Abstract
Peptidoglycan hydrolases are an effective new source of antimicrobials. A chimeric fusion protein of the Ply187 endopeptidase domain and LysK SH3b cell wall binding domain is a potent agent against Staphylococcus aureus in four functional assays.
Keywords: Bacteriophage, endolysin, CHAP domain, SH3b cell wall binding domain, Staphylococcus aureus
Introduction
Staphylococcus aureus is a pathogen that causes a broad spectrum of human and animal diseases and has adapted to antibiotic selective pressures resulting in a high prevalence of multi-drug resistant strains (de Lencastre et al., 2007). The spread of these antibiotic-resistant strains is a threat to public health and a critical concern to health care providers worldwide. Thus, the search for novel antimicrobials against pathogens that are refractory to resistance development is essential.
Phage endolysins are cell wall hydrolases that are produced near the end of the phage lytic cycle to help the nascent phage escape the infected host. Endolysins are ideally suited as antimicrobials for several reasons as described previously (Loessner, 2005, Donovan et al., 2009). It has been postulated that phage endolysins have co-evolved with their host such that they target cell wall bonds that are believed difficult for the host cell to alter, and thus bacterial resistance to phage endolysins is unlikely (Fischetti, 2005). Due to the absence of an outer membrane as in Gram-negative bacteria, endolysins are able to access the Gram-positive peptidoglycan ‘from without’ and lyse these bacteria when exposed externally.
The S. aureus bacteriophage 187 endolysin (Ply187) gene was initially reported by Loessner et al. (Loessner et al., 1999). Ply187 consists of 628 amino acids with a calculated molecular mass of 71.6 kDa. Typically, endolysins from a Gram-positive background have a modular structure with an N-terminal catalytic domain for peptidoglycan hydrolysis and a C-terminal cell wall binding domain (CBD) (Loessner, 2005). However, the Pfam domain database indicates that the amino terminus of Ply187 harbors a cysteine, histidine-dependent amidohydrolase/peptidases (CHAP) domain (Bateman & Rawlings, 2003, Rigden et al., 2003) and the C-terminus contains a glucosaminidase domain with no known CBD (Loessner et al., 1999) (Fig. 1A). CBDs are essential for some endolysins’ lytic activity and often determine specificity (Baba & Schneewind, 1996, Lu et al., 2006, Loessner et al., 2002, Grundling & Schneewind, 2006, Sass & Bierbaum, 2007). Ply187 is, thus, somewhat unique among lysins of Gram-positive phages in that it lacks a CBD. The Ply187 CHAP domain shows weak homology (40% identity) with the CHAP domain of other well-characterized lysins, such as LysK, an endolysin from phage K (O’Flaherty et al., 2005). Previous experimental data indicates that cloned full-length Ply187 is nearly inactive while C-terminal truncated Ply187 (1-157aa; Ply187AN) is much more active than the full-length protein, suggesting an inhibitory domain at the C-terminus (Loessner et al., 1999). In an effort to improve the Ply187 lytic activity the Ply187AN sequences were fused to the LysK SH3b cell wall binding domain (KSH3b) to generate a chimeric Ply187AN-KSH3b fusion protein (Fig. 1A), similar to work reported from this lab with the streptococcal LambdaSa2 (LSA2) endolysin N-terminal lytic domain (Becker et al., 2009). In a series of functional assays we have demonstrated that this fusion protein is much more active than the parental Ply187AN truncated enzyme.
Figure 1. Constructs utilized, SDS-PAGE and Zymogram analysis of endolysin constructs.
