SUMMARY
The analysis of exonic DNA from prostate cancers has identified recurrently mutated genes, but the spectrum of genome-wide alterations has not been profiled extensively in this disease. We sequenced the genomes of 57 prostate tumors and matched normal tissues to characterize somatic alterations and to study how they accumulate during oncogenesis and progression. By modeling the genesis of genomic rearrangements, we identified abundant DNA translocations and deletions that arise in a highly interdependent manner. This phenomenon, which we term “chromoplexy”, frequently accounts for the dysregulation of prostate cancer genes and appears to disrupt multiple cancer genes coordinately. Our modeling suggests that chromoplexy may induce considerable genomic derangement over relatively few events in prostate cancer and other neoplasms, supporting a model of punctuated cancer evolution. By characterizing the clonal hierarchy of genomic lesions in prostate tumors, we charted a path of oncogenic events along which chromoplexy may drive prostate carcinogenesis.
INTRODUCTION
Though often curable at early stages, clinically advanced prostate cancer causes over 250,000 deaths worldwide annually (Jemal et al., 2011). Identifying prostate cancers that require aggressive treatment and gaining durable control of advanced disease comprise two pressing public health needs. A deeper understanding of the molecular genetic changes that occur during the development of invasive and metastatic tumors may provide useful insights into these problems.
Genetic studies of prostate cancer have revealed numerous recurrent DNA alterations that dysregulate genes involved in prostatic development, chromatin modification, cell cycle regulation and androgen signaling, among other processes (Baca and Garraway, 2012). Chromosomal deletions accumulate early in prostate carcinogenesis and commonly inactivate tumor suppressor genes (TSGs) such as PTEN, TP53 and CDKN1B (Shen and Abate-Shen, 2010). In addition, recent exome sequencing of localized and castration-resistant prostate cancer has identified base-pair mutations in genes such as SPOP, FOXA1 and KDM6A, which implicate a range of deregulated cellular processes in prostate tumor development (Barbieri et al., 2012; Grasso et al., 2012; Kumar et al., 2011).
Structural genomic rearrangements also play a critical role in prostate carcinogenesis. Roughly half of prostatic adenocarcinomas overexpress an oncogenic ETS transcription factor gene (most commonly ERG) due to somatic fusion with a constitutively active or androgen-regulated promoter (Tomlins et al., 2007; Tomlins et al., 2005). In addition, disruptive rearrangements may inactivate TSGs such as PTEN or MAGI2 (Berger et al., 2011). Interestingly, analysis of prostate cancer genomes has revealed complex “chains” of rearrangements, which may result when broken DNA ends are shuffled and re-ligated to one another in a novel configuration (Berger et al., 2011). In theory, these DNA-shuffling events could simultaneously dysregulate multiple cancer genes, but the prevalence and consequences of rearrangement chains could not be assessed with the small panel of tumors sequenced.
Given the importance of structural genomic alterations in prostate cancer genesis and progression, we performed whole genome sequencing (WGS) and DNA copy number profiling of 57 prostate cancers to define a spectrum of oncogenic events that may operate during prostate tumor development. Through computational modeling of rearrangements and copy number alterations, we inferred that the chromosomal disarray in a typical tumor may accumulate over a handful of discrete events during tumor development. We employ the term “chromoplexy” to describe this putative phenomenon of complex genome restructuring (from the Greek pleko, meaning to weave or to braid). These complex rearrangement events occur in the majority of prostate cancers and may commonly inactivate multiple tumor-constraining genes in a coordinated fashion. This knowledge informs a model for punctuated tumor evolution relevant to prostate cancer and possibly other malignancies.
RESULTS
The Landscape of Genomic Rearrangement in Prostate Cancer
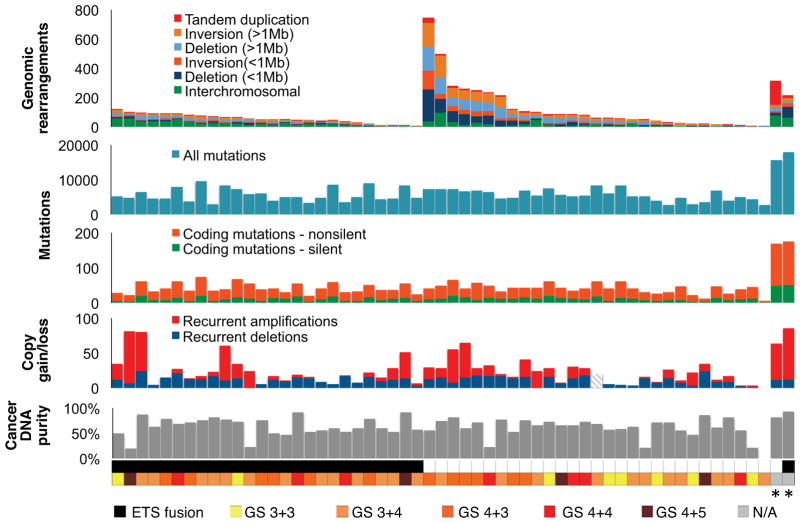
We sequenced the genomes of 55 primary prostate adenocarcinomas and two neuroendocrine prostate cancer (NEPC) metastases that developed following castration-based therapy, along with paired normal tissue. We selected treatment-naïve adenocarcinomas across a range of clinically relevant tumor grades and stages (Gleason score 6 through 9; pathological stage pT2N0 through pT4N1; Table S1). Roughly 1.68×1013 sequenced base pairs aligned uniquely to the hg19 human reference genome (Table S2). Sequencing of tumor and normal DNA to mean coverage depths of 61x and 34x, respectively, revealed 356,136 somatic base-pair mutations, with an average of 33 non-silent exonic mutations per primary tumor (Figure 1 and Table S3A). We profiled somatic DNA copy number alterations (SCNAs) with high-density oligonucleotide arrays (Table S3B). Additionally, we conducted transcriptome sequencing on 20 tumors, along with matched benign prostate tissue for 16 cases.
Figure 1. Somatic Alterations in 57 Prostate Tumor Genomes.
WGS was conducted on 55 prostate adenocarcinomas and two lung metastases from neuroendocrine prostate cancers (NEPC, *) along with paired normal DNA to detect somatic rearrangements and mutations. Gains and losses of DNA copy number at sites of recurrent SNCAs were detected with Affymetrix SNP 6.0 arrays (recurrent SCNAs were not assessed for sample P07-144, hatched lines). Bottom, cancer DNA purity was evaluated by assessing allelic ratios from sequence reads covering heterozygous single-nucleotide polymorphisms at sites of chromosomal deletion (Supplemental Experimental Procedures). ETS gene fusions (ERG, ETV1) were detected by sequencing and validated by fluorescence in situ hybridization (FISH).
