which are defined by the cytokine profile secreted by the cells. The secreted cytokines then help mediate the immune response and help dictate the nature of the inflammatory response. The most common helper T cell subtypes are Th1, Th2, and Th17.
The factors that mediate the polarization of a T cell into a particular subtype are complex, but may be partly determined by the strength of the TCR signal, and thus could be influenced by CARMA1 activity.89 There are few reports investigating the function of CARMA1 in CD4+ T cell subtype commitment. Reports from our group and others have confirmed the relevance of CARMA1 in allergic airway inflammation, a process highly dependent on the generation of Th2 effector cells.109, 145, 146 Furthermore, a mouse line with a hypoactive mutant form of CARMA1 develops spontaneous allergic disease, probably due to an increased propensity for T cells to form Th2 cells and decreased Treg development.120 Most recently, it has been shown that CARMA1 directs polarization towards a Th2 phenotype via its regulation of JunB and GATA3 transcription factors.73 A role of CARMA1 in Th1 mediated inflammatory processes has not been well established. Studies suggest that Th1 polarization is impaired in CARMA1-deficient CD4+ T cells, but the defect may not be as profound as with Th2 cells.106, 147 However, these studies have indicated that CARMA1 induced NF-κB activation is essential for the induction of Th17 differentiation. These authors indicate a relatively selective role for CARMA1 in Th17 differentiation, that is independent of CARMA1-mediated survival and proliferative responses.106 Interestingly, in contrast to other studies, they suggest that reported defects in Th1 and Th2 differentiation in CARMA1-deficient T cells are largely due to a defect in cell cycle progression. Altogether, these observations confirm the requirement of CARMA1 for T cell differentiation, although the importance of CARMA1 may differ under different polarizing conditions.
2. NKT/CD8αα/NK Cells
As mentioned, CARMA1-deficient mice have impaired development of Tregs presumably due to effects on the strength of TCR signaling. NKT and CD8αα T cells are T cell subtypes that are also selected by the nature and strength of the TCR signals.122, 148 However, NKT cell development is not dependent on CARMA1 expression and there are actually increased numbers of CD8αα T cells in the gut of CARMA1-deficient mice.122 NK cells also develop in CARMA1-deficient animals (unpublished observations), however, CARMA1 seems to be necessary for NK cell-mediated effector functions through TAK1.149
IV. CARMA1 role in non-TCR Signaling Pathways
TCR signaling has thus far remained the most extensively investigated mechanism that leads to the formation of the CBM complex in T cells. Recently, it has been shown that co-stimulatory molecules such as OX40 can also induce assembly and recruitment of CBM complexes to lipid rafts in T cells in an antigen-independent manner.11 TCR independent engagement of OX40 with OX40L from antigen presenting cells led to the assembly of a complex of proteins consisting of CARMA1, Bcl10, MALT1, TRAF2, RIP and IKK.11 Furthermore, formation of the OX40 signalosome that contained the CBM complex has been shown to be essential for the prolonged survival of antigen-experienced T cells in the absence of the antigen. It has been well known that OX40 signaling contributes to the formation of memory T cells.150, 151 Therefore, the association of CBM signalosome to OX40 in the absence of antigen signals suggests that this could be an important mechanism that guides effector T cells into the memory T cell phase.
There are additional examples for the association of CBM complexes with T cell co-stimulatory molecules. For example, engagement of the T cell co-stimulatory molecule CD26 with its APC-expressed ligand caveolin-1 resulted in the association of CARMA1 to the cytoplasmic tail of CD26, and induced T-cell proliferation, IL-2 production, and NF-κB activation in a TCR-dependent manner.152 Furthermore, ligation of CD26 by caveolin-1 recruits a complex consisting of CD26, CARMA1, Bcl10, and IKKβ to lipid rafts.152 The serine/threonine kinase Akt has been shown to play a role in CD28 mediated NF-κB activation.153–156 It appears that Akt binds to CARMA1 and cooperates with other upstream signaling elements to help induce NF-κB activity.10, 46 CARMA1 has also been shown to be necessary for optimal IL-2 signaling via CD25.132 Although the mechanisms for this interaction are not known, it appears that CARMA1-deficient CD4+ T cells have defective JAK1 and STAT5 phosphorylation. Overall these data suggest that co-stimulatory pathways may help fine-tune TCR signaling responses by modulating CARMA1 providing further sensitivity and specificity in the pathway.
V. Role of CARMA1 in disease
Given the essential role of CARMA1 for lymphocyte survival, proliferation, and activation, it is not surprising that studies have found that changes in CARMA1 activity are associated with lymphoproliferative disorders. The most well-established association is with gain of function mutations and B cell lymphomas, most notably diffuse large B cell lymphoma.14 Other studies have linked persistent CARMA1 activity to congenital B cell lymphocytosis and to T cell lymphomas/leukemias.16, 157 Most of these mutations and variants are associated with enhanced CARMA1-mediated NF-κB signaling or CARMA1 overexpression.158, 159 Analysis suggests that these mutations often disrupt inhibitory domains on CARMA1 that normally prevent binding to other proteins.158 These data suggest that CARMA1 could be a potential therapeutic target in certain T and B cell lymphoma/leukemia and may have a role in other lymphoproliferative disorders.
