Abstract
As part of an ongoing effort to expand the genetic alphabet for in vitro and eventually in vivo applications, we have synthesized a wide variety of predominantly hydrophobic unnatural base pairs exemplified by d5SICS-dMMO2 and d5SICS-dNaM. When incorporated into DNA, the latter is replicated and transcribed with greater efficiency and fidelity than the former, however previous optimization efforts identified the para and methoxy-distal meta positions of dMMO2 as particularly promising for further optimization. Here, we report the stepwise optimization of dMMO2 via the synthesis and evaluation of eighteen novel para-derivatized analogs of dMMO2, followed by further derivatization and evaluation of the most promising analogs with meta substituents. Subject to size constraints, we find that para substituents can optimize replication via both steric and electronic effects and that meta methoxy groups are unfavorable while fluoro substituents can be beneficial or deleterious depending on the para substituent. In addition, we find that improvements in the efficiency of unnatural triphosphate insertion translate most directly into higher fidelity replication. Importantly, we identify multiple, unique base pair derivatives that when incorporated into DNA are well replicated. The most promising, d5SICS-dFEMO, is replicated under some conditions with greater efficiency and fidelity than d5SICS-dNaM. These results clearly demonstrate the generality of hydrophobic forces for the control of base pairing within DNA, provide a wealth of new SAR data, and importantly identify multiple new candidates for eventual in vivo evaluation.
1. Introduction
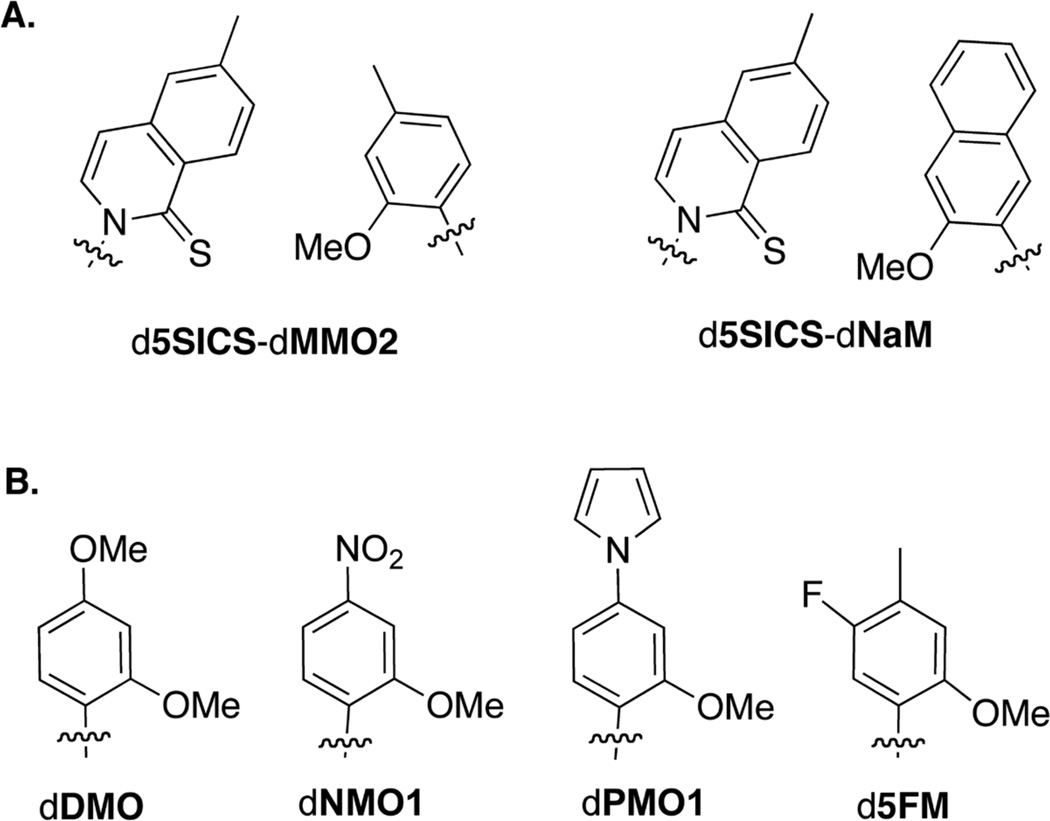
With the long term goal of expanding the genetic code, we1–4 and others5–7 have worked towards the identification of unnatural nucleotides that stably pair within duplex DNA as well as during replication and transcription, and thus constitute an unnatural base pair. We have identified a class of unnatural base pairs, exemplified by d5SICS-dMMO2 and d5SICS-dNaM (Figure 1A), that are both efficiently replicated2,8,9 and efficiently transcribed.10 From a conceptual perspective, this efficient replication and transcription are of particular interest because they are mediated only by hydrophobic and packing forces between nucleobases that have no structural homology to their natural counterparts. Overall, d5SICS-dNaM is replicated and transcribed more efficiently than d5SICS-dMMO2, and is also the only unnatural base pair shown to be efficiently replicated in a sequence-independent manner during PCR;2 however, the individual steps of replication are not equally efficient. For example, incorporation of dMMO2TP opposite d5SICS is less efficient than incorporation of dNaMTP, but continued extension of a primer terminating with dNaM by incorporation of the next correct triphosphate is slower than that of a primer terminating with dMMO2. While past SAR studies have demonstrated that replication is most limited by the synthesis of the strand containing dMMO2 or dNaM,8–11 the relative contributions of efficient unnatural triphosphate incorporation and extension to the overall efficiency and fidelity are not well understood. Thus, both dMMO2 and dNaM remain promising partners for d5SICS, but the simpler and more atom-economical scaffold of dMMO2 makes it a particularly promising scaffold for further optimization.
Figure 1.
(a) Unnatural base pairs d5SICS-dMMO2 and d5SICS-dNaM. (b) dMMO2 derivatives, dDMO, dNMO1, dPMO1, and d5FM. Only nucleobase analogs shown with sugar and phosphate omitted for clarity.
Previous structure-activity relationship (SAR) data indicate that the ortho methoxy group of the dMMO2 scaffold is necessary for efficient replication,8,12,13 and that substituents at the adjacent meta position are not well tolerated.14–16 Thus modification at the para- and remaining meta-position of the dMMO2 scaffold appears to be most promising for optimization. Previous SAR studies also suggest that modifications at the para position generally have larger effects, for example, dDMOTP, dNMO1TP, and dPMO1TP (Figure 1B) are inserted opposite d5SICS more efficiently than dMMO2TP,9,17 but those at the meta position can also be beneficial, for example, after incorporation of the corresponding triphosphate, d5FM (Figure 1B) is more efficiently extended than dMMO2.10 Nonetheless, all of the resulting unnatural pairs are still replicated significantly less efficiently than d5SICS-dNaM.
Nowhere has the optimization of synthetic molecules for biological function been more successful than in medicinal chemistry, which traditionally relies on the synthesis of derivatives in conjunction with efficient assays for the rapid identification of the most promising compounds and the elucidation of SAR data for additional optimization efforts. To emulate this approach, herein we report an optimized set of divergent synthetic strategies to access derivatives of dMMO2TP, as well as their efficient analysis via pre-steady state kinetics and PCR assays. We synthesized a small library of novel para–derivatized dMMO2 analogs that when combined with dDMO, dNMO1, and dPMO1, provide a much more complete survey of the potential of this site for optimization. Several of the most promising analogs were then further derivatized with meta fluorine or methoxy substituents, whose characterization along with d5FM provides an initial analysis of the effects of simultaneous meta- and para-derivatization. A wealth of SAR data was generated and several well replicated derivative base pairs were identified, including d5SICS-dFEMO, which under some conditions is replicated better than d5SICS-dNaM. These results further demonstrate the robustness and generality of hydrophobic and packing forces for the control of DNA replication and also further validate the dMMO2 scaffold as a partner for d5SICS. Moreover, several of the newly identified unnatural base pairs are not only well replicated but also have varying physicochemical properties that may eventually facilitate replication in vivo.
2. Results
2.1. Design and synthesis of para-substituted derivatives of dMMO2
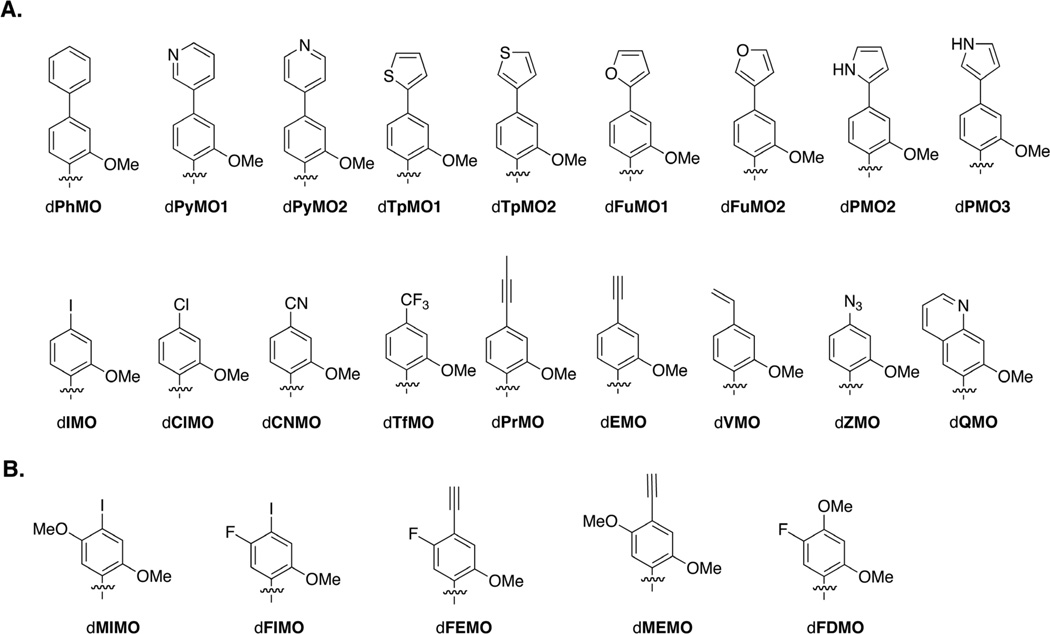
We first designed eighteen para-derivatized dMMO2 analogs (Figure 2A), which when combined with the previously reported analogs, dDMO, dNMO1, and dPMO1, provide a rather complete survey of steric and electronic effects. Along with dPMO1, the bis-aromatic analogs dPhMO, dPyMO1, dPyMO2, dTpMO1, dTpMO2, dFuMO1, dFuMO2, dPMO2, and dPMO3 were designed to explore the effects of annular substituents and the dIMO and dClMO derivatives were designed to alter nucleobase bulk and electronics. The remainder of the analogs, dPrMO, dEMO, dVMO, dCNMO, dZMO, and dQMO and dTfMO, were designed to help deconvolute the contributions of sterics and electrostatics.