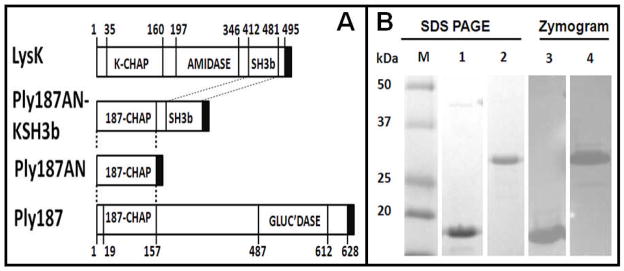
A. Schematic of constructs. Black box = pET21a derived 6 × His tag; GLUC’DASE = glucosaminidase domain. B. SDS-PAGE and zymogram analysis of Ply187 constructs. The proteins migrate as expected for their predicted molecular weights: Ply187AN (Lanes 1 and 3): 18.9kDa, Ply187AN-KSH3b (Lanes 2 and 4): 30.6 kDa. 4μg of each Ni-NTA purified protein was loaded per lane. M = excess prestained Kaleidoscope protein standards (Bio-Rad).
Materials and Methods
Strains
All Gram positive strains used to determine lysin target range are described in Table 1. All plasmid constructs were created and cryopreserved in E. coli DH5α (Invitrogen, Carlsbad, CA) and expressed from freshly transformed E. coli BL21 (DE3) (Invitrogen, Carlsbad, CA) as described below.
Table 1.
Susceptibility of multiple bacterial strains to lysis by the parental enzyme LysK and the fusion protein Ply187AN-KSH3b
| Strain | Source | Susceptibilitya
|
|
|---|---|---|---|
| LysK | Ply187AN-KSH3b | ||
| S. aureus Newman | b | ++ | ++(+) |
| S. aureus MN8 | b | ++(+) | +++ |
| S. aureus SA113 | b | ++ | +++ |
| S. aureus Reynolds CP5 | b | ++(+) | +++ |
| S. aureus Newbould (305) | ATCC 29740 | ++(+) | +++ |
| S. aureus SA019 | c | ++(+) | +++ |
| S. aureus SA020 | c | ++(+) | ++(+) |
| S. aureus SA021 | c | ++ | ++ |
| S. aureus SA026 | c | ++ | ++(+) |
| S. aureus NRS382 (MRSA) | NRS 382 | +(+) | ++(+) |
| S. aureus NRS383 (MRSA) | NRS 383 | ++ | ++(+) |
| S. aureus NRS384 (MRSA) | NRS 384 | ++ | +++ |
| S. aureus NRS385 (MRSA) | NRS 385 | ++ | ++(+) |
| Staphylococcus chromogenes | d | ++(+) | ++(+) |
| Staphylococcus epidermidis | d | ++ | +++ |
| Staphylococcus hyicus | d | +++ | +++ |
| Staphylococcus simulans | d | ++(+) | +++ |
| Staphylococcus warneri | d | ++(+) | +++ |
| Staphylococcus xylosus | d | +++(+) | +++(+) |
| Streptococcus agalactiae | ATCC 27541 | − | − |
| Streptococcus dysgalactiae | e | − | + |
| Streptococcus uberis | f | − | − |
| Listeria monocytogenes Petite ScottA | ATCC 49594 | − | − |
| Rhodococcus equi | g | − | − |
| Lactobacillus amylovorus 4540 | h | − | − |
| Lactobacillus reuteri 14171 | h | − | − |
| E. coli H5 | i | − | − |
| E. coli DH5α | Invitrogen | − | − |
| Salmonella enteritidis | ATCC13076 | − | − |
| Klebsiella pneumoniae | j | − | − |
smallest amount of protein in a volume of 10 μl causing a lysis zone after overnight incubation: ++++, 0.1 pmol; +++, 1 pmol; ++, 10 pmol; +, 100 pmol. “(+)” represents a faint lysis zone; “− “, no lysis zone at the highest amount tested (100 pmol). Scores represent averaged results from two separate experiments.