CARMA1 has also been linked to other diseases. Recently a case report has demonstrated that CARMA1-deficiency in humans leads to profound immunodeficiency,17 and a recent genome-wide association study has identified a susceptibility locus for atopic dermatitis within the CARMA1 gene.18, 160 These data are especially intriguing since the “Unmodulated” mutant mouse strain that carries a CARMA1 single nucleotide variant that decreases TCR-induced NF-κB signaling spontaneously develops allergic dermatitis.25, 120 Furthermore, Th2 polarization of T cells and the development allergic airway inflammation is critically dependent on CARMA1 expression in mouse models.109, 145, 146 These data suggest that CARMA1 activity may be an important factor for allergy and could be targeted to modulate allergic sensitization. Another genome-wide association study has also linked allelic variation in CARMA1 to inflammatory bowel disease,18 and given new data on the role of CARMA1 in Th1- and Th17-type inflammation,106, 147, 161 there may also be a role for CARMA1 in other inflammatory diseases such as organ transplant rejection and autoimmunity.
VI. Conclusion
The primary function of scaffold proteins is to facilitate the formation of protein complexes that then work to amplify signaling pathways. The unique expression patterns and localization of these proteins also provide cell- and pathway-specific activity. We propose that in T cells CARMA1 has as its primary function the amplification of TCR signaling providing a mechanism to enhance signals from pMHC complexes and to fine-tune TCR-mediated signaling in response to an antigen. Prior research has demonstrated that CARMA1 plays an important role in T cell activation, survival, differentiation, effector/memory cell function, and in the pathogenesis of lymphoproliferative and inflammatory diseases. Thus, comprehensively defining its mechanisms of action and regulation could reveal novel therapeutic targets for T cell-mediated diseases.
Abbreviations
- TCR
T cell receptor
- pMHC
peptide-major histocompatibility complex
- APC
antigen presenting cells
- BCR
B cell receptor
References
- 1.Floyd TL, Koehn BH, Kitchens WH, Robertson JM, Cheeseman JA, Stempora L, Larsen CP, Ford ML. Limiting the amount and duration of antigen exposure during priming increases memory T cell requirement for costimulation during recall. J Immunol. 2011 Feb 15;186(4):2033–41. doi: 10.4049/jimmunol.1003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalia V, Sarkar S, Ahmed R. Fine-tuning CD4+ central memory T cell heterogeneity by strength of stimulation. Eur J Immunol. 2008 Jan;38(1):15–9. doi: 10.1002/eji.200738044. [DOI] [PubMed] [Google Scholar]
- 3.Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E. Different T cell receptor signals determine CD8+ memory versus effector development. Science. 2009 Jan 23;323(5913):502–5. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 4.Teitell MA, Pandolfi PP. Molecular genetics of acute lymphoblastic leukemia. Annu Rev Pathol. 2009;4:175–98. doi: 10.1146/annurev.pathol.4.110807.092227. [DOI] [PubMed] [Google Scholar]
- 5.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011 May 6;332(6030):680–6. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito T, Yokosuka T, Hashimoto-Tane A. Dynamic regulation of T cell activation and co-stimulation through TCR-microclusters. FEBS Lett. 2010 Dec 15;584(24):4865–71. doi: 10.1016/j.febslet.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Montecalvo A, Kane LP. Regulation of NF-kappaB induction by TCR/CD28. Immunol Res. 2011 Aug;50(2–3):113–7. doi: 10.1007/s12026-011-8216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Matsumoto R, You Y, Che T, Lin XY, Gaffen SL, Lin X. CD3/CD28 costimulation-induced NF-kappaB activation is mediated by recruitment of protein kinase C-theta, Bcl10, and IkappaB kinase beta to the immunological synapse through CARMA1. Mol Cell Biol. 2004 Jan;24(1):164–71. doi: 10.1128/MCB.24.1.164-171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004 May;4(5):348–59. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Phong B, Wilson DC, Hirsch R, Kane LP. Akt fine-tunes NF-kB-dependent gene expression during T cell activation. J Biol Chem. 2011 Aug 23; doi: 10.1074/jbc.M111.259549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.So T, Soroosh P, Eun SY, Altman A, Croft M. Antigen-independent signalosome of CARMA1, PKCtheta, and TNF receptor-associated factor 2 (TRAF2) determines NF-kappaB signaling in T cells. Proc Natl Acad Sci U S A. 2011 Feb 15;108(7):2903–8. doi: 10.1073/pnas.1008765108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blonska M, Lin X. NF-kappaB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011 Jan;21(1):55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tampella G, Baronio M, Vitali M, Soresina A, Badolato R, Giliani S, Plebani A, Lougaris V. Evaluation of CARMA1/CARD11 and Bob1 as candidate genes in common variable immunodeficiency. J Investig Allergol Clin Immunol. 2011;21(5):348–53. [PubMed] [Google Scholar]
- 14.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harbor Perspect Biol. 2010 Sep;2(9):a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S, Matsumoto T, Yada S, Hirahashi M, Suekane H, Yao T, Goda K, Iida M. Overexpression of caspase recruitment domain (CARD) membrane-associated guanylate kinase 1 (CARMA1) and CARD9 in primary gastric B-cell lymphoma. Cancer. 2005 Nov 1;104(9):1885–93. doi: 10.1002/cncr.21421. [DOI] [PubMed] [Google Scholar]
