Abstract
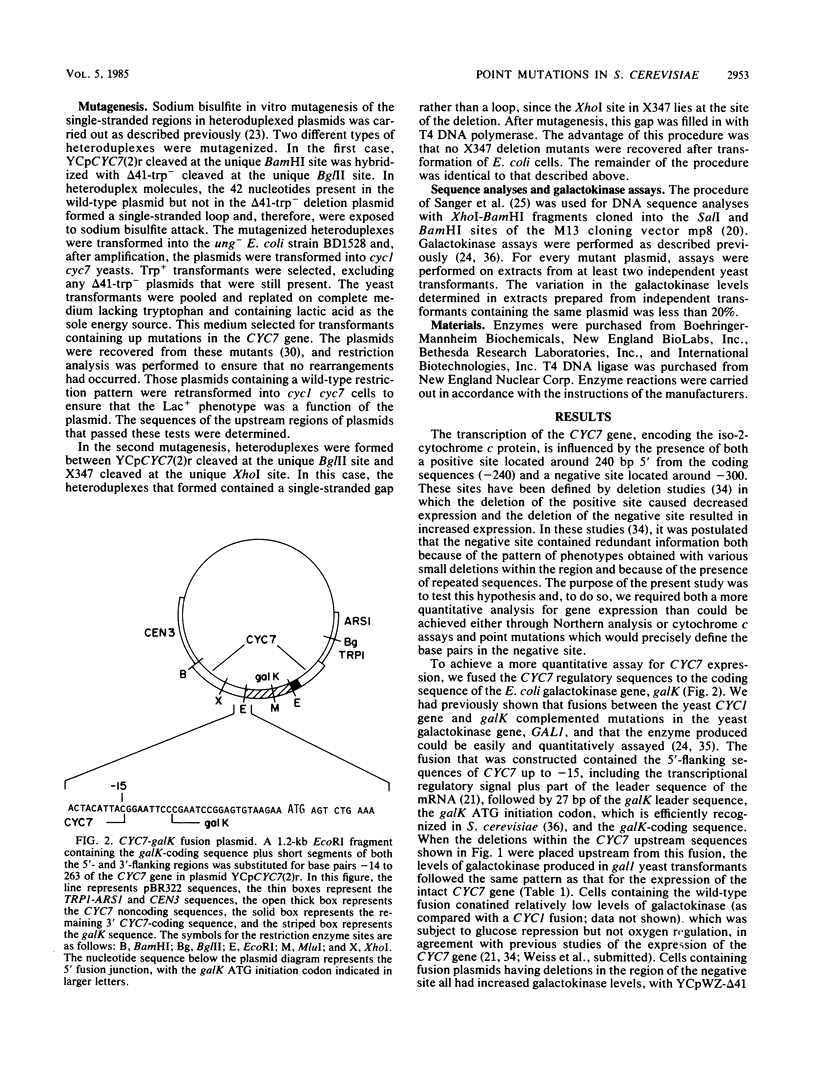
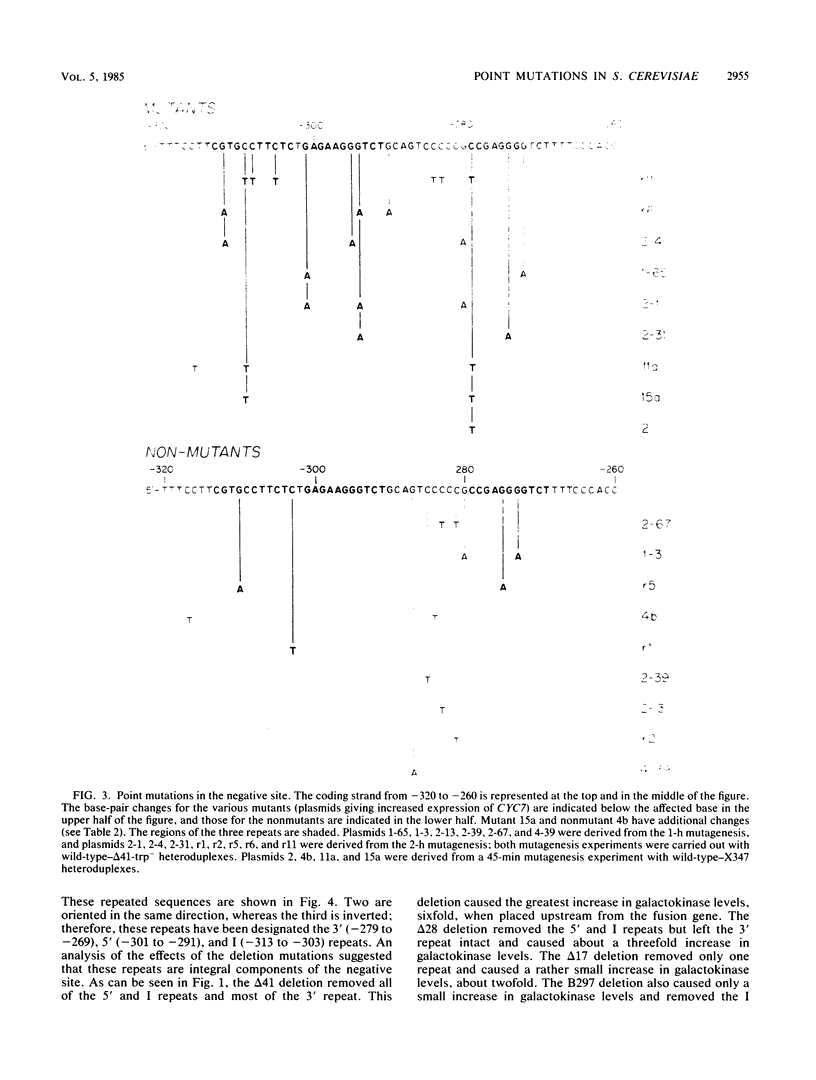
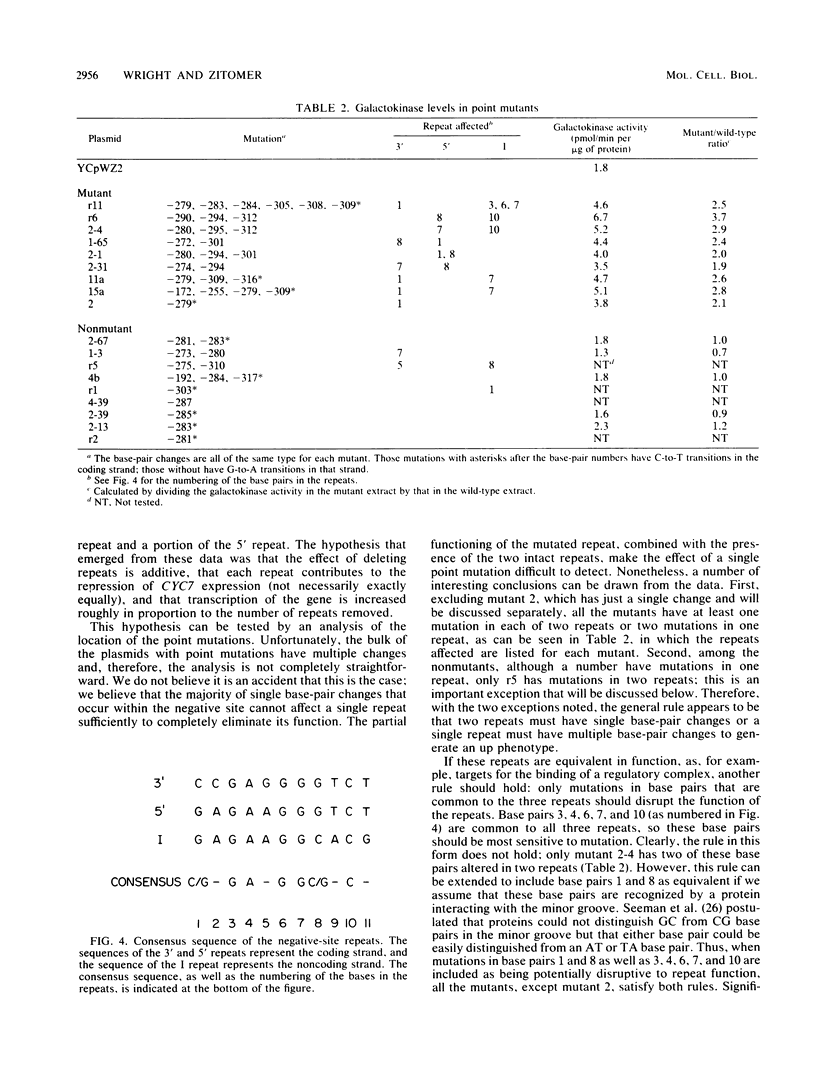
The transcription of the CYC7 gene of Saccharomyces cerevisiae, encoding the iso-2-cytochrome c protein, is controlled by two upstream regulatory elements, a positive element and a negative element. The nature of the DNA sequences in the negative element were investigated in a two-part approach. The first involved the construction of a CYC7-galK fusion gene which placed the coding sequence of the Escherichia coli galactokinase gene under the regulation of the CYC7 upstream sequences. This fusion allowed the quantitation by galactokinase enzyme assays of the effects on gene expression of a variety of previously isolated deletion mutations within the negative site. The results suggested that the negative site contained three related sequences. This hypothesis was tested in the second part of these studies, the selection of point mutations within the region of the negative site which led to increased CYC7 expression. Point mutations were introduced by a technique which induced mutations within a localized region at high efficiency. All but one of the mutations involved more than a single base-pair change. The mutations followed the pattern that multiple base-pair changes occurred in one repeat or single base-pair changes occurred in two repeats, with the exception of one mutant, which had a single base-pair change in one repeat. This pattern of mutations and the base pairs that were altered strongly supported the hypothesis that the repeats are integral elements of the negative site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Clavilier