Figure 2.
(a) Eighteen mono para-substituted analogs of dMMO2. (b) Five meta, para-di-substituted analogs of dMMO2. Only nucleobase analogs shown with sugar and phosphate omitted for clarity.
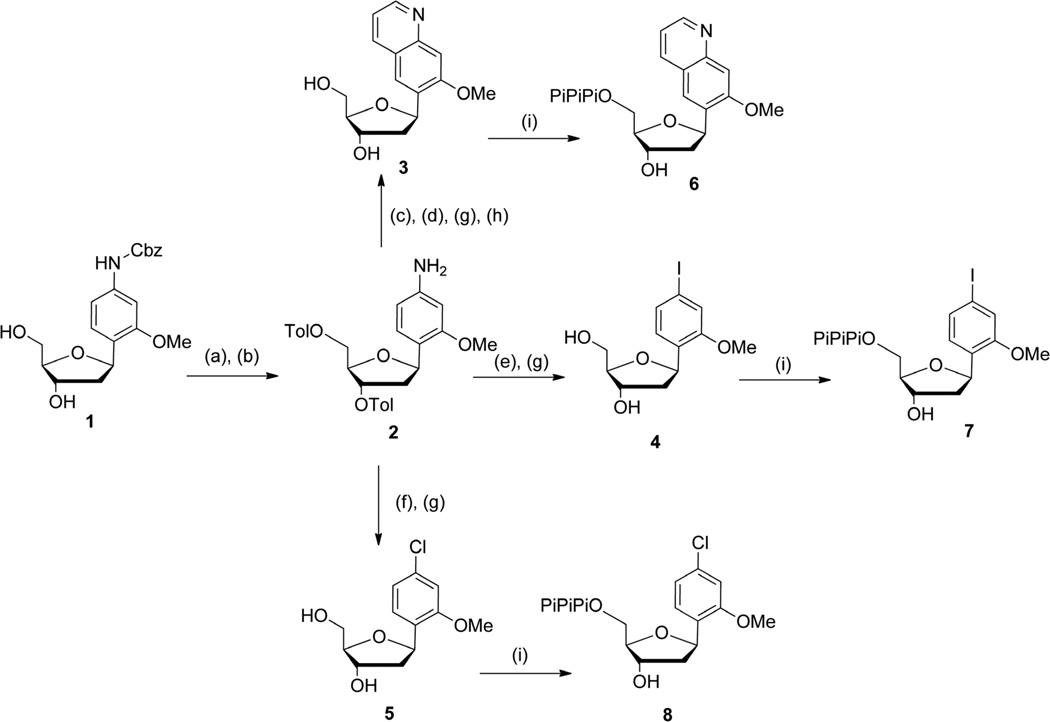
The unnatural nucleotides analogs were synthesised as shown in Schemes 1 – 5. dQMO, dIMO and dClMO triphosphates were obtained from the previously reported precursor 19 (Scheme 1).18 Briefly, hydroxyl group protection followed by hydrogenation afforded compound 2, which was then sulfonated,19 coupled to acrolein via conjugate addition, acidified to form the quinoline ring, and finally deprotected with sodium methoxide to provide dQMO (3) in good yield. Toward dIMO (4) and dClMO (5), 2 was subjected to Sandmeyer iodination and chlorination, respectively, and then deprotected. Free nucleosides 3 – 5 were converted to the corresponding triphosphates 6 – 8 under Ludwig conditions,20 and purified by anion exchange chromatography followed by HPLC. The purity of each triphosphate was confirmed by 31P NMR, HPLC, and MALDI-TOF MS (Supporting Information).
Scheme 1.
Conditions: (a) Toluyl chloride, pyridine, rt, 15 h, 59%; (b) 10% Pd/C, H2, EtOAc, NEt3, rt, 1 h, 91%; (c) (1) TsCl, pyr, CH2Cl2, rt, 40 min; (2) acrolein, NEt3, MeOH, 0 °C → rt, 20 min, 85%; (d) HCl 3N, THF, 80 °C, 40 min, 80%; (e) HCl aq 6M, NaNO2, KI, THF, 0 °C → rt, 2h, 55%; (f) HCl aq 6M, NaNO2, CuCl, THF, 0 °C → 40 °C, 5h, 23%; (g) MeONa 30% in MeOH, MeOH:CH2Cl2 8:2, 5 °C → rt, 30 min − 1 h: 3, 90%; 4, 86%; 5, 96%; (h) tBuOK, THF, 70 °C, 3 h, 78%; (i) proton sponge, POCl3, PO(OMe)3, −15 °C ° −10 °C, 3 h then Bu3N, (Bu3NH)2H2P2O7 in DMF, −10 °C → 0 °C, 30 min then TEAB buffer (0.5M), rt, 10 min: 6, 27%; 7, 48%; 8, 57%.
Scheme 5.
Conditions: (a) I2, H5IO6, MeOH, 70 °C, 5 h, 79%; (b) Pd(OAc)2, AsPh3, nBu3N, DMF, 70 °C, 15 h; (c) TBAF 1 M in THF, 0 °C → rt, 1 h, 27% 2 steps; (d) NaBH(OAc)3, AcOH, CH3CN, −4 °C, 45 min, 88%; (e) proton sponge, POCl3, PO(OMe)3, −15 °C →−10 °C, 3 h then Bu3N, (Bu3NH)2H2P2O7 in DMF, −10 °C → 0 °C, 30 min then TEAB buffer (0.5M), rt, 10 min, 27%; (f) Pd(OAc)2, TPPTS, CuI, triethylsilylacetylene, Et3N, H2O:ACN 2:1, 65 °C, 30 min; (g) NH4OH 30%, rt, 1 h, 40% 2 steps.
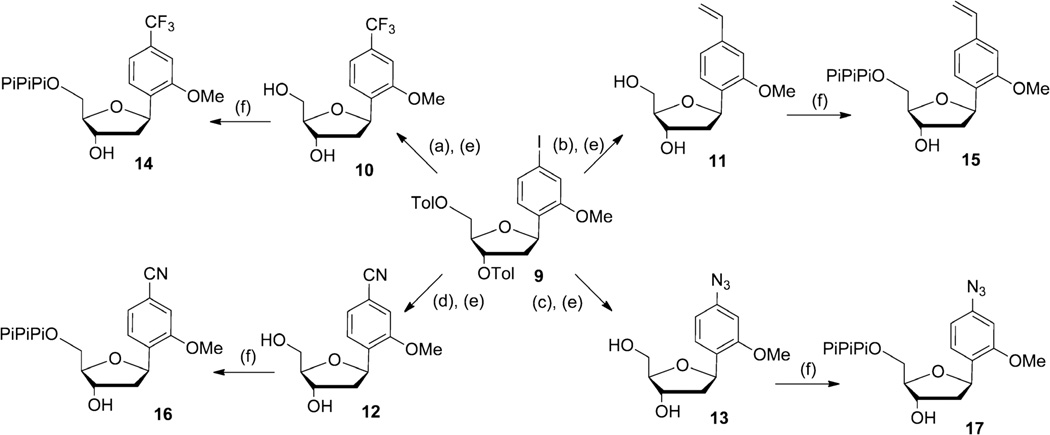
Nucleotides dTfMO, dVMO, dCNMO and dZMO were obtained from the toluyl protected intermediate 9 as shown in Scheme 2. Potassium (trifluoromethyl)trimethoxyborate was used as a source of CF3 nucleophiles for the copper-catalyzed trifluoromethylation,21 and deprotection yielded dTfMO (10). Toward dVMO, we found that Suzuki-Miyaura cross-coupling with vinyltrifluoroborate,22 palladium cross-coupling with vinylaluminium reagent23 or vinyltriethoxysilane,24 or Stille cross-coupling with vinyltributyltin25 all resulted in the conversion of the aromatic iodide (9) to its vinyl analog with good yields. Because the Stille cross-coupling generated cleaner crude material, we proceeded with this route, and the dVMO (11) nucleoside was obtained after deprotection. Palladium-catalyzed cyanation of the aryl iodide (9) using potassium hexacyanoferrate (II) in water and under microwave irradiation, followed by deprotection yielded dCNMO (12).26 It is noteworthy that with this particular substrate, palladium-catalyzed cyanation in organic solvent using zinc cyanide failed to give any desired product and only low yields were obtained with copper cyanide. Toward dZMO, the aromatic iodide of 9 was subjected to a mild CuI/diamine catalyzed Ulmann type coupling with aqueous sodium azide. The reaction proceeded cleanly to completion and deprotection then provided dZMO (13) in good yield. Free nucleosides 10 – 13 were converted to the corresponding triphosphates 14 – 17 and purified as described above.
Scheme 2.
Conditions: (a) CuI, 70 °C, 16 h, 1,10-phenanthroline, KCF3B(OMe)3, DMSO, 70%; (b) Pd2dba3, CuI, AsPh3, vinyltributyltin, dioxane, 50 °C, 2 h, 72%; (c) K4Fe(CN)6, Pd(OAc)2, KF, TBAB, H2O, microwave−150 °C, 15 min, 55%; (d) NaN3, CuI, N,N’-dimethylethylenediamine, 90 °C, 45 min, 74%; (e) MeONa 30% in MeOH, MeOH/CH2Cl2 8/2, 5 °C → rt, 45 min − 1 h: 10, 85%; 11, 90%; 12, 79%; 13, 98%; (f) proton sponge, POCl3, PO(OMe)3, −15 °C →−10 °C, 3 h then Bu3N, (Bu3NH)2H2P2O7 in DMF, −10 °C → 0 °C, 30 min then TEAB buffer (0.5M), rt, 10 min: 14, 20%; 15, 36%; 16, 42%; 17, 25%.