- 16.Oshiro A, Tagawa H, Ohshima K, Karube K, Uike N, Tashiro Y, Utsunomiya A, Masuda M, Takasu N, Nakamura S, Morishima Y, Seto M. Identification of subtype-specific genomic alterations in aggressive adult T-cell leukemia/lymphoma. Blood. 2006 Jun 1;107(11):4500–7. doi: 10.1182/blood-2005-09-3801. [DOI] [PubMed] [Google Scholar]
- 17.Stepensky P, Keller B, Buchta M, Kienzler AK, Elpeleg O, Somech R, Cohen S, Shachar I, Miosge LA, Schlesier M, Fuchs I, Enders A, Eibel H, Grimbacher B, Warnatz K. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J Allergy Clin Immunol. 2013 Feb;131(2):477–85. doi: 10.1016/j.jaci.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 18.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH International IBDGC. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012 Nov 1;491(7422):119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Mendoza A, Suga H, Ruiz-Trillo I. Evolution of the MAGUK protein gene family in premetazoan lineages. BMC Evol Biol. 2010;10:93. doi: 10.1186/1471-2148-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–45. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat Immunol. 2002 Sep;3(9):830–5. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 22.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006 Nov;6(11):799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 23.Tanner MJ, Hanel W, Gaffen SL, Lin X. CARMA1 coiled-coil domain is involved in the oligomerization and subcellular localization of CARMA1 and is required for T cell receptor-induced NF-kappaB activation. J Biol Chem. 2007 Jun 8;282(23):17141–7. doi: 10.1074/jbc.M700169200. [DOI] [PubMed] [Google Scholar]
- 24.Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S, Bron C, Tschopp J, Thome M. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002 Sep;3(9):836–43. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 25.Jun JE, Wilson LE, Vinuesa CG, Lesage S, Blery M, Miosge LA, Cook MC, Kucharska EM, Hara H, Penninger JM, Domashenz H, Hong NA, Glynne RJ, Nelms KA, Goodnow CC. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003 Jun;18(6):751–62. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 26.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002 Oct 1;21(19):5184–94. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, Rawlings DJ. Phosphorylation of the CARMA1 Linker Controls NF-kappaB Activation. Immunity. 2005 Dec;23(6):561–74. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Bouchier-Hayes L, Martin SJ. CARD games in apoptosis and immunity. EMBO Rep. 2002 Jul;3(7):616–21. doi: 10.1093/embo-reports/kvf139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007 Jan;14(1):10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- 30.Blonska M, Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol Rev. 2009 Mar;228(1):199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton K, Dixit VM. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr Biol. 2003 Jul 15;13(14):1247–51. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 32.Park JH, Bae JY, Park HH. Self-oligomerization of the CARD domain prevents complex formation in the CARMA1 signalosome. Int J Mol Med. 2013 May;31(5):1280–7. doi: 10.3892/ijmm.2013.1307. [DOI] [PubMed] [Google Scholar]
- 33.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001 Apr 13;276(15):11877–82. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 34.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004 Feb 6;5(2):170–6. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 35.Woolfson DN, Bartlett GJ, Bruning M, Thomson AR. New currency for old rope: from coiled-coil assemblies to alpha-helical barrels. Curr Opin Struct Biol. 2012 Aug;22(4):432–41. doi: 10.1016/j.sbi.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Che T, You Y, Wang D, Tanner MJ, Dixit VM, Lin X. MALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-kappaB activation. J Biol Chem. 2004 Apr 16;279(16):15870–6. doi: 10.1074/jbc.M310599200. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Wang D, Lin X. Phosphorylation of CARMA1 Plays a Critical Role in T Cell Receptor-Mediated NF-kappaB Activation. Immunity. 2005 Dec;23(6):575–85. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signalling. Nat Rev Immunol. 2009 Jan;9(1):47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 39.Shambharkar PB, Blonska M, Pappu BP, Li H, You Y, Sakurai H, Darnay BG, Hara H, Penninger J, Lin X. Phosphorylation and ubiquitination of the IkappaB kinase complex by two distinct signaling pathways. Embo J. 2007 Apr 4;26(7):1794–805. doi: 10.1038/sj.emboj.7601622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004 May 7;14(3):289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 41.Medeiros RB, Burbach BJ, Mueller KL, Srivastava R, Moon JJ, Highfill S, Peterson EJ, Shimizu Y. Regulation of NF-kappaB activation in T cells via association of the adapter proteins ADAP and CARMA1. Science. 2007 May 4;316(5825):754–8. doi: 10.1126/science.1137895. [DOI] [PubMed] [Google Scholar]