Jean C. Lee, Channing Laboratory, Brigham and Women’s Hospital, Boston, MA, USA
Yasunori Tanji, Tokyo Institute of Technology, Yokohama, Japan; bovine mastitis isolates
Max Paape, ABBL, ANRI, ARS, USDA, Beltsville, MD, USA; bovine mastitis isolates
W.D. Schultze, BARC Dairy, Beltsville, MD, USA; isolated from a clinical case
Strain 0140; Dr. A.J. Bramley, Compton Laboratory, Newbury, United Kingdom; isolated from a clinical case
Strain 33701; Steve Giguere, College of Veterinary Medicine, University of Georgia, Athens, GA, USA
Ken Bischoff, ARS, Peoria, Ill, USA.
Manan Sharma, EMFSL, ANRI, ARS, USDA, Beltsville, MD, USA
Strain K-6; E.J. Carroll, Dept. Vet Med, University of California, Davis, CA, USA; cow 2612 clinical case
Construction of expression vectors
The Ply187 protein sequence is available (Y07740) through GenBank. The truncated Ply187 N-terminal domain (Ply187AN; 1-157aa) was commercially synthesized based on published sequence (Loessner et al 1999). To enhance the heterologous expression of Ply187 endolysin in E. coli, the sequences encoding the Ply187AN were converted to an E. coli codon bias, commercially synthesized and subcloned into pUC57 with engineered 5′ NdeI (CATATG; ATG = start of translation) and 3′ XhoI (CTCGAG; codes for aa’s LE) restriction enzyme sites (Genscript; Piscataway, NJ). These sites did not introduce translational stop codons and were used to subclone the protein coding sequences into pET21a for expression and purification in E. coli B21 (DE3) (EMD Biosciences, San Diego, CA). Subcloning of the Ply187AN construct into the pET21a expression vector was via conventional means, effectively adding six C-terminal His codons to the protein coding sequences, using a strategy similar to previous constructs from the Donovan lab (Becker et al 2009). Similarly, the chimeric Ply187AN (Ply187AN-KSH3b) was fused to the LysK SH3b by subcloning the Ply187AN NdeI-XhoI DNA fragment harboring all coding sequences into a similarly digested pre-constructed pET21a-KSH3b vector described previously (Becker et al 2009). Recombinant LysK was used in this work as a positive control (Becker et al 2009).
Recombinant protein expression and purification
For protein expression, E. coli B21 (DE3) (EMD Biosciences) expression cultures were grown at 37°C in Luria-Bertani (LB) broth containing ampicillin (100μg/ml) to an OD600nm of 0.4 – 0.6, chilled on ice for 30 min., induced by addition of isopropyl-b-D-thiogalactopyranoside (IPTG) to a final concentration of 1mM, and grown at 10°C for 20 hrs. E. coli harvested from 500 ml cultures were lysed via sonication and His-tagged proteins isolated using nickel-chromatography Ni-NTA (Qiagen, Valencia, CA) as described previously (Becker et al 2009). Briefly, wash and elution profiles were empirically determined to be, 10 ml of 10 mM imidazole, 20 ml of 20 mM imidazole and elution with 0.5 ml of 250 mM imidazole in the same phosphate-buffered saline (PBS) (50 mM NaH2PO3, 300 mM NaCl, pH8.0) with 30% glycerol to prevent precipitation of the purified protein. All samples were then desalted with Zeba desalting column (Pierce, Rockford, IL) equilibrated in 2X PBS buffer and filter sterilized. Sterilized protein preparation was stored at 4°C in 2X PBS buffer until the time of assay.
SDS-PAGE and zymogram analysis of Ply187 constructs
4ug per lane of each construct and Kaleidoscope protein standards (Bio-Rad) were analyzed using 15% SDS-PAGE. Zymograms were prepared identically as the SDS PAGE gels, but with 300 ml culture equivalents of mid log phase S. aureus cells (OD600nm of 0.4–0.6). The SDS-PAGE and zymogram were running simultaneously. SDS gels were Coomassie stained and zymograms were washed in excess water for 1 hr and incubated at room temperature in PBS, resulting in areas of clearing in the turbid gel where a lytic protein had localized.