To identify genomic rearrangements, we analyzed paired-end sequencing reads that map to the reference genome in unexpected orientations using the dRanger algorithm (Berger et al., 2011). We observed 5596 high-confidence rearrangements that were absent from normal DNA in both this cohort and an extended panel of 172 non-cancerous genome sequences (Figure 1 and Table S3C). We validated 113 rearrangements by re-sequencing and/or PCR amplification of tumor and normal DNA (Table S3C). We did not discover novel recurrent gene fusions, but observed several singleton events that may lead to overexpression of oncogenes. For example, sense-preserving fusions joined NRF1 to BRAF (PR-4240) and CRKL to the ERK-2 kinase gene MAPK1 (P04-1084; Figure S1A), leaving the kinase domains of BRAF and MAPK1 primarily intact. Several genes underwent recurrent disruptive rearrangements with potential biological consequence, such as PTEN, RB1, GSK3B and FOXO1 (Figure S1 and Table S4). Thus, rearrangement of these genes may contribute to the development of localized prostate cancer.
DNA Deletions and Rearrangements Reveal Signatures of Complex Genome-Restructuring Events
Rearrangements involving cancer gene loci often occurred in the context of a “chain”, in which the two rearrangement breakpoints map to the reference genome near breakpoints from other rearrangements (Figure 2A, left). Such characteristic breakpoint distributions were observed in our initial study of seven prostate cancer genomes (Berger et al., 2011) and appear to reflect collections of broken DNA ends that are shuffled and ligated to one another in an aberrant configuration. Given the involvement of prostate cancer genes in rearrangement chains, we set out to survey chained rearrangements systematically to clarify their prevalence and potential biological consequences.
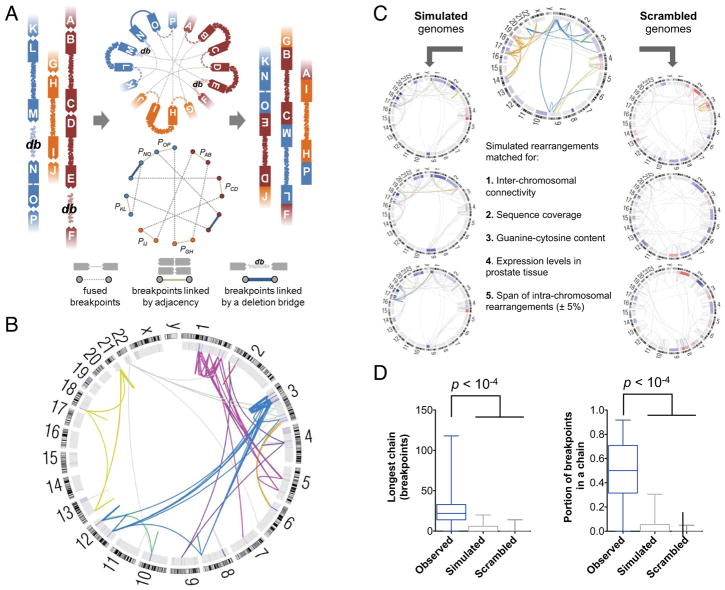
Figure 2. Integrated Analysis of Genomic Deletions and Rearrangements Reveals Signatures of Concurrent Alterations.
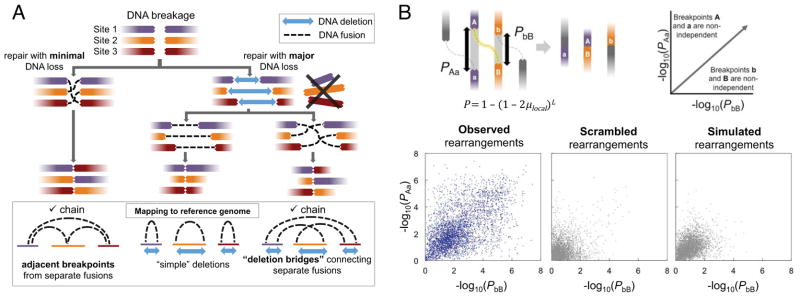
(A) Three scenarios by which multiple DNA double-strand breaks may be repaired. Concerted repair with minimal loss of DNA (left) results in fusion breakpoints that map to adjacent positions in the reference genome. Loss of DNA at sites of double-strand breaks may result in simple deletions (middle) or “deletion bridges” (right) that span breakpoints from distinct fusions on the reference genome. Adjacent breakpoints or deletion bridges may provide evidence for chained rearrangements.
(B) For the two breakpoints of each rearrangement (labeled A and B), the probability P of a second independently generated breakpoint (a or b) falling within the observed distance (L) was assessed based on the expected local rate of rearrangements (μlocal). The x- and y- coordinates represent the negative log of P for the two breakpoints in each fusion. Rearrangements near the upper right corner of the plot are unlikely to have arisen independently of other rearrangements. Observed rearrangements are compared to simulated and scrambled data.
See also Figure S2.
We first determined whether additional chains could be identified by integrative analysis of chromosomal deletions and rearrangements. Although rearrangement chains may arise with minimal loss of genetic material, substantial DNA deletions were often evident at the fusion junctions of chained rearrangements (Figure 2A, right). When these deletions are overlaid with somatic rearrangement locations on the reference genome, the deletions create “bridges” that span the sequence between breakpoints from two different fusions (Figure 2A, bottom right). In all informative tumors in our cohort, the breakpoints at either end of a deletion were more often fused to novel partners rather than to each other (thus creating “deletion bridges”, rather than “simple deletions”; Figure S2A). Importantly, this observation indicates that the many rearrangements demonstrating DNA loss near a breakpoint may be linked by deletion bridges to additional rearrangements in a chain.