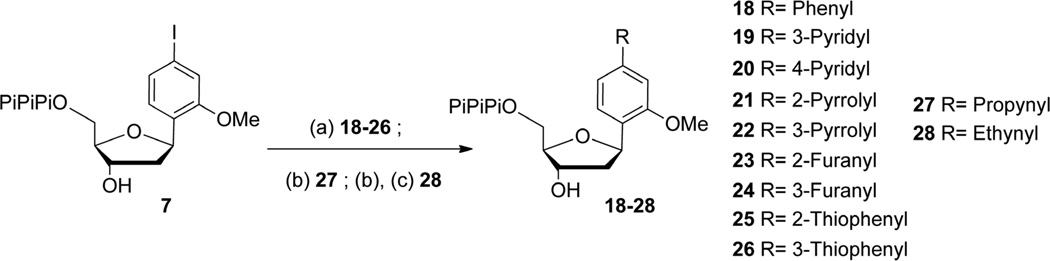
The triphosphates of dPhMO, dPyMO1, dPyMO2, dTpMO1, dTpMO2, dFuMO1, dFuMO2, dPMO2, dPMO3, dPrMO, and dEMO were readily obtained from the unprotected triphosphate 7 using aqueous Sonogashira or Suzuki-Miyaura cross-coupling (Scheme 3). dPhMO to dPMO3 (18 – 26) were obtained using a previously reported approach involving aqueous palladium cross-coupling in the presence of a water soluble sulfonated triphenylphosphine ligand (TPPTS) and cesium carbonate with quantitative conversion of the aromatic amine.27–32 Reaction time and temperature were optimized to avoid triphosphate degradation. dPrMO triphosphate (27) was obtained using aqueous copper catalyzed Sonogashira coupling in presence of TPPTS, triethylamine and a large excess of propyne gas. The dEMO triphosphate (28) was obtained similarly by coupling triethylsilylacetylene and freeing the alkyne with ammonia. Each triphosphate was purified as described above.
Scheme 3.
Conditions: (a) Pd(OAc)2, TPPTS, Cs2CO3, boronic acid derivative, H2O:ACN 2:1, 70 °C, 30 min, >70%; (b) Pd(OAc)2, TPPTS, CuI, NEt3, H2O/ACN 2/1, 30 min, 27 propyne 70 °C, 70%, 28 triethylsilylacetylene 55 °C 65%; (c) NH4OH 30%, rt, 1 h, 28, 60% 2 steps.
2.2. Initial pre-steady-state kinetic analysis of para modified derivatives
In previous work, we employed steady-state kinetics to analyze the various steps that contribute to the replication of DNA containing an unnatural base pair, including the rate at which the unnatural base pair is synthesized (by incorporation of an unnatural triphosphate opposite its cognate base in a template), and the rate at which the nascent primer terminus is extended by incorporation of the next correct natural triphosphate. While such experiments are time intensive, they provided critical information about the synthesis of the unnatural base pairs, which for the early and less efficiently replicated analogs was required for optimization. In contrast, replication of the current candidates is very efficient and under steady-state conditions limited by product dissociation,33 rendering the steady-state kinetics data less helpful for the optimization of processive synthesis. Thus, we developed a higher throughput pre-steady state assay that is based on determining under a fixed set of conditions the amount of a dMMO2TP analog and dCTP that are added to a 23mer primer opposite their cognate nucleotides in a 45mer template (containing d5SICS at position 24 and dG at position 25) by the Klenow fragment of E. coli DNA polymerase I (Kf). The percent incorporation (%incorporation) of the unnatural triphosphate was defined as the ratio, [24mer+25mer]/[23mer+24mer+25mer], and the percent extension (%extension) was defined as the ratio, [25mer]/[24mer+25mer], determined in the presence of saturating concentrations of unnatural triphosphate.
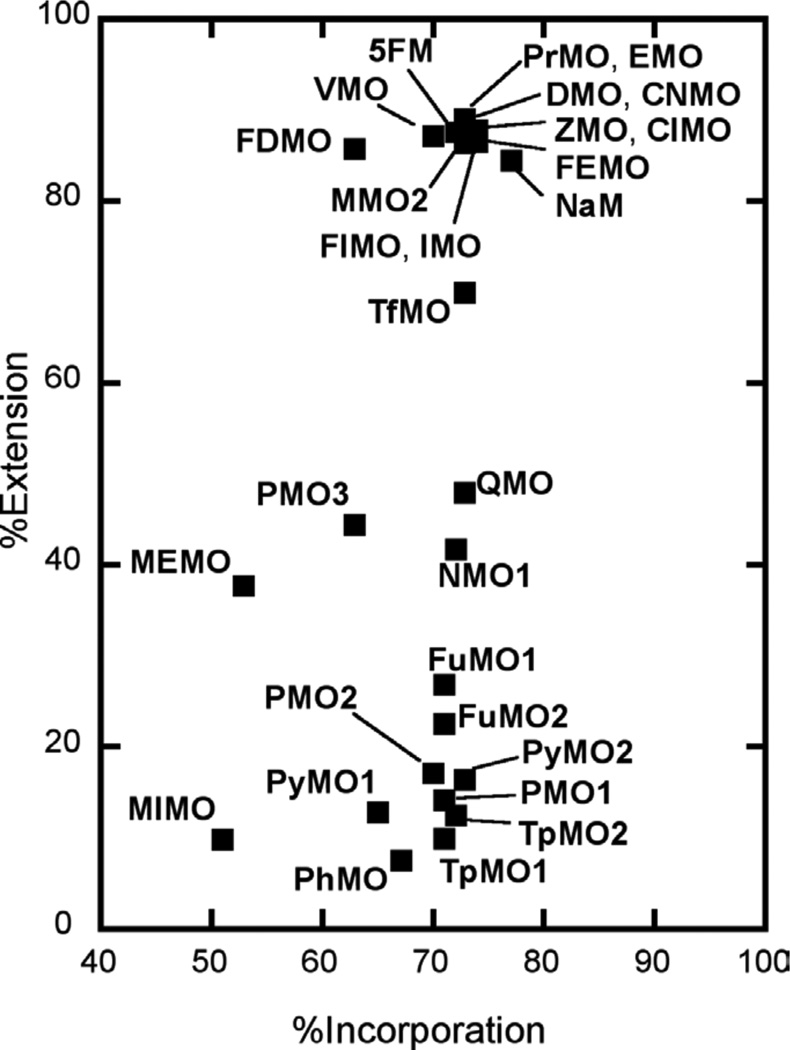
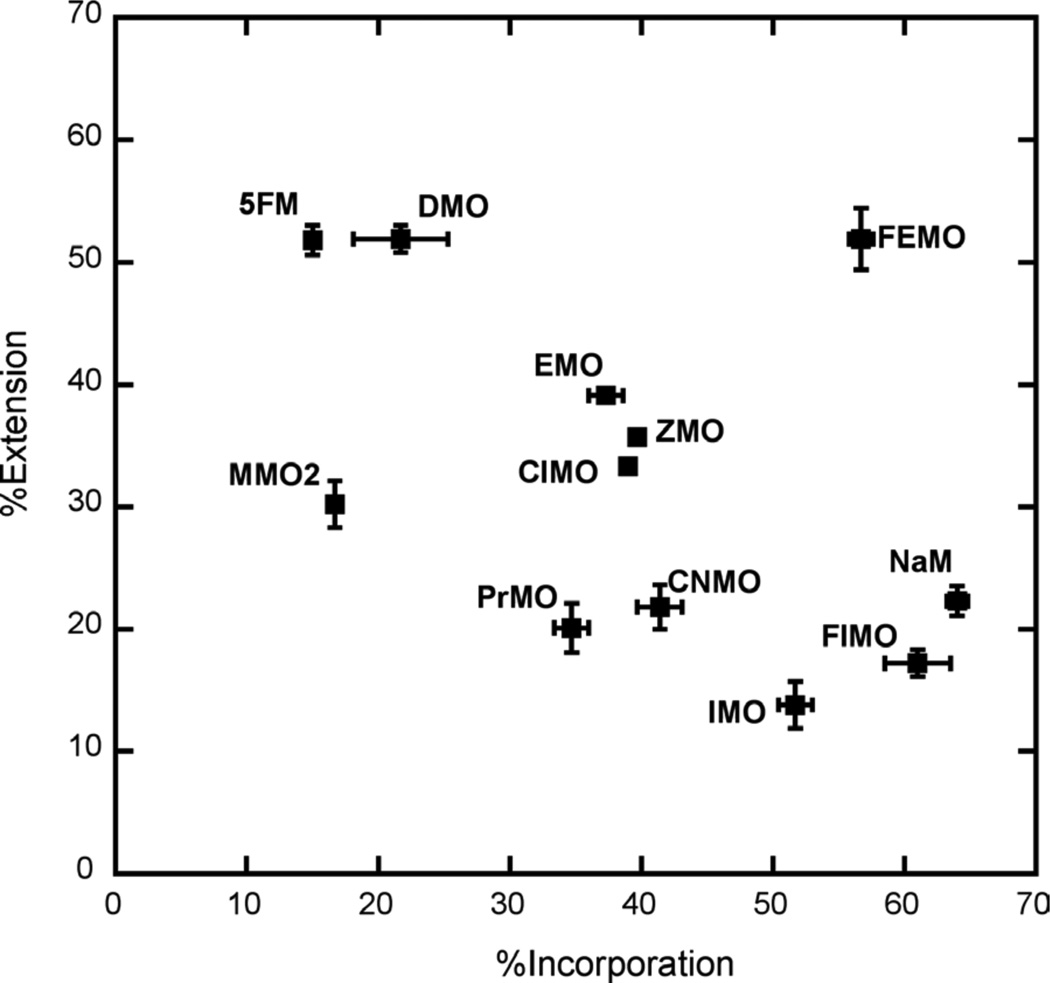
We first explored DNA synthesis with relatively high concentrations of unnatural triphosphate and dCTP (20 µM each; Figure 3 and Table S1) and with reaction times of 10 s. Under these conditions, all of the reactions, including those with dMMO2TP and dNaMTP, showed similar accumulation of 24mer, confirming that incorporation is fast relative to extension and that 20 µM of the unnatural triphosphate is sufficient for saturation (further confirmed with reactions run with 50 µM unnatural triphosphate, data not shown). In contrast, very different %extension values were observed in each reaction. With dMMO2TP or dNaMTP at the primer terminus, the %extension is 85%. Nine derivatives paired opposite d5SICS are extended significantly less efficiently, including dPhMO, dTpMO1, dPyMO1, dTpMO2, dPMO1, dPyMO2, dPMO2, dFuMO2, and dFuMO1. The four derivatives dNMO1, dPMO3, dQMO, and dTfMO are extended more efficiently, but still significantly less efficiently than dMMO2TP or dNaMTP. Interestingly, the %extension of eight derivatives, including dVMO, dIMO, dClMO, dCNMO, dZMO, dDMO, dPrMO, and dEMO, is slightly greater than that of either dMMO2TP or dNaMTP.