- 42.Lee KY, D’Acquisto F, Hayden MS, Shim JH, Ghosh S. PDK1 nucleates T cell receptor-induced signaling complex for NF-kappaB activation. Science. 2005 Apr 1;308(5718):114–8. doi: 10.1126/science.1107107. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara H, Maeda S, Watarai H, Kurosaki T. IkappaB kinase beta-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J Exp Med. 2007 Dec 24;204(13):3285–93. doi: 10.1084/jem.20070379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara H, Yasuda T, Aiba Y, Sanjo H, Hamadate M, Watarai H, Sakurai H, Kurosaki T. PKC beta regulates BCR-mediated IKK activation by facilitating the interaction between TAK1 and CARMA1. J Exp Med. 2005 Nov 21;202(10):1423–31. doi: 10.1084/jem.20051591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishiguro K, Green T, Rapley J, Wachtel H, Giallourakis C, Landry A, Cao Z, Lu N, Takafumi A, Goto H, Daly MJ, Xavier RJ. Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-kappaB activation. Mol Cell Biol. 2006 Jul;26(14):5497–508. doi: 10.1128/MCB.02469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayan P, Holt B, Tosti R, Kane LP. CARMA1 is required for Akt-mediated NF-kappaB activation in T cells. Mol Cell Biol. 2006 Mar;26(6):2327–36. doi: 10.1128/MCB.26.6.2327-2336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao G, Li Z, Molinero L, Alegre ML, Ying H, Sun Z, Penninger JM, Zhang J. T-cell receptor-induced NF-kappaB activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol Cell Biol. 2008 Apr;28(7):2470–80. doi: 10.1128/MCB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner D, Brechmann M, Rohling S, Tapernoux M, Mock T, Winter D, Lehmann WD, Kiefer F, Thome M, Krammer PH, Arnold R. Phosphorylation of CARMA1 by HPK1 is critical for NF-kappaB activation in T cells. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14508–13. doi: 10.1073/pnas.0900457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, Davis RE, Lenz G, Anderson DE, Arnoult D, Vazquez A, Sakai K, Zhang J, Meng Z, Veenstra TD, Staudt LM, Lenardo MJ. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature. 2009 Mar 5;458(7234):92–6. doi: 10.1038/nature07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Song R, Gao Y, Li Y, Wang S, Liu HY, Wang Y, Hu YH, Shu HB. Protein kinase C-delta negatively regulates T cell receptor-induced NF-kappaB activation by inhibiting the assembly of CARMA1 signalosome. J Biol Chem. 2012 Jun 8;287(24):20081–7. doi: 10.1074/jbc.M111.335463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J. 2007 Nov 14;26(22):4634–45. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, Li Y, Hu YH, Song R, Gao Y, Liu HY, Shu HB, Liu Y. STUB1 is essential for T-cell activation by ubiquitinating CARMA1. Eur J Immunol. 2013 Jan 15; doi: 10.1002/eji.201242554. [DOI] [PubMed] [Google Scholar]
- 53.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004 Sep 15;18(18):2195–224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 54.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006 Oct 30;25(51):6758–80. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 55.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 56.Zhou H, Wertz I, O’Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004 Jan 8;427(6970):167–71. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 57.Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008 Mar;9(3):263–71. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 58.Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, Fasel N, Thome M. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008 Mar;9(3):272–81. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 59.Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, Guzzardi M, Decaillet C, Grau M, Dorken B, Lenz P, Lenz G, Thome M. Malt1-dependent RelB cleavage promotes canonical NF-{kappa}B activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci U S A. 2011 Aug 30;108(35):14596–601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J, Salmena L, Hakem R, Straus S, Lenardo M. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005 Mar 4;307(5714):1465–8. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- 61.Kawadler H, Gantz MA, Riley JL, Yang X. The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol Cell. 2008 Aug 8;31(3):415–21. doi: 10.1016/j.molcel.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 63.Dong C, Yang DD, Tournier C, Whitmarsh AJ, Xu J, Davis RJ, Flavell RA. JNK is required for effector T-cell function but not for T-cell activation. Nature. 2000 May 4;405(6782):91–4. doi: 10.1038/35011091. [DOI] [PubMed] [Google Scholar]
- 64.Dong C, Yang DD, Wysk M, Whitmarsh AJ, Davis RJ, Flavell RA. Defective T cell differentiation in the absence of Jnk1. Science. 1998 Dec 11;282(5396):2092–5. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 65.Yang DD, Conze D, Whitmarsh AJ, Barrett T, Davis RJ, Rincon M, Flavell RA. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity. 1998 Oct;9(4):575–85. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]
- 66.Weiss L, Whitmarsh AJ, Yang DD, Rincon M, Davis RJ, Flavell RA. Regulation of c-Jun NH(2)-terminal kinase (Jnk) gene expression during T cell activation. J Exp Med. 2000 Jan 3;191(1):139–46. doi: 10.1084/jem.191.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blonska M, Pappu BP, Matsumoto R, Li H, Su B, Wang D, Lin X. The CARMA1-Bcl10 signaling complex selectively regulates JNK2 kinase in the T cell receptor-signaling pathway. Immunity. 2007 Jan;26(1):55–66. doi: 10.1016/j.immuni.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002 May;4(5):E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 69.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003 Nov;3(11):859–68. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 70.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988 Dec 2;55(5):875–85. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 71.Chauhan D, Kharbanda SM, Rubin E, Barut BA, Mohrbacher A, Kufe DW, Anderson KC. Regulation of c-jun gene expression in human T lymphocytes. Blood. 1993 Mar 15;81(6):1540–8. [PubMed] [Google Scholar]