Plate lysis assay
Plate lysis assays are described previously (Becker et al, 2009). In brief, purified proteins for each construct were sterilized and diluted in sterile PBS buffer with 30% glycerol. 10μl containing 10, 1 or 0.1 μg of test protein were spotted onto a freshly spread lawn of mid-log phase (OD600 ~ 0.4–0.6) of either S. aureus Newman or S aureus 305 cells that had air-dried for 10 min on tryptic soy agar plates. The spotted plates were air-dried for 10 min in laminar flow hood, and incubated overnight in a 37 C environment. Scoring of the cleared spots occurred with 20 h of plating the cells.
Turbidity reduction assay
Turbidity reduction assays are described previously (Becker et al, 2009). In brief, staphylococcal strain S. aureus Newman grown to logarithmic phase (OD600nm =0.4–0.6) at 37°C in Brain Heart infusion broth (DIFCO, Franklin Lakes, NJ) was performed in a 96-well dish and analyzed in a plate reader. Rates of lysis that occur with 0.2 ml of S. aureus cells at an OD600nm = 1.0 in a 96 well plate were determined every 20 seconds by a plate reader [ΔOD600nm] for five minutes. Peaks in OD are determined on a sliding scale and reported results are within the linear range of the assay. Results are presented as Specific activities (ΔOD600nm/μg/min).
Minimal inhibitory concentration assay
A classical microdilution broth method for determination of the MIC was used (Jones et al 1985) with modifications as described previously (Becker et al 2009). All MIC values represent four assays with 3 replicates in each assay.
Milk Assays
Fresh milk samples obtained from the USDA BARC, Beltsville, MD dairy herd was pasteurized for 30 minutes at 63°C. The milk was brought to 37°C and was spiked with exponentially growing (OD600nm = 0.4 – 0.6) cells of the mastitis causing S. aureus strain Newbould 305 at a concentration of ~1 × 103 cfu/ml. Immediately after inoculation, purified enzyme was added at a concentration of 8 μM, and the milk was incubated at 37°C without shaking for 3 hours. Samples were taken immediately before and immediately after addition of enzyme, as well as after 30, 60, 120, and 180 minutes, and the CFUs determined by direct plating on duplicate TSA plates. A sample spiked with lysin buffer alone, no enzyme served as a control. The absence of CFUs in pasteurized milk was verified by direct plating on TSA plates.
Results
SDS PAGE and Zymogram analysis of Ply187-derived lysins
Nickel chromatography purified proteins were analyzed using 15% SDS-PAGE and Kaleidoscope protein standards (Bio-Rad, Hercules, CA) (Fig. 1B), with or without 300 ml culture equivalents of mid log phase S. aureus cells (OD600nm = 0.4–0.6) embedded in the gel as described previously (Becker et al., 2009), to verify the absence of co-purifying lytic contaminants. Coomassie-stained SDS-PAGE of each purified protein CHis-Ply187AN and C-His-Ply187AN-KSH3b indicated that the two constructs were able to be expressed in E. coli and purified at greater than 95% purity (Becker et al., 2009). Zones of lysis on the zymogram gel run in parallel with the SDS-PAGE indicate that the predicted protein in each preparation is the only protein with staphylolytic activity (Fig. 1B).