Figure 3.
Values of %incorporation and %extension with 10 s reaction times and 20 µM dMMO2 analog/20 µM dCTP.
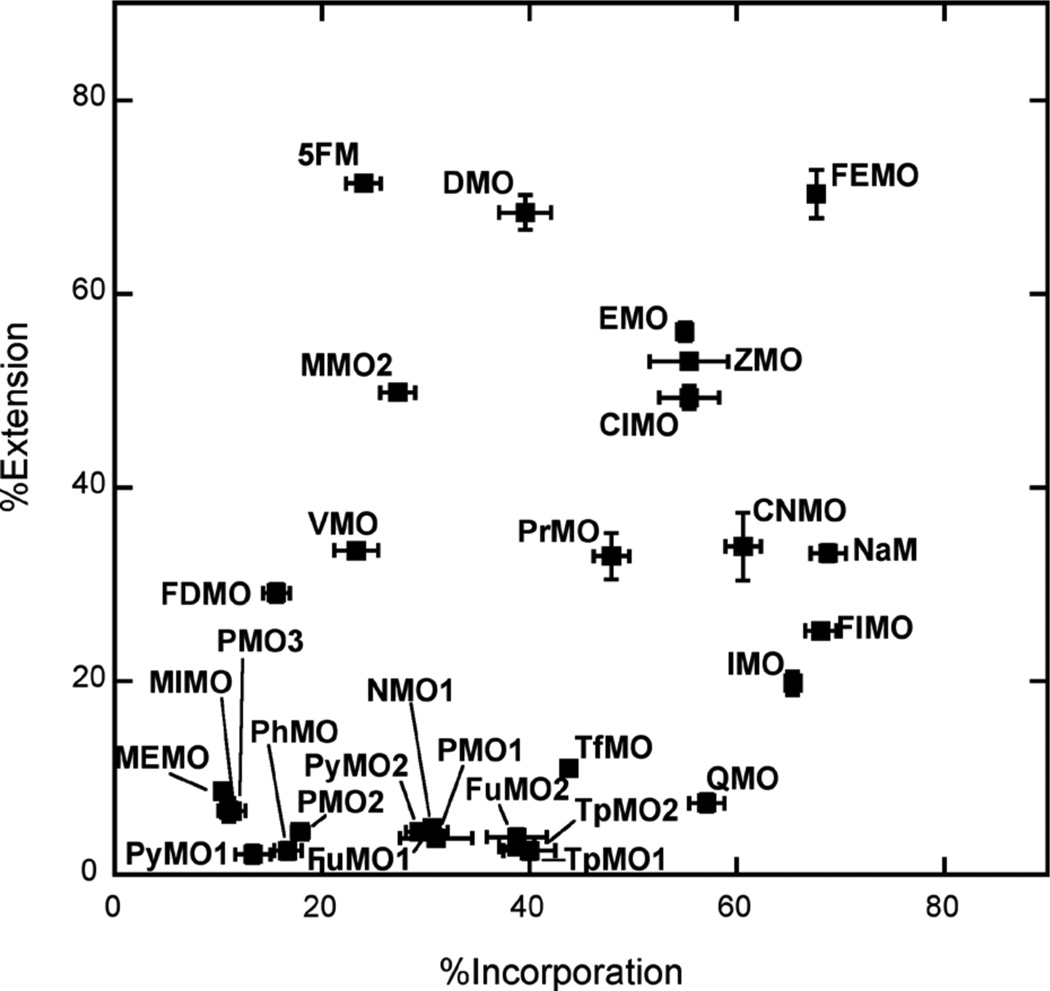
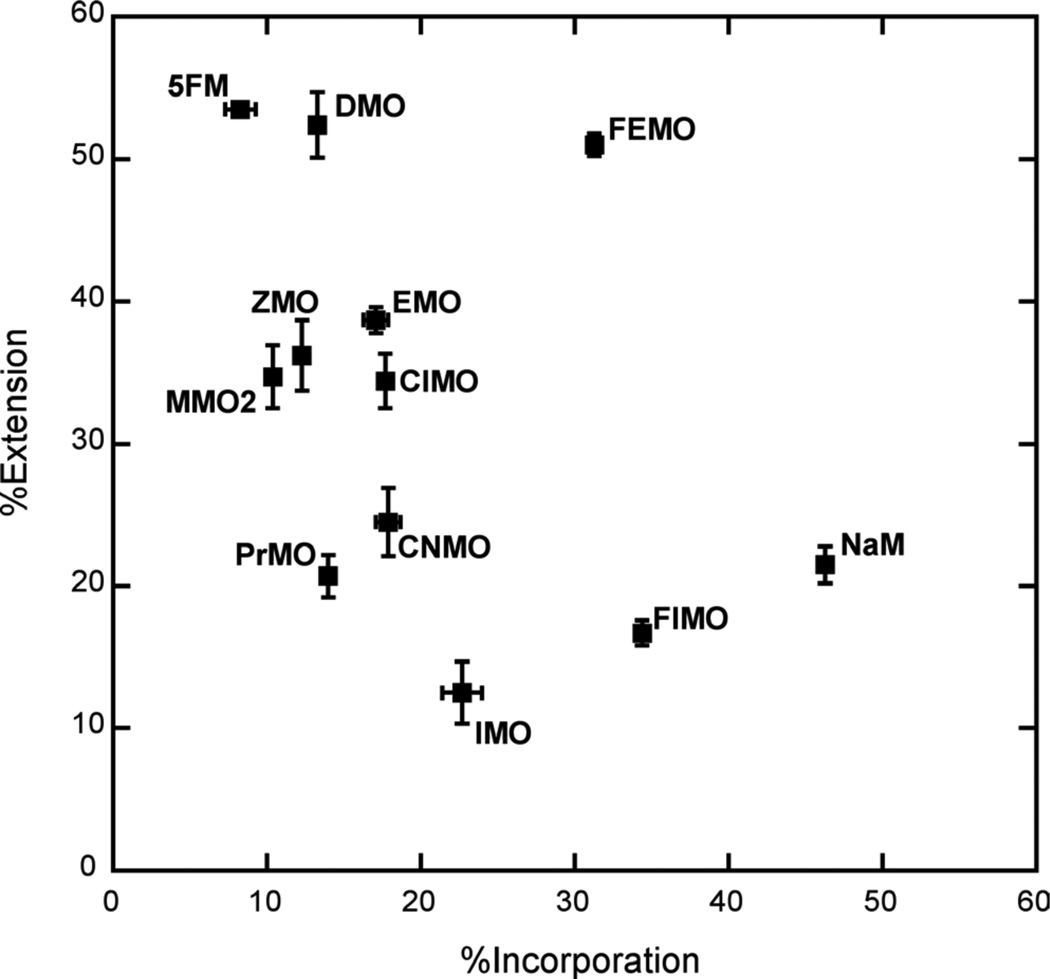
To further differentiate the unnatural triphosphates, we examined DNA synthesis in the presence of lower concentrations of triphosphates (for incorporation, 1 µM for both unnatural triphosphates and dCTP, and for extension, 20 µM unnatural triphosphate and 1 µM dCTP; Figure 4 and Table S2. Under these conditions, the %incorporation values for dMMO2TP and dNaMTP are 27% and 69%, respectively. As expected, a much broader range of incorporation efficiencies were observed with the different analogs (12% to 65%) than at high triphosphate concentrations. Five of the analogs are incorporated less efficiently than dMMO2TP, including dPMO2TP, dPMO3TP, dPyMO1TP, dPhMOTP, and dVMOTP, and sixteen are inserted better, including, dPyMO2TP, dFuMO1TP, dPMO1TP, dNMO1TP, dTpMO1TP, dFuMO2TP, dTpMO2TP, dDMOTP, dTfMOTP, dPrMOTP, dEMOTP, dClMOTP, dZMOTP, dQMOTP, dCNMOTP, and dIMOTP. While dQMOTP incorporation is more efficient than dMMO2TP incorporation, it is less efficient than dNaM incorporation, demonstrating that the added nitrogen substituent is not beneficial. Most interestingly, under these conditions the %incorporation values for dEMOTP, dClMOTP, dZMOTP, dQMOTP, dCNMOTP, and dIMOTP approach that for dNaMTP.
Figure 4.
Values of %incorporation and %extension with 10 s reaction times and with 1 µM dMMO2 analog/1 µM dCTP for the incorporation reactions and 20 µM dMMO2 analog/1 µM dCTP for the extension reactions. Error bars shown are standard deviations determined from three independent experiments.
At the reduced dCTP concentration, the %extension values for dMMO2 or dNaM paired opposite d5SICS are 50% and 33%, respectively. Again, a wide variety of extension efficiencies were observed for the different derivatives (Figure 4), with fourteen significantly to moderately lower than dNaM, including, dPhMO, dPyMO1, dTpMO2, dPyMO2, dTpMO1, dFuMO1, dFuMO2, dPMO2, dPMO1, dNMO1, dPMO3, dQMO, dTfMO, and dIMO, and three similar to dNaM, including dPrMO, dCNMO, and dVMO. Interestingly, dClMO, dZMO, and dEMO paired opposite d5SICS are extended with efficiencies similar to dMMO2, while dDMO is extended more efficiently.
2.3. More stringent pre-steady-state kinetic analysis of the most promising para modified derivatives
Based on the preliminary analysis described above, the seven para substituted derivatives, dPrMO, dEMO, dIMO, dClMO, dCNMO, dZMO, and dDMO, were selected for further analysis under more stringent conditions. We first measured DNA synthesis with shorter reaction times (5 s), and with unnatural triphosphate and dCTP concentrations maintained at 1 µM to characterize unnatural triphosphate incorporation and at 20 µM and 1 µM, respectively, to characterize extension (Figure 5 and Table S3). Under these conditions, the %incorporation values for dMMO2TP and dNaMTP are 17% and 64%, respectively, and the %extension values for the corresponding unnatural primer termini are 30% and 23%, respectively. For each of the derivative triphosphates, the %incorporation is greater than that for dMMO2TP, with dIMOTP exhibiting the highest value of 52%. Three derivatives are extended less efficiently than dNaM, including dIMOTP, dPrMO, and dCNMO; dZMO is extended with an efficiency between dNaM and dMMO2; and dClMO, dEMO, and dDMO are actually extended more efficiently than either dMMO2 or dNaM.
Figure 5.