We next considered whether rearrangements in a chain might arise independently of one another, for instance, at loci that are predisposed toward fusion due to DNA secondary structure or nuclear proximity (Burrow et al., 2010; De and Michor, 2011). To investigate this, we created a probabilistic model for the independent generation of detectible rearrangements across the genome (Figure S2B). Using this model, we calculated the probability that any pair of neighboring DNA breakpoints X and Y would arise independently of each other (PXY) based on (1) their reference genome distance and (2) the local rate of rearrangements observed in our tumor panel (Figure 2B). As a control, we created ten simulated genomes for each tumor, with rearrangement locations matched for chromosome, local gene expression levels, sequence guanine/cytosine content and DNA replication timing, among other factors (Supplemental Experimental Procedures). In addition, we generated “scrambled” genomes by combining rearrangements from distinct tumors, preserving locus-specific effects that may promote double strand breakage. The observed rearrangements, but not the simulated or scrambled data, showed marked deviation from the independent model (Figure S2C) and statistical enhancement of chain-like patterns (Figure 2B). For 50% of rearrangements, the reference genome locations of both breakpoints were nearer to breakpoints of additional rearrangements than would be expected by chance (p < 10−4 for observed versus simulated or scrambled PXY values). To the extent that our model correctly predicts the genomic distribution of independent rearrangements, these results suggest that rearrangement chains are unlikely to arise from independent events, thus raising the hypothesis that they occur by a coordinated process.
“Chromoplexy” Generates Chained Chromosomal Rearrangements and Deletions
Having identified chained patterns of rearrangements that may result from interdependent alterations, we created an algorithm called ChainFinder to search for such events systematically (Figures 3A and S3). ChainFinder employs a statistically based search rooted in graph theory to identify genomic rearrangements and associated deletions that deviate significantly from our independent model described above, and thus appear to have arisen in an interdependent fashion (Supplemental Experimental Procedures).
Figure 3. The ChainFinder Algorithm.
(A) ChainFinder creates a graph representation of genomic breakpoints that may be linked in chains by somatic fusions, statistical adjacency or deletion bridges. ChainFinder assigns two neighboring breakpoints to the same chain if the p-value for their independent generation (P) is rejected with a false-discovery rate below 10−2. For each cycle (closed path) within the graph, all scenarios are considered where one or more rearrangements in the cycle could have arisen independently. All rearrangements in a cycle are assigned to the same chain if every such scenario is rejected with a family-wise error rate below 10−2 across all scenarios. Please see the Supplemental Experimental Procedures for additional details.
(B) Circos plot of chained rearrangements in a prostate adenocarcima (P09-1042). Rearrangements depicted in the same color arose within the same chain; fusions in gray were not assigned to a chain. The inner ring depicts copy number gains and losses in blue and red, respectively.
(C) The false positive rate of ChainFinder was assessed using simulated and scrambled genomes based on observed rearrangements.
(D) For observed, simulated and scrambled genomes, the longest chain was compared along with the portion of breakpoints in any chain. Median values, middle quartiles, and range are indicated.
See also Figure S3.
We used ChainFinder to survey our panel of prostate tumors for rearrangement chains. Strikingly, this analysis revealed numerous chains involving widely variable numbers of rearrangements. Some chains involved only three fusions, while others contained more than forty rearrangements that wove together five or more chromosomes (Table S5A; Figure 3B and S3C). We have termed the putative process of genomic restructuring that produces these complex chains “chromoplexy” (from the Greek pleko, meaning “to weave” or “to braid”). Chromoplectic chains of five or more rearrangements (ten or more breakpoints) were detected in 50 out of 57 tumors (88%; Table S5B and Figure S3C), while 36 tumors (63%) contained two or more such chains. Overall, 39% of rearrangements participated in chains, while ChainFinder detected chains in only 2.8% and 0.2% of rearrangements from simulated or scrambled genomes, respectively (Figure 3C–D). Thus, our statistical analysis of breakpoint distributions suggests that chromoplexy frequently generates multiple structural alterations in a coordinated fashion.
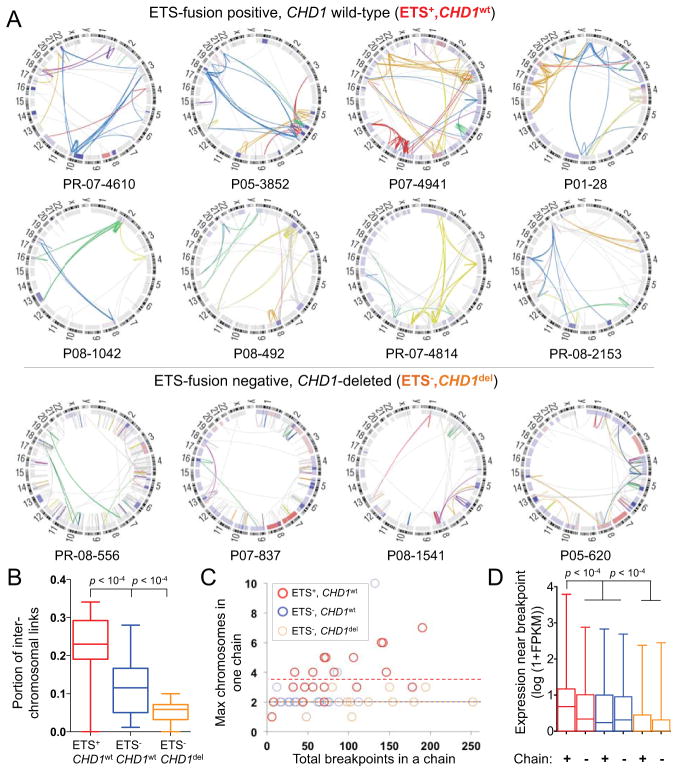
We noted profound phenotypic differences in chromoplexy in subsets of prostate cancers. Chromoplexy in tumors harboring oncogenic ETS fusions (ETS+) produced significantly more inter-chromosomal rearrangements than ETS− tumors (p < 10−4) and involved a greater maximum number of chromosomes in a single event (p = 0.009; Figure 4A–C). Interestingly, oncogenic ERG fusions frequently arose in the setting of chromoplexy (15 of 26 cases, 58%). Given that fusion of TMPRSS2 and ERG occurs in the setting of androgen receptor-driven transcription (Haffner et al., 2010), the intricate chains in ETS+ tumors could reflect DNA injury at transcriptional hubs occupied by loci from multiple chromosomes. Consistent with this possibility, chromoplexy in ETS-positive nuclei primarily affected regions of the genome that are highly expressed in prostate tumors (Figure 4D) and that co-localize in interphase nuclei (Figure S4A). Thus, chromoplexy in ETS+ tumors appears to reflect a distinct process of genome restructuring that may be coupled to transcriptional processes.
Figure 4. Manifestations of Chromoplexy Vary by ETS Fusion Status.
(A) Circos plots of rearrangement chains in representative tumors, grouped by the presence of ETS rearrangements and CHD1 disruption. Rearrangements in the same chain are depicted in one color. Rearrangements in gray were not assigned to a chain. The inner ring shows copy number gain and loss in red and blue, respectively.