Chimeric Ply187AN-KSH3b lysin is active in killing live S. aureus in plate lysis, turbidity reduction and MIC assays
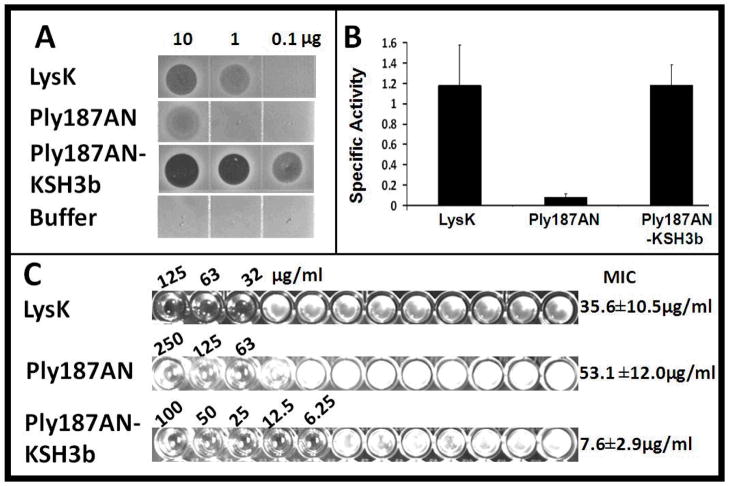
To evaluate the fusion constructs’ potential as an antimicrobial agent, we verified and quantitated the lytic activity of Ply187AN-KSH3b, the parental truncation (Ply187AN), and a strong antimicrobial endolysin, LysK (Becker et al., 2009, O’Flaherty et al., 2005) in three different antimicrobial assays. The plate lysis assay results in Fig. 2A demonstrate that both 10 μg and 1 μg LysK (0.2 and 0.02 μmol, respectively; molecular weight (MW): 55.8 kD) produce a zone of clearing, indicating that 1 μg LysK in 10 μl of buffer is effective at eliminating the S. aureus lawn, consistent with previous reports (Becker et al., 2009, O’Flaherty et al., 2005). In contrast, only 10 μg of Ply187AN (0.5 μmol; MW: 18.9 kD) produced a zone of clearing in the plate lysis assay, indicating that Ply187AN is less effective than LysK in this assay. Surprisingly, Ply187AN-KSH3b produces a zone of clearing at 10,1μg, and 0.1 μg (0.3, 0.03, and 0.003 μmol, respectively; MW: 30.6 kD), indicating that Ply187AN-SH3b is more active than Ply187AN and LysK. All bacterial strain susceptibility tests are described in Table 1. The staphylococcal strains included bovine mastitis isolates, MRSA strains, and coagulase negative staphylococci. Both the fusion protein and LysK were able to lyse all staphylococcal strains tested, with Ply187AN-KSH3b exhibiting higher activity than LysK against many strains when compared on a molar basis. In contrast, both enzymes were inactive against non-staphylococcal strains with the exception of Ply187AN-KSH3b showing weak activity against Streptococcus dysgalactiae.
Figure 2. Fusion of LysK SH3b domain enhances Ply187AN antimicrobial activity in plate lysis, turbidity reduction and MIC assays.
A. Representative plate lysis assay with S. aureus strain Newman. Zones of clearing represent lysis of the lawn. 10 μg and 1 μg LysK is equivalent to 0.2 and 0.02 μmol, respectively; MW: 55.8 kD. 10 μg of Ply187AN is equivalent to 0.5 μmol; MW: 18.9 kD. 10, 1μg and 0.1 μg of Ply187AN-KSH3b is equivalent to 0.3, 0.03, and 0.003 μmol, respectively; MW: 30.6 kD. B. Turbidity reduction assays with S. aureus strain Newman. Results are presented as Specific activities (ΔOD600nm/μmol/min). The assay volume was 200 μl. Error bars represent SEM for three or more independent experiments. C. Representative MIC of LysK and purified Ply187 derivatives. The MIC assays were repeated four times with 3 replicates in each assay.“