Values of %incorporation and %extension with 5 s reaction times and 1 µM dMMO2 analog/1 µM dCTP for the incorporation reactions and 20 µM dMMO2 analog/1 µM dCTP for the extension reactions. Error bars shown are standard deviations determined from three independent experiments.
We next examined synthesis with further reduced concentrations of triphosphates (0.2 µM unnatural triphosphate and 0.5 µM dCTP for incorporation, and 20 µM unnatural triphosphate and 0.5 µM dCTP for extension) (Figure 6 and Table S4). For reference, we note that even under these challenging conditions, the %incorporation and %extension of a dC-dG base pair remain above 90%. Under these incorporation conditions, the %incorporation values for dMMO2TP and dNaMTP are 10% and 45%, respectively. Again, the %incorporation for each derivative triphosphate is intermediate between those of dMMO2TP and dNaMTP, with dIMOTP being the greatest. Under these extension conditions, the pairs formed between d5SICS and dNaMTP or dMMO2TP are extended with %extensions of 22% and 35%, respectively. Two derivatives, dIMO and dPrMO, are extended less efficiently than dNaM, while dCNMO and dClMO are inserted with efficiencies intermediate between those of dNaM and dMMO2, and lastly three derivatives, dZMO, dEMO, and most notably dDMO, are extended more efficiently than dMMO2.
Figure 6.
Values of %incorporation and %extension with 10 s reaction times and 0.2 µM dMMO2 analog/0.5 µM dCTP for incorporation reactions and 20 µM dMMO2 analog/0.5 µM dCTP for extension reactions. Error bars shown are standard deviations determined from three independent experiments.
2.4. Design, synthesis, and analysis of five meta, para di-substituted derivatives
Based on the above described data and the potential for generating illuminating SAR data, five para substituted derivatives were selected for further derivatization with a meta fluoro or methoxy substituent, generating dFIMOTP, dMIMOTP, dFEMOTP, dMEMOTP, and dFDMOTP (Figure 2B). Due to its analogous substitution pattern, we also included the previously reported d5FMTP derivative in the current analysis (Figure 1B).
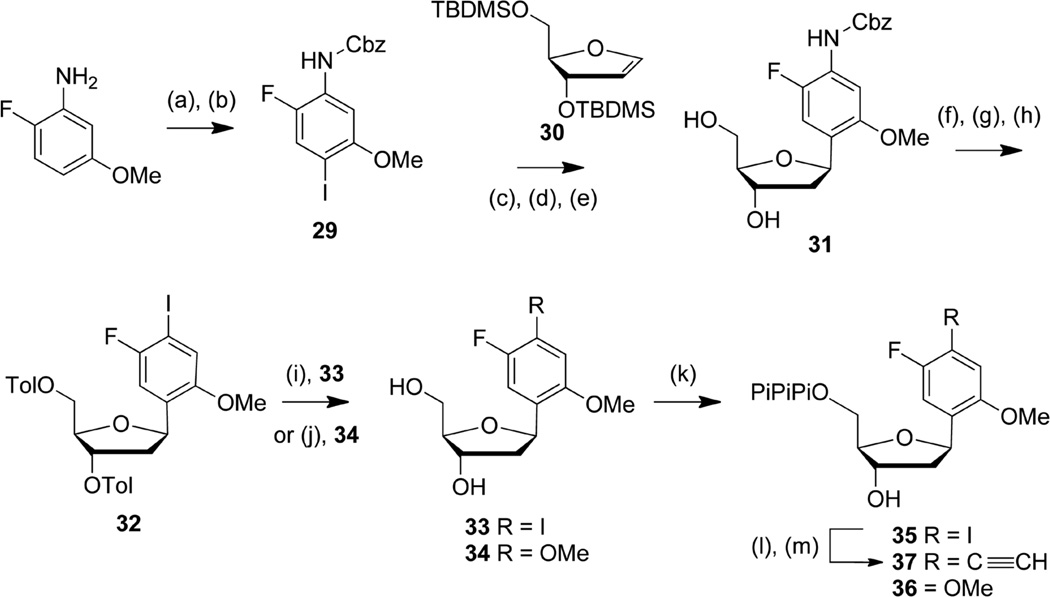
dFIMO, dFDMO, and dFEMO were synthesized as shown in Scheme 4. First, commercially available 2-fluoro-5-methoxyaniline was protected and iodinated in the presence of a silver salt in a non-protic solvent to afford the anisidine 29. The modified nucleoside 31 was then obtained in three steps via Heck coupling of 29 and the 2’-deoxyribose glycal 30, followed by sugar deprotection and selective reduction of the resulting 3’ keto group. Hydroxyl groups were protected with toluyl groups and the Cbz group was removed by hydrogenation. dFIMO (33) was prepared from 31 via a Sandmeyer iodination followed by sugar deprotection. We note that due to the inherent instability of the aryl diazonium intermediate, efficient iodination required the simultaneous addition of sodium nitrite and iodine salts. Analog dFDMO (34) was obtained from 31 via a copper-catalyzed coupling in neat methanol in the presence of 1,10-phenanthroline and cesium carbonate.34 Efficient product formation required 6 h at 110 °C and microwave irradiation, and even under these optimized conditions, a small amount of the reduced 3-fluoroanisole nucleoside byproduct was consistently detected. During the course of the reaction, the toluyl groups were removed, and dFDMO (34) was obtained after silica gel purification. Free nucleosides 33 – 34 were converted to the corresponding triphosphates 35 – 36 and purified as described above. The dFEMO triphosphate (37) was obtained from the dFIMO triphosphate (35) using aqueous copper catalyzed Sonogashira coupling in the presence of triethylsilylacetylene, followed by removal of the triethylsilyl protecting group as described above.
Scheme 4.
Conditions: (a) CBz-Cl, NaHCO3, THF, rt, 1 h, 84%; (b) I2, Ag2SO4, ACN, −20 °C, 40 min, 96%; (c) Pd(OAc)2, AsPh3, nBu3N, DMF, 70 °C, 15 h; (d) TBAF 1 M in THF, 0 °C → rt, 4 h, 54% 2 steps; (e) NaBH(OAc)3, AcOH, CH3CN, −4 °C, 1 h, 91%; (f) Toluyl chloride, pyridine, rt, 3 h, 88%; (g) 10% Pd/C, H2, EtOAc, NEt3, rt, 1 h, 70%; (h) NaNO2, KI, HCl aq 6 M, THF, 0 °C → rt, 2h, 40%; (i) MeONa 30% in MeOH, MeOH:CH2Cl2 8:2, rt, 15 min, 92%; (j) CuI, 1,10-phenanthroline, Cs2CO3, MeOH, microwave−110 °C, 6 h, 46%; (k) proton sponge, POCl3, PO(OMe)3, −15 °C →−10 °C, 3 h then Bu3N, (Bu3NH)2H2P2O7 in DMF, −10 °C → 0 °C, 30 min then TEAB buffer (0.5M), rt, 10 min: 35, 16%; 36, 20%; (l) Pd(OAc)2, 3,3′,3″-phosphinidynetris(benzenesulfonic acid) trisodium salt (TPPTS), CuI, triethylsilylacetylene, Et3N, H2O:ACN 2:1, 65°C, 30 min; (m) NH4OH 30%, rt, 1 h, 50% 2 steps.
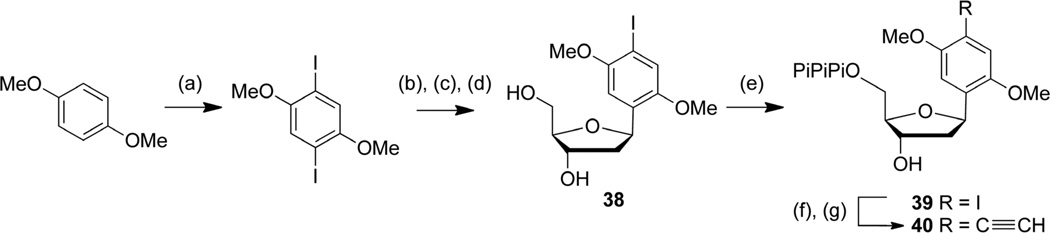
The dMIMO and dMEMO analogs were synthesized from the commercially available 2,4-dimethoxybenzene via diiodination, as previously reported35 (Scheme 5). The modified nucleoside 38 was then obtained in three steps via Heck coupling with the 2’-deoxyribose glycal 30, followed by sugar deprotection and selective reduction. Free nucleoside 38 was then converted to the corresponding triphosphate 39 as described above. The dMEMO triphosphate (40) was obtained from 39 via an aqueous copper catalyzed Sonogashira coupling in presence of triethylsilylacetylene followed by triethylsilyl deprotection.
The incorporation and extension of the resulting six meta, para-disubstituted derivatives were examined under each of the pre-steady-state assay conditions described above (Figures 3 – 6). We found that methoxy substitution in both cases examined (dMIMO and dMEMO) significantly decreases both the %incorporation and %extension, while the effects of fluoro substitution are more variable. In the case of dFDMOTP, the fluoro substituent dramatically reduces both %incorporation and %extension (relative to dDMOTP). With dFIMOTP, we found that the fluoro substituent increases incorporation efficiency, but has little effect on extension (relative to dIMOTP), while with d5FMTP, it has little effect on incorporation but significantly increases extension (relative to dMMO2TP). Finally, with dFEMOTP, the fluoro substituent significantly increases the efficiency of both incorporation and extension. Importantly, under these pre-steady-state conditions, including both unnatural triphosphate incorporation and extension, d5SICS-dFEMO is more efficiently replicated than d5SICS-dNaM.