(B) Rearrangement chains in ETS+ tumors contain a greater proportion of inter-chromosomal fusions than chains in ETS− tumors. In (B) and (D), box plots indicate median values, middle quartiles, and range.
(C) The maximum number of chromosomes involved in a single rearrangement chain (y-axis), grouped by ETS status. The total number of breakpoints in chains in each tumor is depicted on the x-axis to allow comparison of tumors with similar degree of detectable chromoplexy.
(D) ETS+ chromoplexy breakpoints are enriched near DNA that is highly expressed in 16 prostate tumor transcriptomes.
See also Figure S4.
In contrast, chromoplexy in a subset of ETS-negative cancers resembled chromothripsis (Rausch et al., 2012; Stephens et al., 2011), a process of chromatin shattering yielding extensive DNA rearrangement, often of one or two focal chromosomal regions. In particular, seven ETS− tumors contained up to seven-fold more rearrangements than the whole-cohort average (Figure S4B). These tumors harbored focal deletions or disruptive rearrangements involving the chromatin-modifying enzyme gene CHD1, a putative tumor-suppressor gene that may regulate genomic stability (Huang et al., 2011; Liu et al., 2012). The rearrangements in CHD1del tumors were predominantly intra-chromosomal both within chains (p = 2×10−4) and overall (p = 4×10−4; Figure S4C). Moreover, the rearrangements in CHD1del samples arose in late-replicating DNA with low guanine and cytosine content (Figure S4B), generally corresponding to gene-poor heterochromatin. An extended cohort of 199 prostate adenocarcinomas revealed that CHD1 loss was associated with an increased number of recurrent SCNAs (p = 1.5×10−8) (Figure S4C). Given the postulated roles of CHD1 in genome stability and maintenance of chromatin architecture (Gaspar-Maia et al., 2009), these findings raise the possibility that CHD1 deletion may contribute to the distinctive patterns of genomic instability observed in CHD1del tumors.
We investigated whether chromoplexy is unique to prostate cancer by analyzing a panel of 59 additional tumor genomes including melanoma, non-small cell lung cancer, head and neck squamous cell carcinoma, and breast adenocarcinoma (Table S5B and Figure S3C). Every tumor type demonstrated multiple instances of chains involving 5 or more rearrangements. Thus, a small number of chromoplectic events may account for the wide array of rearrangements and deletions in several common cancers.
Chromoplexy Commonly Dysregulates Cancer Genes
To assess the role of chromoplexy in prostate cancer development, we examined the genomic regions altered by deletion or disruptive rearrangements in the context of chains. Using a list of 17 potential prostate tumor suppressor genes from the KEGG database (Kanehisa et al., 2012), we found that 26 of the 57 tumors (46%) have either deletion or rearrangement of at least one gene in a chain of three or more rearrangements (Table S5C). Inclusion of the TMPRSS2-ERG fusion and 10 putative prostate cancer genes added 9 more samples. Several cancer genes were recurrently deleted or rearranged by chromoplexy, including PTEN (9 cases), NKX3-1 (8 cases), CDKN1B (3 cases), TP53 (4 cases), and RB1 (2 cases). Thus, chromoplexy may conceivably influence prostate carcinogenesis by disrupting tumor suppressor genes and creating oncogenic fusions.
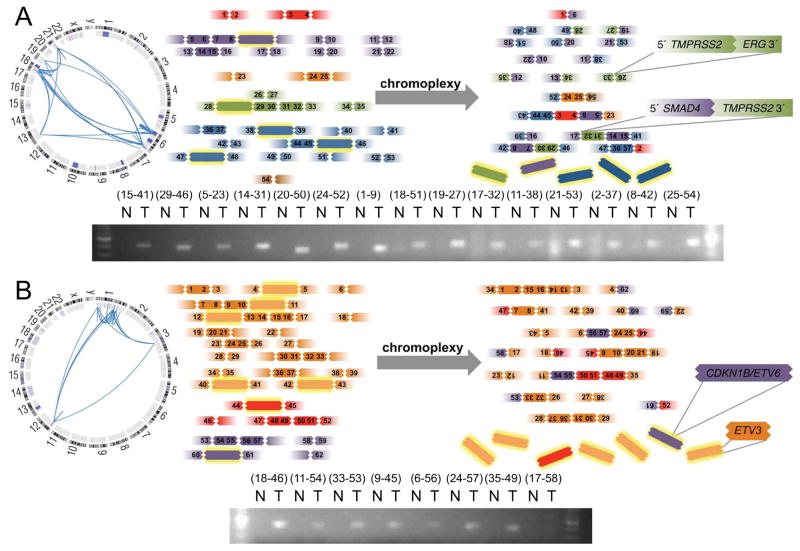
The concurrent shuffling and deletion of multiple regions across the genome that appears to underlie chromoplexy could simultaneously inactivate tumor suppressor genes that are geographically distant from each other (i.e. on separate chromosomes). We noted several examples where multiple cancer genes were apparently disrupted by a single instance of chromoplexy. For instance, a chain of 27 rearrangements across 6 chromosomes included the TMPRSS2-ERG fusion (21q) as well as a disruptive rearrangement of the prostate tumor suppressor gene SMAD4 (18q) (Ding et al., 2011) (Figures 5A and S5). In a second example, the adjacent CDKN1B/ETV6 tumor suppressor genes (12p) and the ETV3 locus (1q) were lost in the context of deletion bridges within one chain (Figure 5B). Additional instances of chromoplexy disrupted interacting genes in the same pathway: for instance, co-deletion of PIK3R1 (5q) with PTEN (10q) and TP53 (17p) with CHEK2 (22q) occurred in two chains (Table S5C). Thus, chromoplexy may simultaneously dysregulate multiple cancer genes across the genome. Such events may provide selective advantages to incipient cancer cells, particularly given that the loss of some TSGs promotes prostate cancer only in the context of specific accompanying molecular lesions (Chen et al., 2005).
Figure 5. Chromoplexy May Coordinately Dysregulate Multiple Cancer Genes.
(A) Chromoplectic chain of 27 somatic rearrangements across 6 chromosomes in tumor P05-3852, involving fusion of TMPRSS2 and ERG and disruptive rearrangement of SMAD4.
(B) The putative tumor suppressor genes CDKN1B, ETV6 and ETV3 were lost in the context of deletion bridges in a 25-rearrangement chain affecting 3 chromosomes in PR-05-3595. In both panels, selected rearrangements were assessed by PCR of tumor and normal DNA.