To further quantify the degree of lytic enhancement obtained from the fusion of the Ply187 CHAP domain to the KSH3b domain we compared the staphylolytic activities in both turbidity reduction (Fig. 2B) and minimal inhibitory concentration (MIC) assays (Fig. 2C). A standardized turbidity assay modified from (Donovan et al., 2006) with staphylococcal strains grown to logarithmic phase (OD600nm=0.4–0.6) at 37° C in Brain Heart infusion broth (DIFCO, Franklin Lakes, NJ) was performed in a 96-well dish and analyzed in a plate reader as described previously (Becker et al., 2009). The specific activity for LysK, Ply187AN and Ply187AN-KSH3b is 1.2±0.4, 0.08±0.03, and 1.2±0.2 OD600nm μmol−1min−1, respectively. Consistent with the plate lysis assay, Ply187AN was only about 10% as active as LysK. However, the addition of the KSH3b domain to the Ply187 CHAP domain yields a ten-fold increase in specific activity (Fig. 2B). The resulting activity of Ply187AN-KSH3b is similar to that of the recombinant LysK protein. A classical microdilution broth method for determination of the MIC was used (Jones et al 1985) with modifications as described previously (Becker et al., 2009) to determine the MIC for each construct. In the MIC assay, LysK inhibits growth of S. aureus Newman at concentrations of 35.6 ± 10.5 μg ml−1, corresponding to 0.6 ± 0.2 μmol ml−1, which is comparable to previous results (Becker et al 2009). The MIC for Ply187AN is 53.5 ± 12.0 μg ml−1 (2.8 ± 0.6 μmol ml−1). Similar to the prior two antimicrobial assays, Ply187AN-KSH3b is more active than Ply187AN, with an MIC of only 7.6 ± 2.9 μg ml−1 (0.24 ± 0.09 μmol ml−1), indicating that this CHAP domain yields a ~ten-fold increase in specific activity compared to Ply187AN. The fusion MIC is five-fold lower than that of LysK’s (and only 2-fold lower when compared on a molar basis). We know that LysK is a very potent antibacterial, showing higher activities in turbidity reduction assays than lysostaphin (Becker et al., 2009). These data indicate that Ply187AN-KSH3b is a potent staphylolytic agent.
Chimeric Ply187AN-KSH3b lysin is active in pasteurized cows milk
The lytic activity of Ply187-AN-KSH3b, Ply187-AN, and LysK were determined via serial diluting plating of timed samples of milk inoculated with S. aureus, and either a lytic enzyme or enzyme buffer alone (Fig. 3). Immediately after addition of enzyme, Ply187-AN-KSH3b samples showed immediate eradication of all CFUs at time zero and remained undetectable throughout the 3-hour period. Ply187-AN and LysK showed no significant difference from the buffer alone control at any time point.
Figure 3. Fusion of LysK SH3b domain enhances Ply187AN antimicrobial activity in pasteurized cow milk.
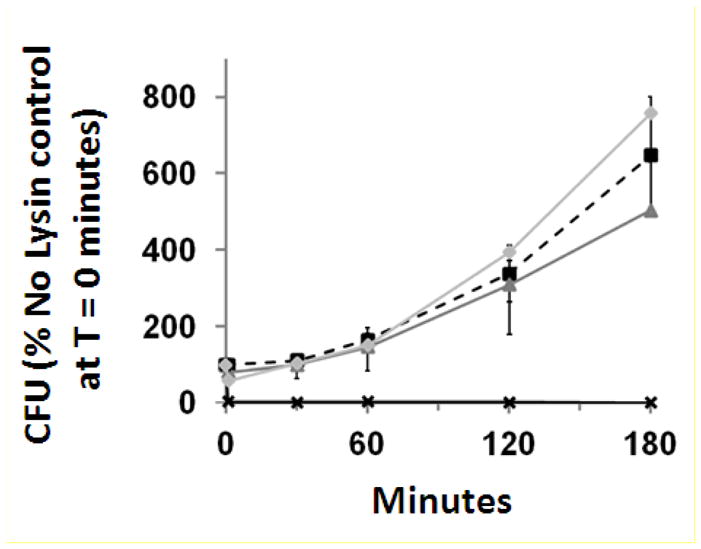
At five time points over a time period of 3 hours, CFU counts increased dramatically in inoculated milk samples following the addition of enzyme storage buffer [
 ], Ply187AN [
], Ply187AN [
 ], and LysK [
], and LysK [
 ]. However, following the addition of Ply187AN-KSH3b [✖], the CFU counts were reduced to, and stayed, at 0% of the enzyme storage buffer control at time point zero, indicating effective killing. Error bars = SEM.