- 72.Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, D’Acquisto F, Ghosh S, Yeh WC, Baier G, Rottapel R, Penninger JM. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003 Jun;18(6):763–75. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 73.Blonska M, Joo D, Zweidler-McKay P, Zhao q, Lin X. CARMA1 controls Th2 cell-specific cytokine expression through regulating JunB and GATA3 transcription factors. J Immunol. 2012;188(7) doi: 10.4049/jimmunol.1102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moreno-Garcia ME, Sommer K, Haftmann C, Sontheimer C, Andrews SF, Rawlings DJ. Serine 649 phosphorylation within the protein kinase C-regulated domain down-regulates CARMA1 activity in lymphocytes. J Immunol. 2009 Dec 1;183(11):7362–70. doi: 10.4049/jimmunol.0902438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eitelhuber AC, Warth S, Schimmack G, Duwel M, Hadian K, Demski K, Beisker W, Shinohara H, Kurosaki T, Heissmeyer V, Krappmann D. Dephosphorylation of Carma1 by PP2A negatively regulates T-cell activation. Embo J. 2011 Feb 2;30(3):594–605. doi: 10.1038/emboj.2010.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frischbutter S, Gabriel C, Bendfeldt H, Radbruch A, Baumgrass R. Dephosphorylation of Bcl-10 by calcineurin is essential for canonical NF-kappaB activation in Th cells. Eur J Immunol. 2011 Aug;41(8):2349–57. doi: 10.1002/eji.201041052. [DOI] [PubMed] [Google Scholar]
- 77.Palkowitsch L, Marienfeld U, Brunner C, Eitelhuber A, Krappmann D, Marienfeld RB. The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-kappaB activation. J Biol Chem. 2011 Mar 4;286(9):7522–34. doi: 10.1074/jbc.M110.155895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brechmann M, Mock T, Nickles D, Kiessling M, Weit N, Breuer R, Muller W, Wabnitz G, Frey F, Nicolay JP, Booken N, Samstag Y, Klemke CD, Herling M, Boutros M, Krammer PH, Arnold R. A PP4 holoenzyme balances physiological and oncogenic nuclear factor-kappa B signaling in T lymphocytes. Immunity. 2012 Oct 19;37(4):697–708. doi: 10.1016/j.immuni.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 79.Moreno-Garcia ME, Sommer K, Shinohara H, Bandaranayake AD, Kurosaki T, Rawlings DJ. MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-kappaB activity. Mol Cell Biol. 2010 Feb;30(4):922–34. doi: 10.1128/MCB.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kojo S, Elly C, Harada Y, Langdon WY, Kronenberg M, Liu YC. Mechanisms of NKT cell anergy induction involve Cbl-b-promoted monoubiquitination of CARMA1. Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17847–51. doi: 10.1073/pnas.0904078106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012 Jun 29;36(6):947–58. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dushek O. Elementary steps in T cell receptor triggering. Front Immunol. 2011;2:91. doi: 10.3389/fimmu.2011.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Call ME, Wucherpfennig KW. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol Immunol. 2004 Apr;40(18):1295–305. doi: 10.1016/j.molimm.2003.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhns MS, Badgandi HB. Piecing together the family portrait of TCR-CD3 complexes. Immunol Rev. 2012 Nov;250(1):120–43. doi: 10.1111/imr.12000. [DOI] [PubMed] [Google Scholar]
- 85.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoder J, Pham C, Iizuka YM, Kanagawa O, Liu SK, McGlade J, Cheng AM. Requirement for the SLP-76 adaptor GADS in T cell development. Science. 2001 Mar 9;291(5510):1987–91. doi: 10.1126/science.1057176. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W, Irvin BJ, Trible RP, Abraham RT, Samelson LE. Functional analysis of LAT in TCR-mediated signaling pathways using a LAT-deficient Jurkat cell line. Int Immunol. 1999 Jun;11(6):943–50. doi: 10.1093/intimm/11.6.943. [DOI] [PubMed] [Google Scholar]
- 88.Finco TS, Kadlecek T, Zhang W, Samelson LE, Weiss A. LAT is required for TCR-mediated activation of PLCgamma1 and the Ras pathway. Immunity. 1998 Nov;9(5):617–26. doi: 10.1016/s1074-7613(00)80659-7. [DOI] [PubMed] [Google Scholar]
- 89.Corse E, Gottschalk RA, Allison JP. Strength of TCR-peptide/MHC interactions and in vivo T cell responses. J Immunol. 2011 May 1;186(9):5039–45. doi: 10.4049/jimmunol.1003650. [DOI] [PubMed] [Google Scholar]
- 90.Corse E, Gottschalk RA, Krogsgaard M, Allison JP. Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol. 2010;8(9) doi: 10.1371/journal.pbio.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jameson SC, Carbone FR, Bevan MJ. Clone-specific T cell receptor antagonists of major histocompatibility complex class I-restricted cytotoxic T cells. J Exp Med. 1993 Jun 1;177(6):1541–50. doi: 10.1084/jem.177.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jenkins MR, Tsun A, Stinchcombe JC, Griffiths GM. The strength of T cell receptor signal controls the polarization of cytotoxic machinery to the immunological synapse. Immunity. 2009 Oct 16;31(4):621–31. doi: 10.1016/j.immuni.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wherry EJ, McElhaugh MJ, Eisenlohr LC. Generation of CD8(+) T cell memory in response to low, high, and excessive levels of epitope. J Immunol. 2002 May 1;168(9):4455–61. doi: 10.4049/jimmunol.168.9.4455. [DOI] [PubMed] [Google Scholar]