Clonal Evolution Reveals Paths of Prostate Cancer Progression
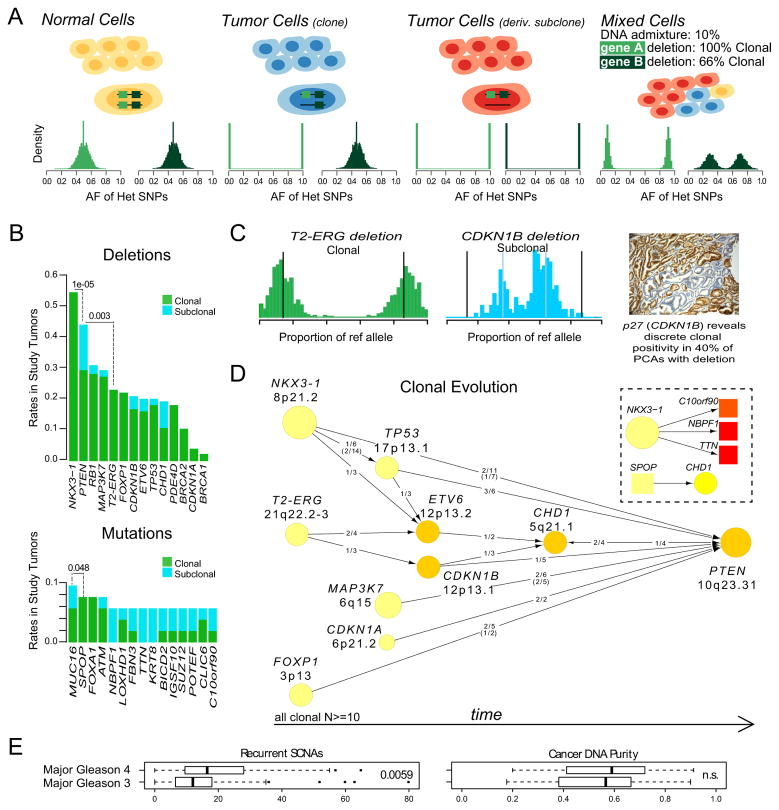
To provide additional insight into the genomic evolution of prostate tumors, we analyzed the clonal status of mutations and deletions in our cohort. Using an approach related to previously described methods (Carter et al., 2012; Nik-Zainal et al., 2012), we exploited the extensive germline SNP genotype data provided by WGS to assess tumor purity and the clonal status of genomic lesions (Figures 6A and S6). Our estimates of tumor purity based on WGS matched those produced by ABSOLUTE analysis of SNP array data (Carter et al., 2012) (R2 = 0.99; p<10−4), with the exception of two samples where admixed normal DNA was detected only from sequencing data (Table S1).
Figure 6. Clonality and Evolution of Prostate Cancer.
(A) Schematic representation of the clonality assessment. The allelic fractions (AFs) of sequencing reads covering heterozygous SNPs were analyzed in order to assess the clonality of somatic DNA alterations. A hypothetical tumor is shown, composed of normal cells, a caner clone and a derivative subclone. The histograms indicate the expected SNP AFs within two deleted genes, A and B. The subclonal deletion of B yields a distinct distribution of AFs compared to the clonal deletion of A.
(B) Selected deletions (top) and mutations (bottom) were classified as clonal or subclonal. Proportion test p-value is listed for the indicated comparisons. Independent samples (Barbieri et al., 2012) are included for support.
(C) Example of clonal (TMPRSS2-ERG) and subclonal (CDKN1B) deletions from the same tumor. Histograms show the proportion of sequencing reads containing the reference allele for heterozygous SNPs in the deleted regions. A representative immunohistochemical stain for the CDKN1B protein p27 shows discrete subclonal positivity in prostate cancer.
(D) Patterns of tumor evolution were inferred based on clonality estimates. Arrows indicate the direction of clonal-subclonal hierarchy between genes that are deleted in the same sample in multiple cases. Deleted genes are represented by circles with size and color intensity reflecting the frequency of overall deletions and subclonal deletions, respectively. Ratios along the arrows indicate the number of samples demonstrating directionality of the hierarchy out of samples with deletion of both genes (ratios in parentheses refer to additional samples; Barbieri et al., 2012). The inset shows a similar analysis of point mutations (Barbieri et al., 2012).
(E) The number of recurrent SCNAs and cancer DNA purity were compared across tumors with major Gleason pattern 4 versus 3.
See also Figure S6.
We first compared the clonality of deletions involving prostate cancer genes, reasoning that lesions that arise early in tumorigenesis or that foster rapid outgrowth would tend to be clonal, while late-arising deletions would more often be subclonal. Several common deletions were strictly clonal, including NKX3-1 and the 3Mb region of chromosome 21q that is frequently deleted to produce the TMPRSS2-ERG fusion (Perner et al., 2006) (Figure 6B and Figure S6). These events are among the earliest detectible alterations in prostate cancer and are frequently observed in prostatic intraepithelial neoplasia (PIN), a prostate cancer precursor lesion (Emmert-Buck et al., 1995; Perner et al., 2007). By contrast, deletions of PTEN were often subclonal (p = 10−5 for comparison with NKX3-1 deletion clonality), as were CDKN1B deletions (Figure 6C). This finding suggests that PTEN and CDKN1B inactivation promotes the early progression of prostate cancer, consistent with the association of these events with higher-stage disease (Barbieri et al., 2012; Halvorsen et al., 2003).
We next used our clonality assessments to deconvolve the sequence of oncogenic events that gives rise to a typical prostate tumor. Reasoning that clonal alterations must originate prior to subclonal alterations within the same tumor, we examined pairs of genes that were deleted in the same sample across multiple tumors to determine the directionality of the clonal-subclonal hierarchy (Figure 6D). Where possible, we confirmed these relationships in independent exome-sequenced tumors. A “consensus path” of progression emerged, beginning with events including deletion of NKX3-1 or FOXP1 and fusion of TMPRSS2 and ERG. These lesions may disrupt normal prostate epithelial differentiation (Bhatia-Gaur et al., 1999; Sun et al., 2008) and effect other oncogenic perturbations. Thereafter, lesions in CDKN1B or TP53 accumulate; these alterations may lead to enhanced proliferation, genomic instability and/or evasion of apoptosis. Finally, loss of PTEN may provide a gating event in the development of aggressive prostate cancers. A similar assessment of point mutation clonality (Figure 6B, lower) revealed higher overall rates of subclonal events, with the exception of early mutations as in SPOP and FOXA1. Together, these results imply that prostate carcinogenesis favors the dysregulation of cancer genes in defined sequences, as has been suggested by studies of developing tumors in colon cancer (Fearon and Vogelstein, 1990).