]. However, following the addition of Ply187AN-KSH3b [✖], the CFU counts were reduced to, and stayed, at 0% of the enzyme storage buffer control at time point zero, indicating effective killing. Error bars = SEM.
Discussion
Although the bacterial SH3b domain is readily identified in multiple domain databases, it is still poorly understood at the level of the site of binding. SH3b domains often determine an endolysin’s specificity (Baba & Schneewind, 1996, Lu et al., 2006, Grundling & Schneewind, 2006, Sass & Bierbaum, 2007, Becker et al., 2009). Low et al. have proposed a model to explain the role of the SH3b domain in lysin lytic activity (Low et al., 2005). This model suggested that the SH3b domain folds back, binds to and inhibits the lytic domain until it recognizes and binds to peptidoglycan when it releases the lytic domain allowing digestion of the peptidoglycan. Although believed to play a role in substrate recognition and binding specificity, its role must be empirically determined. In deletion experiments, Horgan et al., (Horgan et al., 2009) suggested that deletion of the LysK SH3b domain enhanced LysK CHAP domain enzymatic activity while Becker et al (Becker et al., 2009) showed that fusion of the LysK SH3b domain to the LysK CHAP domain was necessary for CHAP domain activity. Other labs have demonstrated that lysostaphin SH3b domain bind to the S. aureus pentaglycine bridge (Baba & Schneewind, 1996, Lu et al., 2006, Grundling & Schneewind, 2006). Despite the lack of a consensus binding site, SH3b domains and other CBDs have been used for decades to develop novel antimicrobials (Diaz et al., 1991, Diaz et al., 1990, Croux et al., 1993, Lopez et al., 1997, Sheehan et al., 1996). Recently, the Fischetti group used a non-SH3b cell wall binding domain to generate a chimeric staphylolytic lysin using the Twort phage endolysin CHAP domain (Daniel et al., 2010). Our lab used staphylococcal SH3b domains in fusions with the streptococcal phage LambdaSA2 endolysin endopeptidase domain to shift the activity from Streptococcus-specificity to an enzyme that recognized both streptococcal and staphylococcal cell walls (Becker et al., 2009). Despite the numerous CBD fusion constructs reported, the Ply187-KSH3 fusion in this study represents the first time that the activity of a lytic domain from an endolysin that naturally lacks a cell wall binding domain was enhanced by adding a known cell wall binding domain.
In conclusion, we have generated a potent chimeric Ply187AN-KSH3b protein by fusing the CHAP endopeptidase domain of endolysin Ply187 from phage 187 and SH3b CBD of LysK from phage K. This chimeric Ply187AN-KSH3b is a more effective antimicrobial than the full length Ply187, the Ply187 truncation (Ply187AN) and also outperforms the known high activity lysin, LysK, in three out of four functional assays.
Acknowledgments
This work was supported in part by Viridax, Inc. Baco Raton, FL, as well as NIH grant1RO1AI075077-01A1; NRI grant 2007-35204-18395 and US State Dept funds supporting a US-Pakistani and US Russian collaboration. All awards are to DMD. Mentioning of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture.