L., Péré-Aubert G., Somlo M., Slonimski P. P. Réseau d'interactions entre des génes non liés : régulation synergique ou antagoniste de la synthèse de l'iso-1-cytochrome c, de l'iso-2-cytochrome c et du cytochrome b2. Biochimie. 1976;58(1-2):155–172. doi: 10.1016/s0300-9084(76)80366-5. [DOI] [PubMed] [Google Scholar]
- Clavilier L., Péré G., Slonimski P. P. Mise en évidence de plusieurs loci indépendants impliqués dans la synthèse de l'iso-2-cytochrome c chez la levure. Mol Gen Genet. 1969;104(2):195–218. doi: 10.1007/BF00272801. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Downie J. A., Stewart J. W., Brockman N., Schweingruber A. M., Sherman F. Structural gene for yeast iso-2-cytochrome c. J Mol Biol. 1977 Jun 25;113(2):369–384. doi: 10.1016/0022-2836(77)90147-4. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Weiss J. L., Walthall D. A., Zitomer R. S. Modulator sequences mediate oxygen regulation of CYC1 and a neighboring gene in yeast. Proc Natl Acad Sci U S A. 1983 Jan;80(1):151–155. doi: 10.1073/pnas.80.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Zitomer R. S. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6129–6133. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A., Yost H. J., Casadaban M. J. Role of an upstream regulatory element in leucine repression of the Saccharomyces cerevisiae leu2 gene. Nature. 1984 Feb 23;307(5953):740–742. doi: 10.1038/307740b0. [DOI] [PubMed] [Google Scholar]