Next, we investigated whether chromoplexy might continue after cancer initiation, and thereby contribute to the progression of a tumor down an oncogenic path. Interestingly, several chains appeared to involve strictly subclonal deletion bridges (Figure S7A), indicating that tumors may sustain multiple rounds of chromoplexy. Together with the observation that chromoplexy may affect both early and late genes in the consensus path (e.g., ERG and PTEN) these findings suggest that chromoplexy also occurs in tumor subclones that emerge later during cancer evolution.
Prostate Cancer Genomic Derangement Increases with Histological Grade
Finally, we considered whether tumors with high-grade histology (indicative of high clinical risk) might occupy positions further along the consensus path. To this end, we quantified recurrent SCNAs in each genome by counting amplifications and deletions that overlapped with regions of significant SNCAs identified by GISTICv2 analysis (e.g., the TP53 and PTEN loci) across 199 tumors reported here and in a previous study (Barbieri et al., 2012; Mermel et al., 2011). Tumors with predominantly Gleason score (GS) 4 histology were significantly enriched for recurrent SCNAs compared to GS 3 tumors (p = 0.0059; Figure 6E) beyond the overall extent of SCNAs, despite similar purity of cancer DNA and mutational burden between the two groups. Altogether, these findings suggest that structural alterations affecting cancer genes, many of which result from chromoplexy, may contribute to the aggressive clinical behavior of high-grade prostate tumors.
DISCUSSION
We have characterized somatic alterations across the genomes of 57 prostate tumors. By systematically profiling rearrangements and copy number alterations, we identified chromoplexy as a common process by which multiple geographically-distant genomic regions may be disrupted at once. Like other classes of complex genomic alterations (Stephens et al. 2011; Forment et al. 2012), chromoplexy was inferred from computational modeling, and its mechanistic underpinnings will need to be addressed experimentally. Chromoplexy is evident in several solid tumor types and in the majority of prostate cancers. In multiple instances, chromoplexy altered more than one cancer gene coordinately. In the future, systematic assessment of chromoplexy from WGS data could reveal groups of cancer gene alterations that confer a selective advantage when sustained all at once, but activate tumor-suppressing safeguards if sustained individually.
Although chained rearrangements could theoretically arise over multiple cellular generations by a “sequential-dependent” mechanism, where the occurrence of each subsequent event depends on the presence of a prior event (Figure S7B), such a mechanism seems unlikely. In particular, a sequential-dependent model fails to account for the many complete or “closed” chains we detected. For a closed chain to arise in a sequential-dependent manner, multiple junctions from ancestral somatic fusions would have to be re-broken precisely and fused to each other (Figure S7B) to complete the chain. Even if breakpoints in a chain could only fuse to one another, to generate the 121 observed closed chains in a sequential-dependent process would require immensely elevated rates of rearrangement in a focused region of the genome (up to ~103 times the maximum observed rate; Figure S7C–D). While we cannot exclude this possibility, plausible biological mechanism(s) could parsimoniously account for chained rearrangements within a single cell cycle, as discussed below.
A unifying feature of chromoplectic alterations is that they occur in a non-independent fashion; however multiple mechanisms may account for chromoplexy. Along these lines, our analyses have revealed distinctive patterns of chromoplexy in ETS−, CHD1del tumors. Tumors with deletion of CHD1 demonstrated an excess of intrachromosomal chained rearrangements and gene deletions, with DNA breakpoints enriched in GC-poor, late-replicating and non-expressed DNA. Previous reports have proposed that similar patterns may result from major DNA damaging events within heterochromatic nuclear compartments (Drier et al 2013). These tumors showed abundant, clustered rearrangements often affecting only one or two chromosomes with two alternating copy number states, perhaps indicating a chromothripsis-like process.
In contrast, chromoplexy in ETS+ tumors differed in the aggregate from chromothripsis in several critical ways. For example, single events joined DNA from dispersed regions of six or more chromosomes in multiple tumors, whereas chromothripsis frequently involves focal rearrangement of one or two chromosomes (Forment et al., 2012). Overall, chromoplexy appears more prevalent in ETS+ prostate cancer than chromothripsis is in any neoplasm (Stephens et al., 2011, Forment et al., 2012). Chromoplexy frequently involves fewer rearrangements than the “catastrophic” chromothripsis defined by Stephens et al., but may continue throughout tumor development. Our analysis of breakpoint locations in ETS+ tumors suggests that chromoplexy in this setting may be linked to proposed transcriptional DNA-damaging processes (Lin et al., 2009), potentially related to androgen receptor signaling. We stress that this hypothesis awaits experimental validation, which could involve FISH or chromosome conformation capture (3C) before and after inducing a predicted co-localizing event (e.g., testosterone exposure in prostate epithelial cells). Our findings align with the observation that ERG-overexpressing cancer cells accumulate DNA damage and are sensitive to poly ADP-ribose polymerase inhibition (Brenner et al., 2011). Chromoplexy is active prior to ETS gene fusions, however, and generated ERG fusions in many instances.
Whole genome analysis also clarified the chronology of oncogenic events in prostate cancer progression, driven in part by chromoplexy. Genome-wide sequence coverage of germline SNPs allowed us to identify DNA lesions that arose after the founder clone was established. Subsequently, we demonstrated a progression of events within primary tumors that expands upon array-based SCNA co-occurrence studies (Demichelis et al., 2009). A consensus path of tumor evolution begins with events such as loss of NKX3-1 or fusion of TMPRSS2 and ERG. The path proceeds with the loss of CDKN1B, TP53, PTEN, and other progression-associated lesions. We found that the histological grade of cancer may partially reflect its progression down this path.
A Continuum Model for Tumor Evolution
Tumorigenesis is classically understood to progress by a gradual accumulation of oncogenic alterations in the genome of a pre-cancerous cell. This textbook view was recently challenged by the discovery of chromothripsis, in which catastrophic rearrangements are incurred by “shattering” and reassembly of focal regions of the genome (Forment et al., 2012; Rausch et al., 2012; Stephens et al., 2011).