- 94.Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. Cutting edge: TCR ligation triggers digital activation of NF-kappaB. J Immunol. 2010 Oct 15;185(8):4520–4. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999 Oct 29;286(5441):958–61. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 96.Sprent J, Kishimoto H. The thymus and negative selection. Immunol Rev. 2002 Jul;185:126–35. doi: 10.1034/j.1600-065x.2002.18512.x. [DOI] [PubMed] [Google Scholar]
- 97.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O’Brien W, Thome M, Littman DR. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr Biol. 2003 Jul 15;13(14):1252–8. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 98.Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S, Millar DG, Bouchard D, Wakeham A, Ohashi PS, Mak TW. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell. 2001 Jan 12;104(1):33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 99.Takada K, Jameson SC. Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol. 2009 Dec;9(12):823–32. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 100.Sharpe AH. Mechanisms of costimulation. Immunol Rev. 2009 May;229(1):5–11. doi: 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hara H, Bakal C, Wada T, Bouchard D, Rottapel R, Saito T, Penninger JM. The molecular adapter Carma1 controls entry of IkappaB kinase into the central immune synapse. J Exp Med. 2004 Nov 1;200(9):1167–77. doi: 10.1084/jem.20032246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harding FA, McArthur JG, Gross JA, Raulet DH, Allison JP. CD28-mediated signalling co-stimulates murine T cells and prevents induction of anergy in T-cell clones. Nature. 1992 Apr 16;356(6370):607–9. doi: 10.1038/356607a0. [DOI] [PubMed] [Google Scholar]
- 103.Boise LH, Minn AJ, Noel PJ, June CH, Accavitti MA, Lindsten T, Thompson CB. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-XL. Immunity. 1995 Jul;3(1):87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 104.Vella AT, Dow S, Potter TA, Kappler J, Marrack P. Cytokine-induced survival of activated T cells in vitro and in vivo. Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3810–5. doi: 10.1073/pnas.95.7.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manicassamy S, Gupta S, Huang Z, Sun Z. Protein kinase C-theta-mediated signals enhance CD4+ T cell survival by up-regulating Bcl-xL. J Immunol. 2006 Jun 1;176(11):6709–16. doi: 10.4049/jimmunol.176.11.6709. [DOI] [PubMed] [Google Scholar]
- 106.Molinero LL, Cubre A, Mora-Solano C, Wang Y, Alegre ML. T cell receptor/CARMA1/NF-kappaB signaling controls T-helper (Th) 17 differentiation. Proc Natl Acad Sci U S A. 2012 Nov 6;109(45):18529–34. doi: 10.1073/pnas.1204557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Molinero LL, Yang J, Gajewski T, Abraham C, Farrar MA, Alegre M-L. CARMA1 Controls an Early Checkpoint in the Thymic Development of FoxP3+ Regulatory T Cells. J Immunol. 2009 Jun 1;182(11):6736–43. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–79. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 109.Ramadas RA, Roche MI, Moon JJ, Ludwig T, Xavier RJ, Medoff BD. CARMA1 is necessary for optimal T cell responses in a murine model of allergic asthma. J Immunol. 2011 Dec 15;187(12):6197–207. doi: 10.4049/jimmunol.1101348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bevan MJ. Memory T cells as an occupying force. Eur J Immunol. 2011 May;41(5):1192–5. doi: 10.1002/eji.201041377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol Rev. 2006 Jun;211:154–63. doi: 10.1111/j.0105-2896.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 112.Woodland DL, Ely KH, Crowe SR, Tighe M, Brennan JW, Harmsen AG, Cauley LS. Antiviral memory T-cell responses in the lung. Microbes Infect. 2002 Aug;4(10):1091–8. doi: 10.1016/s1286-4579(02)01633-7. [DOI] [PubMed] [Google Scholar]
- 113.Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol. 2009 Mar;9(3):153–61. doi: 10.1038/nri2496. [DOI] [PubMed] [Google Scholar]
- 114.Au-Yeung BB, Fowell DJ. A key role for Itk in both IFN gamma and IL-4 production by NKT cells. J Immunol. 2007 Jul 1;179(1):111–9. doi: 10.4049/jimmunol.179.1.111. [DOI] [PubMed] [Google Scholar]
- 115.Dienz O, Eaton SM, Krahl TJ, Diehl S, Charland C, Dodge J, Swain SL, Budd RC, Haynes L, Rincon M. Accumulation of NFAT mediates IL-2 expression in memory, but not naive, CD4+ T cells. Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7175–80. doi: 10.1073/pnas.0610442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Honma K, Kimura D, Tominaga N, Miyakoda M, Matsuyama T, Yui K. Interferon regulatory factor 4 differentially regulates the production of Th2 cytokines in naive vs. effector/memory CD4+ T cells. Proc Natl Acad Sci U S A. 2008 Oct 14;105(41):15890–5. doi: 10.1073/pnas.0803171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zeng H, Chen Y, Yu M, Xue L, Gao X, Morris SW, Wang D, Wen R. T cell receptor-mediated activation of CD4+CD44hi T cells bypasses Bcl10: an implication of differential NF-kappaB dependence of naive and memory T cells during T cell receptor-mediated responses. J Biol Chem. 2008 Sep 5;283(36):24392–9. doi: 10.1074/jbc.M802344200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Workman CJ, Szymczak-Workman AL, Collison LW, Pillai MR, Vignali DA. The development and function of regulatory T cells. Cell Mol Life Sci. 2009 Aug;66(16):2603–22. doi: 10.1007/s00018-009-0026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schuster M, Glauben R, Plaza-Sirvent C, Schreiber L, Annemann M, Floess S, Kuhl AA, Clayton LK, Sparwasser T, Schulze-Osthoff K, Pfeffer K, Huehn J, Siegmund B, Schmitz I. IkappaB(NS) Protein Mediates Regulatory T Cell Development via Induction of the Foxp3 Transcription Factor. Immunity. 