2.5. PCR analysis
To more fully evaluate replication, DNA containing a dMMO2 analog paired opposite d5SICS was amplified by PCR. Efficiency was characterized by monitoring amplification level and fidelity (defined as unnatural base pair retention per doubling) was determined by amplicon sequencing (Figures S62 – S65). Initial assays were performed with 100 pg of a previously reported DNA template (previously referred to as D6,2,11 where the unnatural base pair is flanked on each side by three randomized natural nucleotides, Supporting Information), 100 µM unnatural triphosphate, and 200 µM of each natural dNTP, a 60 s extension time, and OneTaq polymerase, which is a commercially available mixture of two family A polymerases, exonuclease-negative Taq polymerase and exonuclease-positive DeepVent (Table 1). To facilitate this initial screen, the DNA was subjected to only 14 cycles of amplification, obviating the need for dilutions during the amplification process. Under these conditions, DNA containing dMMO2-d5SICS or d5SICS-dNaM is amplified ~600-fold (which is 2.5-fold lower than the analogous DNA containing a natural dA-dT base pair at the same position) and with fidelities of 97.5% and 99.9%, respectively. DNA containing d5SICS paired opposite one of the ten derivatives dPhMO-dPMO3 is amplified with only modest efficiency and fidelity. DNA containing d5SICS paired opposite any of the remaining derivatives, except dMIMO, dMEMO, and dFDMO, is amplified between 500- and 800-fold, but with variable fidelity. The fidelity with DNA containing dPMO1 is very low, while that with dMIMO, dMEMO, dQMO, d5FM, dDMO, dCNMO, or dPrMO is better, but still less than that with dMMO2. DNA containing dTfMO or dNMO1, or dFDMO is amplified with similar fidelity as that containing dMMO2, while DNA with dVMO, dEMO, dFEMO, dFIMO, dClMO, or dZMO is amplified with higher fidelity than that containing dMMO2. Under these conditions DNA containing d5SICS-dIMO is amplified with a fidelity approaching that of DNA containing d5SICS-dNaM.
Table 1.
PCR amplification and fidelity with OneTaq DNA polymerase.a
| dMMO2 analog | Amplification | Fidelityb |
|---|---|---|
| dPhMO | 3.1×102 | < 90c |
| dPyMO1 | 2.6×102 | < 90c |
| dPyMO2 | 2.2×102 | < 90c |
| dTpMO1 | 0.4×102 | < 90c |
| dTpMO2 | 0.8×102 | < 90c |
| dFuMO1 | 3.3×102 | < 90c |
| dFuMO2 | 1.8×102 | < 90c |
| dPMO2 | 3.0×102 | < 90c |
| dPrMO | 6.0×102 | 97.0 ± 0.3 |
| dEMO | 7.1×102 | 98.48 ± 0.04 |
| dNMO1 | 5.3×102 | 97.41 ± 0.17 |
| dPMO1 | 5.0×102 | 91.57 ± 0.12 |
| dIMO | 6.0×102 | 99.23 ± 0.05 |
| dClMO | 6.3×102 | 98.9 ± 0.3 |
| dCNMO | 6.8×102 | 96.89 ± 0.08 |
| dTfMO | 4.6×102 | 97.2 ± 0.2 |
| dVMO | 5.6×102 | 98.2 ± 0.2 |
| dZMO | 5.4×102 | 98.99 ± 0.07 |
| dQMO | 5.0×102 | 95.7 ± 0.3 |
| dFIMO | 6.3×102 | 98.7 ± 0.2 |
| dMIMO | 3.2×102 | 94.3 ± 0.4 |
| dFEMO | 7.5×102 | 98.6 ± 0.4 |
| dMEMO | 3.7×102 | 95.0 ± 0.8 |
| dFDMO | 4.7×102 | 97.6 ± 0.3 |
| dNaM | 5.4×102 | 99.85 ± 0.13 |
| dMMO2 | 6.2×102 | 97.49 ± 0.01 |
| dDMO | 7.9×102 | 96.6 ± 0.3 |
| d5FM | 6.4×102 | 96.3 ± 0.5 |
| -d | 14×102 | n.d. |
See Materials and Methods for experimental details. Error was determined from three independent experiments.
Fidelity (f) was determined by sequencing (see Materials and Methods) and is defined as the retention of the unnatural base pair per doubling, calculated as R = fn, where R is the retention of the unnatural base pair, n is the number of doublings, calculated as log2(A), and A is the amplification level. Errors for f were propagated from those determined for R.
Unnatural base pair retention was below 50% and the fidelity was thus estimated to be below 90%.
Natural template was amplified without unnatural base pair under identical conditions as a control.
Previously, we reported that an optimal balance between polymerization and 3’–5’ exonuclease activity is important for the high fidelity amplification of DNA containing d5SICS-dNaM.2 To determine if proofreading similarly contributes to the replication of the derivatives explored here, we repeated the amplifications for a subset of the analogs with Taq polymerase alone, under conditions expected to emphasize differences that included both higher amplification (starting with 10 pg of template), and shorter extension times (15 s) (Table 2). Under these conditions, d5SICS-dNaM is amplified with reduced but still reasonable fidelity. However, neither DNA containing dMMO2 nor that containing dPrMO, dNMO1, dTfMO, dVMO, dQMO, dDMO, or d5FM is well amplified. DNA containing dCNMO, dIMO, dClMO, dZMO, or dEMO is better amplified, but still not amplified as well as DNA containing dNaM. However, under these conditions, DNA containing dFEMO or dFIMO is amplified with fidelities approaching that of DNA containing dNaM.
Table 2.
PCR amplification and fidelity with Taq DNA polymerase.a
| dMMO2 analog | Amplification | Fidelity (sequencing)b |
|---|---|---|
| dPrMO | 3.7×103 | < 85c |
| dEMO | 6.0×103 | 93.4 ± 1.4 |
| dNMO1 | 3.4×103 | < 85c |
| dIMO | 4.2×103 | 90.88 ± 0.13 |
| dClMO | 5.9×103 | 91.4 ± 1.1 |
| dCNMO | 5.0×103 | 88 ± 4 |
| dTfMO | 3.0×103 | < 85c |
| dVMO | 2.9×103 | < 85c |
| dZMO | 4.2×103 | 91.69 ± 0.12 |
| dQMO | 2.9×103 | < 85c |
| dFIMO | 4.4×103 | 96.4 ± 0.9 |
| dFEMO | 6.9×103 | 95.8 ± 0.5 |
| dFDMO | 2.2×103 | < 85c |
| dNaM | 3.7×103 | 98.11 ± 0.03 |
| dMMO2 | 2.9×103 | < 85c |
| dDMO | 1.1×103 | < 85c |
| d5FM | 3.2×103 | < 85c |
| -d | 29×103 | n.d. |
See Materials and Methods for experimental details. Error was determined from three independent experiments.
Fidelity (f) was determined by sequencing (see Materials and Methods) and is defined as the retention of the unnatural base pair per doubling, calculated as R = fn, where R is the retention of the unnatural base pair, n is the number of doublings, calculated as log2(A), and A is the amplification level. Errors for f were propagated from those determined for R.
Unnatural base pair retention was below 50% and the fidelity was thus estimated to be below 85%.
Natural template was amplified without unnatural base pair under identical conditions as a control.
With the data supporting the importance of exonuclease activity, we returned to OneTaq-mediated amplification and examined the 1013-fold amplification of a subset of the analogs (Table 3). Under these conditions, DNA containing dNMO1 or dVMO paired opposite d5SICS is not replicated well, DNA containing dCNMO, dClMO, dIMO, dZMO, or dEMO, is better replicated, and DNA containing d5SICS-dFIMO or d5SICS-dFEMO is replicated with a fidelity approaching that of d5SICS-dNaM.
Table 3.
PCR amplification and fidelity with OneTaq DNA polymerase and high amplification.a
| dMMO2 analog | Amplification | Fidelity (sequencing)b |
|---|---|---|
| dEMO | 1.4×1013 | 98.55 ± 0.16 |
| dNMO1 | 1.5×1013 | < 96c |
| dIMO | 1.3×1013 | 98.3 ± 0.4 |
| dClMO | 1.5×1013 | 98.2 ± 0.3 |
| dCNMO | 1.5×1013 | 97.4 ± 0.3 |
| dVMO | 1.5×1013 | < 96c |
| dZMO | 1.5×1013 | 98.4 ± 0.3 |
| dFIMO | 1.1×1013 | 98.74 ± 0.05 |
| dFEMO | 1.5×1013 | 98.77 ± 0.08 |
| dNaM | 0.9×1013 | 99.92 ± 0.02 |
| -d | 2.7×1013 | n.d. |
See Materials and Methods for experimental details. Error was determined from three independent experiments.
Fidelity (f) was determined by sequencing (see Materials and Methods) and is defined as the retention of the unnatural base pair per doubling, calculated as R = fn, where R is the retention of the unnatural base pair, n is the number of doublings, calculated as log2(A), and A is the amplification level. Errors for f were propagated from those determined for R.
Unnatural base pair retention was below 50% and the fidelity was thus estimated to be below 96%.
Natural template was amplified without unnatural base pair under identical conditions as a control.
In the OneTaq system, DNA is mainly replicated by Taq (a family A polymerase36,37), while DeepVent (a family B polymerase36,37) is mainly responsible for proofreading. To explore replication by a family B polymerase alone, PCR amplifications were performed with KOD polymerase and a select set of the analogs (Table 4). KOD clearly replicates d5SICS-dNaM with lower fidelity than either OneTaq or Taq, and replicates the pairs with dIMO and dFIMO with even lower fidelity. However, DNA containing dZMO, dClMO, dEMO, dCNMO, or especially dFEMO paired opposite d5SICS is replicated better than with dNaM paired opposite d5SICS. The d5SICS-dFEMO pair is especially noteworthy, as unlike the other pairs, its replication with the family B polymerase is virtually as efficient and high fidelity as replication with the A family polymerases.0
Table 4.
PCR amplification and fidelity with KOD DNA polymerase.a
| dMMO2 analog | Amplification | Fidelity (sequencing)b |
|---|---|---|
| dEMO | 2.2×102 | 93.8 ± 0.3 |
| dIMO | 1.2×102 | < 85c |
| dClMO | 2.6×102 | 93.10 ± 0.01 |
| dCNMO | 3.0×102 | 95.48 ± 0.07 |
| dZMO | 2.1×102 | 92.5 ± 0.5 |
| dFIMO | 1.3×102 | 87.1 ± 0.8 |
| dFEMO | 4.6×102 | 97.4 ± 0.4 |
| dNaM | 1.7×102 | 91.7 ± 0.2 |
| -d | 52×102 | n.d. |
See Materials and Methods for experimental details. Error was determined from three independent experiments.
Fidelity (f) was determined by sequencing (see Materials and Methods) and is defined as the retention of the unnatural base pair per doubling, calculated as R = fn, where R is the retention of the unnatural base pair, n is the number of doublings, calculated as log2(A), and A is the amplification level. Errors for f were propagated from those determined for R.
Unnatural base pair retention was below 50% and the fidelity was thus estimated to be below 85%.
Natural template was amplified without unnatural base pair under identical conditions as a control.