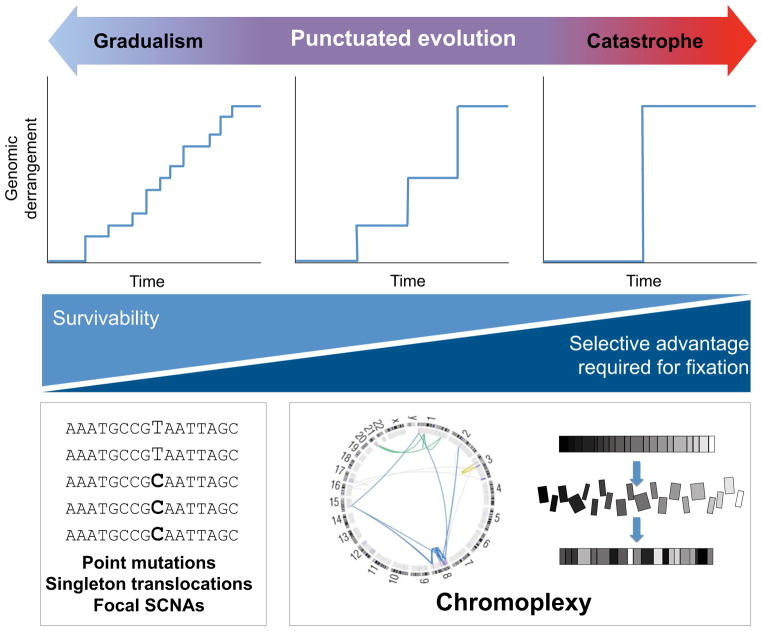
We propose an expanded model for the evolution of prostate cancer, which may also apply to other cancers (Figure 7). As classically understood, passenger and driver alterations can accumulate in a cancer genome gradually over numerous cell divisions, via point mutations, simple translocations and focal copy-number alterations. On the opposite end of the spectrum, extreme instances of chromothripsis can induce massive (albeit relatively localized) DNA damage at once, often with oncogenic consequences (Rausch et al., 2012; Stephens et al., 2011). Between these two extremes lies a broad continuum across which chromoplexy may often restructure cancer genomes. We propose that oncogenic events along this continuum reflect “punctuated” tumor evolution, drawing an analogy from the observation that “punctuated” evolution of species may occur rapidly between periods of relative mutational equilibrium (Gould, 1977). By analogy, a tumor genome may sustain considerable damage over several sequential and punctuated events. Importantly, this framework accords with the observation that chromoplexy events (1) are common, (2) may involve a wide-ranging number of rearrangements, and (3) may continue after cancer-initiating lesions such as NKX3-1 deletion (Figure S7).
Figure 7. A Continuum Model for the Genomic Evolution of Prostate Cancer.
Oncogenic aberrations may accumulate in cancer genomes gradually (left), by punctuated progression (middle) or in a single catastrophic event (right). Chromoplectic rearrangements and deletions induce a modest to large degree of genomic derangement over several successive events. As indicated at bottom, larger-scale rearrangements that affect broader swaths of the genome may be more difficult for a cell to survive, and may tend to require co-occurring oncogenic lesions to become fixed in a tumor.
See also Figure S7.
A cancer might operate at any point along the continuum of progression at a given time. Tumors that develop primarily at the “catastrophic” end may require fewer events and could progress more quickly, because each such event could disrupt multiple cancer-constraining processes. At the same time, catastrophic events that cover diffuse genomic territory are more liable to disrupt essential or beneficial genes, thus imparting a selective disadvantage to (pre)malignant clones that sustain such events. Consequently, the model predicts that survivable chromoplexy (particularly near the catastrophic regime) is likely to involve oncogenic alterations that compensate for the incidental inactivation of essential genes (Figure 7, bottom). This prediction accords with the observation that most tumors show disruption of one or more putative prostate cancer genes within a chain. Moreover, this model raises the possibility that disruption of putative cancer genes by chromoplexy may heighten the probability that such genes represent “driver” events for that particular tumor. If so, this framework may portend important implications for the use of whole-genome sequencing in diagnostic and clinical studies.
In summary, this study highlights the potential for WGS data to capture aspects of the “molecular archeology” of cancer development that are missed by gene- or exome-level sequencing. The characterization of clonal progression and chromoplexy in emerging large panels of cancer genomes may provide insights about cancer initiation and progression, with implications for cancer detection, prevention and therapy.
EXPERIMENTAL PROCEDURES
Sample Acquisition
Prostate tumors were obtained under Institutional Review Board-approved protocols from consented patients undergoing radical prostatectomy or excision of soft-tissue metastases (PR-4240 and PR-7520). Normal DNA derived either from histologically benign prostate tissue or peripheral blood cells. Specimens were collected at Weill Cornell Medical College (WCMC) by A.T. and at various medical centers in Western Australia in conjunction with Uropath Pty LTD (Perth, Australia).
DNA Library Construction and WGS
Tissue cores were extracted from cancerous foci of frozen or paraffin-embedded tumor nodules. After tissue homogenization and lysis, DNA was extracted and assessed for quality (Berger et al., 2011). Following library construction, paired-end sequencing reads of 101 nucleotides were generated with an Illumina GAIIx instrument. Sequencing data were aligned to the hg19 human reference genome using BWA (Li and Durbin, 2009) and processed by the Picard pipelines (http://picard.sourceforge.net).
Detection and Validation of Genomic Alterations
Somatic point mutations, small indels and rearrangements were detected by comparison of tumor and paired normal genome sequences using MuTect (Cibulskis et al. 2013) and Indelocator Sivachenko et al., in preparation) (www.broadinstitute.org/cancer/cga/) and dRanger (Berger et al., 2011), respectively. dRanger was used as described previously, (Berger et al., 2011), except that high-confidence rearrangements required support from four or more high-quality sequencing reads and were filtered against a panel of 176 normal tissue genomes. Somatic fusion breakpoints were located at basepair resolution where possible with the BreakPointer algorithm (Drier et al., 2013). Paired-end reads from rearrangements affecting cancer genes or participating in long chains were inspected manually. A subset of rearrangements was validated by resequencing and/or PCR amplification of tumor and normal DNA.
Chromosomal Copy Number Profiling
Segmented copy number profiles were generated from Affymetrix SNP 6.0 human SNP microarray (Affymetrix, Santa Clara, CA) data as described previously (Barbieri et al., 2012). Sites of significant recurrent copy number alterations were identified by GISTICv2 (Mermel et al. 2011), with a log2 threshold of +/− 0.1 for amplification/deletion signals.
Identification of Chained Rearrangements and Deletions
The ChainFinder algorithm was implemented to detect chromoplexy from the combined analysis of somatic fusion breakpoints and segmented copy number profiles. ChainFinder considers breakpoints as nodes in a graph that are connected by edges corresponding to (a) fusions, (b) deletion bridges or (c) breakpoint adjacency that deviates significantly from the null model of independent breakpoints (Figure S3). Over several steps, the algorithm evaluates potential deletion bridges and adjacently mapping breakpoints to assign rearrangements to chains.