2012 Dec 14;37(6):998–1008. doi: 10.1016/j.immuni.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 120.Altin JA, Tian L, Liston A, Bertram EM, Goodnow CC, Cook MC. Decreased T-cell receptor signaling through CARD11 differentially compromises forkhead box protein 3-positive regulatory versus T(H)2 effector cells to cause allergy. J Allergy Clin Immunol. 2011 Feb 12;127(5):1277–85. e5. doi: 10.1016/j.jaci.2010.12.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barnes MJ, Krebs P, Harris N, Eidenschenk C, Gonzalez-Quintial R, Arnold CN, Crozat K, Sovath S, Moresco EM, Theofilopoulos AN, Beutler B, Hoebe K. Commitment to the regulatory T cell lineage requires CARMA1 in the thymus but not in the periphery. PLoS Biol. 2009 Mar 3;7(3):e51. doi: 10.1371/journal.pbio.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Medoff BD, Sandall BP, Landry A, Nagahama K, Mizoguchi A, Luster AD, Xavier RJ. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur J Immunol. 2009 Jan;39(1):78–84. doi: 10.1002/eji.200838734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chatenoud L, Salomon B, Bluestone JA. Suppressor T cells--they’re back and critical for regulation of autoimmunity! Immunol Rev. 2001 Aug;182:149–63. doi: 10.1034/j.1600-065x.2001.1820112.x. [DOI] [PubMed] [Google Scholar]
- 124.Tang Q, Henriksen KJ, Boden EK, Tooley AJ, Ye J, Subudhi SK, Zheng XX, Strom TB, Bluestone JA. Cutting edge: CD28 controls peripheral homeostasis of CD4+CD25+ regulatory T cells. J Immunol. 2003 Oct 1;171(7):3348–52. doi: 10.4049/jimmunol.171.7.3348. [DOI] [PubMed] [Google Scholar]
- 125.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002 Aug;3(8):756–63. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 126.Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, Naji A, Caton AJ. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001 Apr;2(4):301–6. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 127.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008 Jan;28(1):100–11. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–62. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 129.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009 May;30(5):616–25. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008 May 30;133(5):775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 131.Molinero LL, Yang J, Gajewski T, Abraham C, Farrar MA, Alegre ML. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J Immunol. 2009 Jun 1;182(11):6736–43. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee AJ, Wu X, Cheng H, Zhou X, Cheng X, Sun SC. CARMA1 regulation of regulatory T cell development involves modulation of interleukin-2 receptor signaling. J Biol Chem. 2010 May 21;285(21):15696–703. doi: 10.1074/jbc.M109.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gupta S, Manicassamy S, Vasu C, Kumar A, Shang W, Sun Z. Differential requirement of PKC-theta in the development and function of natural regulatory T cells. Mol Immunol. 2008 Dec;46(2):213–24. doi: 10.1016/j.molimm.2008.08.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, Ploegh HL, Coyle AJ, Rajewsky K. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4566–71. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israel A, Rajewsky K, Pasparakis M. Mature T cells depend on signaling through the IKK complex. Immunity. 2003 Sep;19(3):377–89. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- 136.Daley SR, Hu DY, Goodnow CC. Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-κB. J Exp Med. 2013 Jan 21; doi: 10.1084/jem.20121458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun JB, Czerkinsky C, Holmgren J. Sublingual ‘oral tolerance’ induction with antigen conjugated to cholera toxin B subunit generates regulatory T cells that induce apoptosis and depletion of effector T cells. Scand J Immunol. 2007 Aug-Sep;66(2–3):278–86. doi: 10.1111/j.1365-3083.2007.01975.x. [DOI] [PubMed] [Google Scholar]
- 138.Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005 Jul;115(7):1923–33. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Coombes JL, Maloy KJ. Control of intestinal homeostasis by regulatory T cells and dendritic cells. Semin Immunol. 2007 Apr;19(2):116–26. doi: 10.1016/j.smim.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 140.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008 Jul 18;29(1):114–26. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 141.Kim JM, Rudensky A. The role of the transcription factor Foxp3 in the development of regulatory T cells. Immunol Rev. 2006 Aug;212:86–98. doi: 10.1111/j.0105-2896.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- 142.Zheng Z, Chen G, Joshi S, Brutinel ED, Yahr TL, Chen L. Biochemical characterization of a regulatory cascade controlling transcription of the Pseudomonas aeruginosa type III secretion system. J Biol Chem. 2007 Mar 2;282(9):6136–42. doi: 10.1074/jbc.M611664200. [DOI] [PubMed] [Google Scholar]
- 143.Medoff B, Xavier R. Differences with CARMA1-deficient mice. PLoS Biol. 2009;7:e51. [Google Scholar]