3. Discussion
Following the identification of d5SICS-dMMO2 from a screen of 3600 candidate hydrophobic unnatural base pairs and an initial round of optimization,8 we focused our optimization efforts on the para position of dMMO2. These efforts eventually yielded d5SICS-dDMO17 and d5SICS-dNaM,9,10 with replication of the latter proceeding with the greatest efficiency and highest fidelity, sufficiently so that it is functionally equivalent to a natural base pair for PCR applications.2 However, optimization efforts also suggested that meta substituents of the dMMO2 scaffold, such as fluorine, could optimize replication.10,14 Nonetheless, it remained to be determined just which substituents were optimal, whether substituents at both positions would interact additively or synergistically, and whether substituents might be identified that result in a dMMO2 derivative that when paired with d5SICS is replicated as efficiently as d5SICS-dNaM. To address these questions, we synthesized a diverse set of para-derivatized dMMO2TP analogs that explore a wide variety of structural and physicochemical variations, and we developed pre-steady state and PCR assays for their rapid characterization. Following this initial optimization, several derivatized nucleotides were selected based on their optimized replication or their promise to provide illuminating SAR data for a second phase of diversification via a meta methoxy or fluoro substituent.
3.1 SAR analysis
One of the goals of the present study was to collect SAR data for both the incorporation of a dMMO2TP analog opposite d5SICS, and the extension of the resulting base pair. In previous efforts to optimize dMMO2, we explored several bicyclic derivatives, such as dPMO1, which as a triphosphate under steady-state conditions is inserted opposite d5SICS slightly better than dMMO2TP.9 Large differences in %incorporation were observed with the bicyclic derivatives examined in the current study, with the best inserted being the quinolone derivative, dQMOTP, followed by the thiophene analogs dTpMO1TP and dTpMO2TP, and the furan and pyrrole derivatives, dFuMO1TP, dFuMO2TP, and dPyMO2TP. Clearly heteroatom substitution can have a significant impact, for example, dPhMOTP and dPyMO1TP are inserted much less efficiently than dPyMO2. While large variations were observed in the rates of insertion of the bicyclic derivatives opposite d5SICS, all of them effectively act as chain terminators, due to very poor continued primer extension. This likely results from increased interstrand intercalation between the nucleobases, which may favor triphosphate insertion but mandates deintercalation for continued primer extension.3,10 Thus, this class of derivatives does not appear promising.
To explore the effects of increased aromatic surface area in the absence of a bicyclic nucleobase scaffold, para propynyl, ethynyl, and vinyl substituents were explored with dPrMO, dEMO, and dVMO, respectively. In addition, the effects of altered structure and electronics were explored with dZMO and dCNMO. The vinyl substituent was deleterious for both the incorporation and extension steps of replication. In contrast, all of the remaining substituents significantly increased the efficiency of incorporation, although the increase was less pronounced at lower triphosphate concentrations. Thus, the data suggest that increased aromatic surface area and/or hydrophobicity, possibly subject to certain steric constraints, favor efficient incorporation, and that relative to dNaM, this results from an increase in the affinity of the polymerase for the triphosphate. Relative to dMMO2, the ethinyl and azide substituents have little effect on extension, and the propynyl and cyano groups reduce efficiency, but apparently not due to effects on the binding of dCTP. These effects may result from a combination of steric and electronic factors, both between the pairing nucleobases and with the polymerase. Whatever the origins of the observed effects, with the exception of the vinyl group, these aliphatic and heteroatom-modified para substituents appear to be promising for the optimization of unnatural triphosphate incorporation.
The strongly electron withdrawing para nitro substituent of dNMO1TP had only a small effect on the efficiency of triphosphate incorporation opposite d5SICS, but dramatically reduced extension efficiency of the resulting base pair. In contrast, the weaker electron withdrawing para halogen substituents, especially the iodo substituent, significantly increased incorporation efficiency. In fact, at all but the lowest triphosphate concentrations examined, dIMO is inserted opposite d5SICS almost as efficiently as dNaM. However, relative to dNaM, the effects were somewhat attenuated at the lowest triphosphate concentrations (0.2 µM), again suggesting that the halogenated derivatives bind with an elevated KD. The chloro substituent had little effect on extension, while the iodo decreased it somewhat. As with the aliphatic and heteroatom-derivatized analogs discussed above, halogens appear to be promising para substituents for the optimization of triphosphate incorporation.
In both contexts examined, (dMIMO and dMEMO), a meta methoxy substituent significantly decreased the efficiency of both incorporation and extension. The effects were somewhat smaller at low triphosphate concentrations, suggesting that the methoxy substituents increase the affinity with which both triphosphates bind. In addition, the effects were largely independent of the para substituent. Because any mesomeric effects should increase the electron density of the ortho methoxy group, which at least for extension should be favorable,8,12 the data suggest that the effects may result from forced desolvation of the meta substituent. Regardless, the meta-methoxy substituent is deleterious and will not be included in future optimization efforts.
Very different effects were observed for a meta fluorine in the four contexts examined (dFIMO, dFEMO, d5FM, and dFDMO). In the case of dFDMO (relative to dDMO), the efficiency of both incorporation and extension are reduced, at least in part due to reduced natural and unnatural triphosphate binding. For d5FM (relative to dMMO2), the efficiency of extension is selectively increased, at least in part due to an increased affinity for natural triphosphate binding. For dFIMO (relative to dIMO), the efficiency of incorporation and extension are marginally increased. Finally, for dFEMO (relative to dEMO) the efficiency of both incorporation and extension are increased significantly, at least in part due to increased triphosphate binding. Thus, with an adjacent para methoxy substituent, the meta fluorine substituent is deleterious, but when adjacent to an iodo, methyl, or ethynyl substituent, the meta fluorine substituent is neutral or beneficial. Clearly the effects are not simply related to the size of the substituent. The effects may be rooted in more subtle steric factors or in the unique electron donating ability of the methoxy group. Subtle and difficult to rationalize effects of nucleobase modification have been observed with other analogs.38,39 Whatever the detailed origins of the effects, the data clearly reveal that depending on the nature of the para substituent, a meta fluoro substituent may be distinctly beneficial, especially for the optimization of extension.
The data reveal that several of the para-derivatized dMMO2 derivatives form pairs with d5SICS that are PCR amplified with reasonable efficiency and fidelity. While the effects of meta methoxy substitution were not fully evaluated due to their poor performance, it is clear that just as with the pre-steady state assays, the meta fluoro-substituents of dFIMO and dFEMO improve amplification. When more fully comparing the kinetic and PCR data, an absolute correlation is not expected as the former reflects only one strand context of DNA synthesis. Nonetheless, previous work suggests that the effects of substituents in the context characterized (i.e. incorporation of dMMO2TP analogs opposite d5SICS in the template) tend to be larger than in the opposite context (i.e. with dMMO2 analogs in the template),8–11 and thus strong correlations might persist. This is not the case with amplification efficiency. All of the duplexes examined were amplified with an efficiency within 2-fold of one another, and within ~2–3-fold, 4–8-fold, or 10–40-fold of that containing a natural base pair with OneTaq, Taq, or KOD, respectively. This may result, at least in part, from the relatively long extension times employed (1 min for the OneTaq- and KOD-mediated amplifications). However, there are more significant differences in fidelity. The exact values of amplification fidelity in the cases where it is low are not accurate (due to the experimental challenges of determining the level of unnatural base pair retention when it is very low), and thus we limited our analysis to only those analogs that were generally replicated with higher fidelity and used the data from the higher OneTaq amplification. Interestingly, a clear correlation between %incorporation and fidelity is observed, with correlation coefficients of 0.79, 0.82, 0.51, and 0.65, for the data from Tables 1–4, respectively (Figure S79). Such a correlation is clearly expected in the limit of low or no proofreading activity (3’–5’ exonuclease activity), which suggests that exonucleolytic removal of an unnatural nucleotide at a primer terminus may be inefficient. This conclusion is consistent with the reduced fidelities observed during amplification with Taq alone, and with our previous demonstration that fidelity increased with increases in the ratio of polymerase proofreading to extension activity.2 While this model requires further investigation, the observed correlation suggests that further efforts toward optimization of unnatural base pair replication should focus on improving the rates of triphosphate incorporation.
In agreement with previous results,2 OneTaq appears to be optimal for the replication of DNA containing the unnatural base pairs. While KOD is generally less optimal, with this B family polymerase d5SICS-dFEMO is actually replicated better than d5SICS-dNaM. This may result from the unique mechanism for binding and delivering triphosphates to the KOD active site that is based on electrostatic interactions between the negatively charged triphosphate and basic residues of the polymerase fingers domain.40 Moreover, KOD is highly processive, suggesting that it might have an inherently high affinity for DNA and/or triphosphates,41 possibly allowing some perturbations to be tolerated. However, the other analogs are not as well replicated as d5SICS-dFEMO, suggesting that unique aspects of its structure or physiochemical properties are especially compatible with KOD. Further exploration of the relative replicability of d5SICS-dNaM and d5SICS-dFEMO with different polymerases should not only illuminate the differences in the potential substrate repertoires of different polymerases, but should also help to define the determinants of general replication and facilitate further optimization of the unnatural base pair.
3.2 Progress toward expansion of the genetic alphabet
A primary goal of the present study was to determine if the dMMO2 scaffold could be optimized as a partner for d5SICS. Clearly, this goal was met by the identification of d5SICS-dEMO, d5SICS-dFIMO, and d5SICS-dFEMO, which are significantly better replicated than is d5SICS-dMMO2. In addition, we note that the PCR experiments appear to suggest that the replication of the analogs examined here is not strongly sequence-dependent. This is based on an inspection of the sequencing traces before and after amplification (the three natural nucleotides flanking the unnatural base in the templates employed pair were randomized). However, this data is qualitative and the identification of any replication biases imposed by the unnatural base pairs must await detailed characterization. Future efforts will also focus on the characterization of mutation induced by insertion of an unnatural triphosphate opposite a natural nucleotide. In addition, based on the kinetic and PCR data, it appears that several mono substituted para-derivatives not further explored by derivatization here, including dZMO, dCNMO, and dClMO, merit further exploration as scaffolds, as well. From a conceptual perspective, especially when combined with other reported hydrophobic unnatural base pairs that are well replicated,42,43 the optimizability and apparent robustness of the dMMO2 scaffold attests to the generality of hydrophobic and packing interactions as forces that are capable of controlling the efficient and high fidelity replication of DNA.
An immediate use for replicable unnatural base pairs is the site-specific labeling of DNA within a PCR-amplifiable format for in vitro applications ranging from basic biophysics to SELEX and materials fabrication. The different dMMO2 analogs bear a variety of functional groups that are interesting for such applications. For example, F19 labeling of dFEMO provides an NMR handle for characterization, the azido and cyano groups of dZMO and dCNMO, respectively provide IR probes with unique absorptions,44,45 the iodo group of dIMO provides a handle for bioconjugation via cross-coupling,46 and the azido and alkyne substituents of dZMO, dEMO, and dFEMO provide handles for bioconjugation via click chemistry.47,48 Efforts toward such applications are currently in progress.