First, ChainFinder identifies potential deletion bridges by searching for distinct breakpoints that plausibly correspond to the boundaries of deletion events observed in copy number profiles. Next, a statistical analysis of all nearest-neighbor breakpoint pair distances identifies chain-like distributions of rearrangements. The local rate of expected independent breaks per nucleotide (μ) is calculated for 1Mb genomic windows based on (1) the rearrangement frequency within the window across a panel of tumor genomes and (2) the total number of breaks in the genome under consideration. Given μ, ChainFinder models the probability PXY of observing two independently arising fusion breakpoints within the observed distance L of each other on the reference genome (i.e., the p-value under the null model of independent breaks):
If PXY is rejected with a false-discovery rate of 10−2 (Benjamini, 1995), the corresponding breakpoints are linked in a chain.
The graph is also searched for closed paths (cycles) through nodes and connecting edges. For each cycle, all possible scenarios are considered by which the contained breakpoints could have arisen independently. Breakpoints in the cycle are assigned to the same chain if p values for every scenario can be rejected with a family-wise error rate (FWER) below 10−2.
Lastly, the graph is finalized by assigning additional sets of edges corresponding to deletion bridges that could not be assigned uniquely in the first step. The search maximizes the number of deletion bridges in cycles to find solutions that account most fully for the overlap of fusion breakpoints with boundaries of deletion segments on the reference genome. A complete description of ChainFinder is provided in the Supplemental Experimental Procedures. ChainFinder can be downloaded at www.broadinstitute.org/cancer/cga/chainfinder.
Assessment of Stromal DNA Admixture and Clonality
We used the sequence coverage from germline SNPs at sites of somatic deletion to assess levels of stromal DNA admixture in sequenced samples and to infer the clonal status of mutations and deletions by applying CLONET (CLONality Estimate in Tumors; Supplemental Experimental Procedures). We assessed the allelic fractions of SNP reads within hemizygously-deleted DNA to determine the apparent proportions of DNA from normal cells at the deleted locus. Deletions with the lowest apparent proportions of normal DNA reads were considered clonal. For all other deletions, we estimated the percentage of tumor cells harboring the deletion to infer the clonality of the lesion using simulation-based error estimates. For point mutations, the tumor allelic fraction was corrected for stromal DNA admixture and subclonality was inferred when the corrected fraction differed significantly from the expected value. Lesions present in 80% of cancer cells or less were considered subclonal.
Statistical Analyses
Quantitative comparisons of groups (e.g. numbers of rearrangements or SCNAs) were conducted with the rank-sum Mann-Whitney test, unless indicated otherwise. Box plots indicate median values and middle quartiles.
Supplementary Material
HIGHLIGHTS.
Interdependent DNA rearrangements may coordinately remodel prostate cancer genomes
”Chromoplexy” defines a distinct class of complex structural rearrangements
Multiple prostate cancer genes may be dysregulated coordinately
Clonal evolution reveals paths of prostate cancer progression
Acknowledgments
We thank the members of the Broad Institute Genome Sequencing Platform for their part in this work. This study was supported by the US National Human Genome Research Institute (NHGRI) Large Scale Sequencing Program (U54 HG003067 to the Broad Institute, E.S.L.), the Kohlberg Foundation (L.A.G.), the Starr Cancer Consortium (M.A.R., F.D., A.T., G.G., and L.A.G), the Prostate Cancer Foundation (M.A.R.), US Department of Defense Synergy Awards (PC101020 to F.D., L.A.G. and M.A.R.) and New Investigator Award (PC094516 to F.D.), the Dana-Farber/Harvard Cancer Center Prostate Cancer SPORE (US National Institutes of Health (NIH) P50 CA090381), the US National Cancer Institute, Early Detection Research Network (U01CA111275 and NCI EDRN to F.D. and M.A.R.), the US National Cancer Institute (R01 CA125612 to F.D. and M.A.R.), the Fondazione Trentina per la Ricerca sui Tumori (F.D.) the Swiss Science Foundation (PASMP3_134379/1 to J.-P.T.), the National Institute of General Medical Sciences award number T32GM007753 (S.C.B.) and a US NIH Director’s New Innovator Award (DP2OD002750 to L.A.G.). L.A.G. is an equity holder and consultant in Foundation Medicine, a consultant to Novartis and Millenium/Takeda, and a recipient of a grant from Novartis.
Footnotes
ACCESSION NUMBERS
Binary sequence alignment/map (BAM) files from WGS data, as well as RNAseq and SNP array data were deposited in the database of Genotypes and Phenotypes (dbGaP; phs000447.v1.p1)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baca SC, Garraway LA. The genomic landscape of prostate cancer. Front Endocrinol (Lausanne) 2012;3:69. doi: 10.3389/fendo.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini YaHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc. 1995;57:289–300. [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrow AA, Marullo A, Holder LR, Wang YH. Secondary structure formation and DNA instability at fragile site FRA16B. Nucleic Acids Res. 2010;38:2865–2877. doi: 10.1093/nar/gkp1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol. 2011;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichelis F, Setlur SR, Beroukhim R, Perner S, Korbel JO, Lafargue CJ, Pflueger D, Pina C, Hofer MD, Sboner A, et al. Distinct genomic aberrations associated with ERG rearranged prostate cancer. Gene Chromosome Canc. 2009;48:366–380. doi: 10.1002/gcc.20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, Meyerson M, Beroukhim R, Getz G. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, Zhuang Z, Bostwick DG, Liotta LA, Linehan WM. Allelic loss on chromosome 8p12–21 in microdissected prostatic intraepithelial neoplasia. Cancer Res. 1995;55:2959–2962. [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Eldrige Niles. Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology. 1977;3:115–151. [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9:1474–1479. [PubMed] [Google Scholar]
- Huang S, Gulzar ZG, Salari K, Lapointe J, Brooks JD, Pollack JR. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2011 doi: 10.1038/onc.2011.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA-Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar a, White Ta, MacKenzie aP, Clegg N, Lee C, Dumpit RF, Coleman I, Ng SB, Salipante SJ, Rieder MJ, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci USA. 2011;108 doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lindberg J, Sui G, Luo J, Egevad L, Li T, Xie C, Wan M, Kim ST, Wang Z, et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2012;31:3939–3948. doi: 10.1038/onc.2011.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins Sa, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.