- 144.Barnes MJ, Krebs P, Harris N, Eidenschenk C, Gonzalez-Quintial R, Arnold CN, Crozat K, Sovath S, Moresco EM, Theofilopoulos AN, Beutler B, Hoebe K. Commitment to the Regulatory T Cell Lineage Requires CARMA1 in the Thymus but Not in the Periphery. PLoS Biol. 2009;7(3):e1000051. doi: 10.1371/journal.pbio.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blonska M, Joo D, Zweidler-McKay PA, Zhao Q, Lin X. CARMA1 Controls Th2 Cell-Specific Cytokine Expression through Regulating JunB and GATA3 Transcription Factors. J Immunol. 2012;188(7):3160–8. doi: 10.4049/jimmunol.1102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Medoff BD, Seed B, Jackobek R, Zora J, Yang Y, Luster AD, Xavier R. CARMA1 is critical for the development of allergic airway inflammation in a murine model of asthma. J Immunol. 2006 Jun 15;176(12):7272–7. doi: 10.4049/jimmunol.176.12.7272. [DOI] [PubMed] [Google Scholar]
- 147.Molinero LL, Alegre ML. Role of T cell-nuclear factor kappaB in transplantation. Transplantation Rev. 2012 Jul;26(3):189–200. doi: 10.1016/j.trre.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002 May;2(5):309–22. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 149.Rajasekaran K, Chu H, Kumar P, Xiao Y, Tinguely M, Samarakoon A, Kim TW, Li X, Thakar MS, Zhang J, Malarkannan S. Transforming growth factor-beta-activated kinase 1 regulates natural killer cell-mediated cytotoxicity and cytokine production. J Biol Chem. 2011 Sep 9;286(36):31213–24. doi: 10.1074/jbc.M111.261917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maxwell JR, Weinberg A, Prell RA, Vella AT. Danger and OX40 receptor signaling synergize to enhance memory T cell survival by inhibiting peripheral deletion. J Immunol. 2000 Jan 1;164(1):107–12. doi: 10.4049/jimmunol.164.1.107. [DOI] [PubMed] [Google Scholar]
- 151.Soroosh P, Ine S, Sugamura K, Ishii N. Differential requirements for OX40 signals on generation of effector and central memory CD4+ T cells. J Immunol. 2007 Oct 15;179(8):5014–23. doi: 10.4049/jimmunol.179.8.5014. [DOI] [PubMed] [Google Scholar]
- 152.Ohnuma K, Uchiyama M, Yamochi T, Nishibashi K, Hosono O, Takahashi N, Kina S, Tanaka H, Lin X, Dang NH, Morimoto C. Caveolin-1 triggers T-cell activation via CD26 in association with CARMA1. J Biol Chem. 2007 Feb 6; doi: 10.1074/jbc.M609157200. [DOI] [PubMed] [Google Scholar]
- 153.Bauer B, Krumbock N, Fresser F, Hochholdinger F, Spitaler M, Simm A, Uberall F, Schraven B, Baier G. Complex formation and cooperation of protein kinase C theta and Akt1/protein kinase B alpha in the NF-kappa B transactivation cascade in Jurkat T cells. J Biol Chem. 2001 Aug 24;276(34):31627–34. doi: 10.1074/jbc.M103098200. [DOI] [PubMed] [Google Scholar]
- 154.Kane LP, Andres PG, Howland KC, Abbas AK, Weiss A. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nat Immunol. 2001 Jan;2(1):37–44. doi: 10.1038/83144. [DOI] [PubMed] [Google Scholar]
- 155.Kane LP, Lin J, Weiss A. It’s all Rel-ative: NF-kappaB and CD28 costimulation of T-cell activation. Trends Immunol. 2002 Aug;23(8):413–20. doi: 10.1016/s1471-4906(02)02264-0. [DOI] [PubMed] [Google Scholar]
- 156.Kane LP, Weiss A. The PI-3 kinase/Akt pathway and T cell activation: pleiotropic pathways downstream of PIP3. Immunol Rev. 2003 Apr;192:7–20. doi: 10.1034/j.1600-065x.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- 157.Snow AL, Xiao W, Stinson JR, Lu W, Chaigne-Delalande B, Zheng L, Pittaluga S, Matthews HF, Schmitz R, Jhavar S, Kuchen S, Kardava L, Wang W, Lamborn IT, Jing H, Raffeld M, Moir S, Fleisher TA, Staudt LM, Su HC, Lenardo MJ. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J Exp Med. 2012 Nov 19;209(12):2247–61. doi: 10.1084/jem.20120831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lamason RL, McCully RR, Lew SM, Pomerantz JL. Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry. 2010 Sep 28;49(38):8240–50. doi: 10.1021/bi101052d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fujiwara SI, Yamashita Y, Nakamura N, Choi YL, Ueno T, Watanabe H, Kurashina K, Soda M, Enomoto M, Hatanaka H, Takada S, Abe M, Ozawa K, Mano H. High-resolution analysis of chromosome copy number alterations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified, with single nucleotide polymorphism-typing microarrays. Leukemia. 2008 Oct;22(10):1891–8. doi: 10.1038/leu.2008.191. [DOI] [PubMed] [Google Scholar]
- 160.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, Yamada T, Fujieda S, Tanaka S, Doi S, Miyatake A, Enomoto T, Nishiyama C, Nakano N, Maeda K, Okumura K, Ogawa H, Ikeda S, Noguchi E, Sakamoto T, Hizawa N, Ebe K, Saeki H, Sasaki T, Ebihara T, Amagai M, Takeuchi S, Furue M, Nakamura Y, Tamari M. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012 Nov;44(11):1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 161.Porras DL, Wang Y, Zhou P, Molinero LL, Alegre ML. Role of T-cell-specific nuclear factor kappaB in islet allograft rejection. Transplantation. 2012 May 27;93(10):976–82. doi: 10.1097/TP.0b013e31824d11d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vessichelli M, Ferravante A, Zotti T, Reale C, Scudiero I, Picariello G, Vito P, Stilo R. Neuroepithelial transforming gene 1 (Net1) binds to caspase activation and recruitment domain (CARD)- and membrane-associated guanylate kinase-like domain-containing (CARMA) proteins and regulates nuclear factor kappaB activation. J Biol Chem. 2012 Apr 20;287(17):13722–30. doi: 10.1074/jbc.M111.304436. [DOI] [PMC free article] [PubMed] [Google Scholar]