- McKnight G. L., Cardillo T. S., Sherman F. An extensive deletion causing overproduction of yeast iso-2-cytochrome c. Cell. 1981 Aug;25(2):409–419. doi: 10.1016/0092-8674(81)90059-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Boss J. M., McAndrew S. J., Marr L., Walthall D. A., Zitomer R. S. The molecular characterization of three transcriptional mutations in the yeast iso-2-cytochrome c gene. J Biol Chem. 1982 Jul 10;257(13):7756–7761. [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Nathans D. Local mutagenesis within deletion loops of DNA heteroduplexes. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7214–7217. doi: 10.1073/pnas.79.23.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B. C., Zitomer R. S., Schümperli D., Rosenberg M. The expression in yeast of the Escherichia coli galK gene on CYC1::galK fusion plasmids. Gene. 1983 Nov;25(2-3):249–262. doi: 10.1016/0378-1119(83)90229-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Margoliash E., Parker J., Campbell W. The structural gene for yeast cytochrome C. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1498–1504. doi: 10.1073/pnas.55.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. Negative regulation of STE6 gene expression by the alpha 2 product of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2420–2427. doi: 10.1128/mcb.4.11.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. F., Zitomer R. S. A positive regulatory site and a negative regulatory site control the expression of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1984 Oct;4(10):2023–2030. doi: 10.1128/mcb.4.10.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Walthall D. A., Rymond B. C., Hollenberg C. P. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol. 1984 Jul;4(7):1191–1197. doi: 10.1128/mcb.4.7.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]