A long term goal of the effort to develop unnatural base pairs is the expansion of the genetic alphabet in vivo and the creation of a semi-synthetic organism with increased potential for information storage and retrieval. However, in addition to efficient and high fidelity replication, the demands of the in vivo environment include additional factors, such as substrate uptake, localization within the cell, and off target protein binding. These challenges are similar to those faced in drug discovery, as drug candidates must possess, in addition to suitable biochemical properties, favorable pharmacokinetic properties. Such properties are scaffold-dependent but often unpredictable, and thus similar to efforts to develop any drug, efforts to develop an unnatural base pair that is replicable in vivo will be bolstered by the availability of multiple lead compounds based on different scaffolds. The diversification of the dMMO2 scaffold into several new scaffolds that pair well with d5SICS is in this regard of particular importance.
Materials and Methods
General Synthetic Methods
Synthetic details and compound characterization are provided in the Supporting Information.
Gel-Based incorporation/extension assay
Primer oligonucleotides (Integrated DNA Technologies) were 5’-radiolabeled with T4 polynucleotide kinase (New England Biolabs; Ipswich, MA) and [γ-32P]-ATP (Perkin-Elmer) and annealed to template oligonucleotides10 by heating to 95 °C followed by slow cooling to room temperature. Reactions were initiated by adding a solution of 2× dNTP and dXTP solution (5 µL) to a solution containing polymerase (73.53 nM) and primer:template (40 nM) in 5 µL Klenow reaction buffer (50 mM Tris-HCl, pH 7.5, 10 mM DTT and 50 µg/mL acetylated BSA). After incubation at 25 °C for 5–10 s, reactions were quenched with 20 µL of loading dye (95% formamide, 20 mM EDTA, and sufficient amounts of bromophenol blue and xylene cyanol). Reaction products were resolved by 15% polyacrylamide gel electrophoresis, and gel band intensities corresponding to the extended and unextended primers were quantified by phosphorimaging (Storm Imager, Molecular Dynamics) and Quantity One (Bio-Rad) software. Except for the most permissive conditions, the reported values are the average and standard deviation of three independent determinations (see also Tables S1 – S4).
PCR assay
The synthesis of the DNA duplex used as a template was described previously, where it was referred to as template D6.11 The sequence of the d5SICS template strand is 5’-d-GAAATTAATACGACTCACTA TAGGGTTAAG CTTAACTTTA AGAAGGAGAT TTACTATGGG TCCCGNNN5SICSN NNCGTCTGGT GAATTCCAAG TGCTAGCGCA TGTAATAACC CGGGTCATAG CTGTTTCCTGTGTG-3’, where N is randomized nucleotide and primer regions are underlined. OneTaq and Taq enzymes were obtained from New England Biolabs and KOD Hot Start DNA Polymerase was obtained from Novagen/EMD Millipore Biosciences (Billerica, MA). PCR amplifications were performed in a total volume of 25 µL and with conditions specific for each assay as described in Table S5. After amplification, a 5 µL aliquot was analyzed on a 2% agarose gel to confirm amplicon size (134 bp). The remaining solution was purified by spin-column (DNA Clean and Concentrator-5; Zymo Research, Irvine, CA), quantified by fluorescent dye binding (Quant-iT dsDNA HS Assay kit, Invitrogen), and sequenced on a 3730 DNA Analyzer (Applied Biosystems). Fidelity was determined as the average %retention of the unnatural base pair per doubling as described in the Supporting Information.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (GM 60005 to F.E.R).
Footnotes
Supporting Information Available. Synthetic methods and compound characterization, pre-steady state kinetic assay and data, PCR assay and sequencing data, calculation of PCR fidelity, and analysis of correlation between incorporation efficiency and PCR fidelity. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 1999;121:11585–11586. [Google Scholar]
- 2.Malyshev DA, Dhami K, Quach HT, Lavergne T, Ordoukhanian P, Torkamani A, Romesberg FE. Proc. Natl. Acad. Sci. USA. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A. Nat. Chem. Biol. 2012;8:612–614. doi: 10.1038/nchembio.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Chen F, Alvarado JB, Benner SA. J. Am. Chem. Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaul C, Muller M, Wagner M, Schneider S, Carell T. Nat. Chem. 2011;3:794–800. doi: 10.1038/nchem.1117. [DOI] [PubMed] [Google Scholar]
- 7.Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2009;37:e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavergne T, Malyshev DA, Romesberg FE. Chem. Eur. J. 2012;18:1231–1239. doi: 10.1002/chem.201102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2009;131:14620–14621. doi: 10.1021/ja906186f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda S, Leconte AM, Romesberg FE. J. Am. Chem. Soc. 2007;129:5551–5557. doi: 10.1021/ja068282b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu C, Henry AA, Romesberg FE, Schultz PG. Angew. Chem. Int. Ed. 2002;41:3841–3844. doi: 10.1002/1521-3773(20021018)41:20<3841::AID-ANIE3841>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Seo YJ, Romesberg FE. ChemBioChem. 2009;10:2394–2400. doi: 10.1002/cbic.200900413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang GT, Leconte AM, Romesberg FE. ChemBioChem. 2007;8:1606–1611. doi: 10.1002/cbic.200700308. [DOI] [PubMed] [Google Scholar]
- 16.Hari Y, Hwang GT, Leconte AM, Joubert N, Hocek M, Romesberg FE. ChemBioChem. 2008;9:2796–2799. doi: 10.1002/cbic.200800577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE. Chem. Eur. J. 2010;16:12650–12659. doi: 10.1002/chem.201000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stambasky J, Hocek M, Kocovsky P. Chem. Rev. 2009;109:6729–6764. doi: 10.1021/cr9002165. [DOI] [PubMed] [Google Scholar]
- 19.Tokuyama H, Sato M, Ueda T, Fukuyama T. Heterocycles. 2001;54:105–108. [Google Scholar]
- 20.Ludwig J, Eckstein F. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 21.Knauber T, Arikan F, Roschenthaler GV, Goossen LJ. Chem. Eur. J. 2011;17:2689–2697. doi: 10.1002/chem.201002749. [DOI] [PubMed] [Google Scholar]
- 22.Molander GA, Brown AR. J. Org. Chem. 2006;71:9681–9686. doi: 10.1021/jo0617013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumann H, Kaufmann J, Schmalz HG, Bottcher A, Gotov B. Synlett. 2003:1783–1788. [Google Scholar]
- 24.Alacid E, Najera C. J. Org. Chem. 2008;73:2315–2322. doi: 10.1021/jo702570q. [DOI] [PubMed] [Google Scholar]
- 25.Farina V, Kapadia S, Krishnan B, Wang CJ, Liebeskind LS. J. Org. Chem. 1994;59:5905–5911. [Google Scholar]
- 26.Velmathi S, Leadbeater NE. Tetrahedron Lett. 2008;49:4693–4694. [Google Scholar]
- 27.Hocek M, Pohl R, Klepetáová B. Eur. J. Org. Chem. 2005;2005:4525–4528. [Google Scholar]
- 28.Joubert N, Urban M, Pohl R, Hocek M. Synthesis. 2008:1918–1932. [Google Scholar]
- 29.Urban M, Pohl R, Klepetáová B, Hocek M. J. Org. Chem. 2006;71:7322–7328. doi: 10.1021/jo061080d. [DOI] [PubMed] [Google Scholar]
- 30.Joubert N, Pohl R, Klepetáová B, Hocek M. J. Org. Chem. 2007;72:6797–6805. doi: 10.1021/jo0709504. [DOI] [PubMed] [Google Scholar]
- 31.Stefko M, Slavetinska L, Klepetáová B, Hocek M. J. Org. Chem. 2011;76:6619–6635. doi: 10.1021/jo200949c. [DOI] [PubMed] [Google Scholar]
- 32.Hocek M, Fojta M. Org. Biomol. Chem. 2008;6:2233–2241. doi: 10.1039/b803664k. [DOI] [PubMed] [Google Scholar]
- 33.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 34.Wolter M, Nordmann G, Job GE, Buchwald SL. Org. Lett. 2002;4:973–976. doi: 10.1021/ol025548k. [DOI] [PubMed] [Google Scholar]
- 35.Yi CY, Blum C, Lehmann M, Keller S, Liu SX, Frei G, Neels A, Hauser J, Schurch S, Decurtins S. J. Org. Chem. 2010;75:3350–3357. doi: 10.1021/jo100323s. [DOI] [PubMed] [Google Scholar]
- 36.Filee J, Forterre P, Sen-Lin T, Laurent J. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 37.Kornberg A, Baker TA. DNA Replication, Univ Science Books. 2005 [Google Scholar]
- 38.Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 39.Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Inoue T, Kai Y. J. Mol. Biol. 2001;306:469–477. doi: 10.1006/jmbi.2000.4403. [DOI] [PubMed] [Google Scholar]
- 41.Takagi M, Nishioka M, Kakihara H, Kitabayashi M, Inoue H, Kawakami B, Oka M, Imanaka T. Appl. Environ. Microbiol. 1997;63:4504–4510. doi: 10.1128/aem.63.11.4504-4510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Lee I, Zhou X, Berdis AJ. J. Am. Chem. Soc. 2006;128:143–149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Motea E, Lee I, Berdis AJ. Biochemistry. 2010;49:3009–3023. doi: 10.1021/bi901523y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmermann J, Thielges MC, Seo YJ, Dawson PE, Romesberg FE. Angew. Chem. Int. Ed. 2011;50:8333–8337. doi: 10.1002/anie.201101016. [DOI] [PubMed] [Google Scholar]
- 45.Oh KI, Lee JH, Joo C, Han H, Cho MJ. Phys. Chem. B. 2008;112:10352–10357. doi: 10.1021/jp801558k. [DOI] [PubMed] [Google Scholar]
- 46.Omumi A, Beach DG, Baker M, Gabryelski W, Manderville RA. J. Am. Chem. Soc. 2010;133:42–50. doi: 10.1021/ja106158b. [DOI] [PubMed] [Google Scholar]
- 47.Weisbrod SH, Marx A. Chem. Commun. 2008:5675–5685. doi: 10.1039/b809528k. [DOI] [PubMed] [Google Scholar]
- 48.Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. Int. Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.