References
- Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–4797. [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci. 2003;28:234–237. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- Becker SC, Foster-Frey J, Stodola AJ, Anacker D, Donovan DM. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene. 2009;443:32–41. doi: 10.1016/j.gene.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Becker SC, Dong S, Baker JR, Foster-Frey J, Pritchard DG, Donovan DM. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol Lett. 2009;294:52–60. doi: 10.1111/j.1574-6968.2009.01541.x. [DOI] [PubMed] [Google Scholar]
- Croux C, Ronda C, Lopez R, Garcia JL. Interchange of functional domains switches enzyme specificity: construction of a chimeric pneumococcal-clostridial cell wall lytic enzyme. Mol Microbiol. 1993;9:1019–1025. doi: 10.1111/j.1365-2958.1993.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:1603–1612. doi: 10.1128/AAC.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E, Lopez R, Garcia JL. Chimeric pneumococcal cell wall lytic enzymes reveal important physiological and evolutionary traits. J Biol Chem. 1991;266:5464–5471. [PubMed] [Google Scholar]
- Diaz E, Lopez R, Garcia JL. Chimeric phage-bacterial enzymes: a clue to the modular evolution of genes. Proc Natl Acad Sci U S A. 1990;87:8125–8129. doi: 10.1073/pnas.87.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol. 2007;10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan DM, Becker SC, Dong S, Baker JR, Foster-Frey J, Pritchard DG. Peptidoglycan hydrolase enzyme fusions for treating multi-drug resistant pathogens. Biotech International. 2009;21:6–10. [Google Scholar]
- Donovan DM, Dong S, Garrett W, Rousseau GM, Moineau S, Pritchard DG. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl Environ Microbiol. 2006;72:2988–2996. doi: 10.1128/AEM.72.4.2988-2996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005;13:491–496. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Grundling A, Schneewind O. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J Bacteriol. 2006;188:2463–2472. doi: 10.1128/JB.188.7.2463-2472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan M, O’Flynn G, Garry J, Cooney J, Coffey A, Fitzgerald GF, Ross RP, McAuliffe O. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl Environ Microbiol. 2009;75:872–874. doi: 10.1128/AEM.01831-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RN, Barry AL, Gavan TL, Washington JA., II . Susceptibility tests: microdilution and macrodilution broth procedures. In: Balows A, Hausler JWJ, Shadomy HJ, editors. Manual of clinical microbiology. American Society for Microbiology; Washington D.C: 1985. pp. 972–977. [Google Scholar]
- Loessner MJ, Gaeng S, Scherer S. Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J Bacteriol. 1999;181:4452–4460. doi: 10.1128/jb.181.15.4452-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner MJ, Kramer K, Ebel F, Scherer S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol. 2002;44:335–349. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- Loessner MJ. Bacteriophage endolysins--current state of research and applications. Curr Opin Microbiol. 2005;8:480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Lopez R, Garcia E, Garcia P, Garcia JL. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb Drug Resist. 1997;3:199–211. doi: 10.1089/mdr.1997.3.199. [DOI] [PubMed] [Google Scholar]
- Low LY, Yang C, Perego M, Osterman A, Liddington RC. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J Biol Chem. 2005;280:35433–35439. doi: 10.1074/jbc.M502723200. [DOI] [PubMed] [Google Scholar]
- Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, Sakon J. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J Biol Chem. 2006;281:549–558. doi: 10.1074/jbc.M509691200. [DOI] [PubMed] [Google Scholar]
- O’Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol. 2005;187:7161–7164. doi: 10.1128/JB.187.20.7161-7164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem Sci. 2003;28:230–234. doi: 10.1016/s0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- Sass P, Bierbaum G. Lytic Activity of Recombinant Bacteriophage {phi}11 and {phi}12 Endolysins on Whole Cells and Biofilms of Staphylococcus aureus. Appl Environ Microbiol. 2007;73:347–352. doi: 10.1128/AEM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MM, Garcia JL, Lopez R, Garcia P. Analysis of the catalytic domain of the lysin of the lactococcal bacteriophage Tuc2009 by chimeric gene assembling. FEMS Microbiol Lett. 1996;140:23–28. doi: 10.1111/j.1574-6968.1996.tb08309.x. [DOI] [PubMed] [Google Scholar]