Abstract
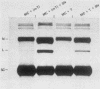
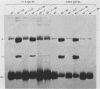
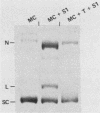
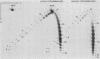
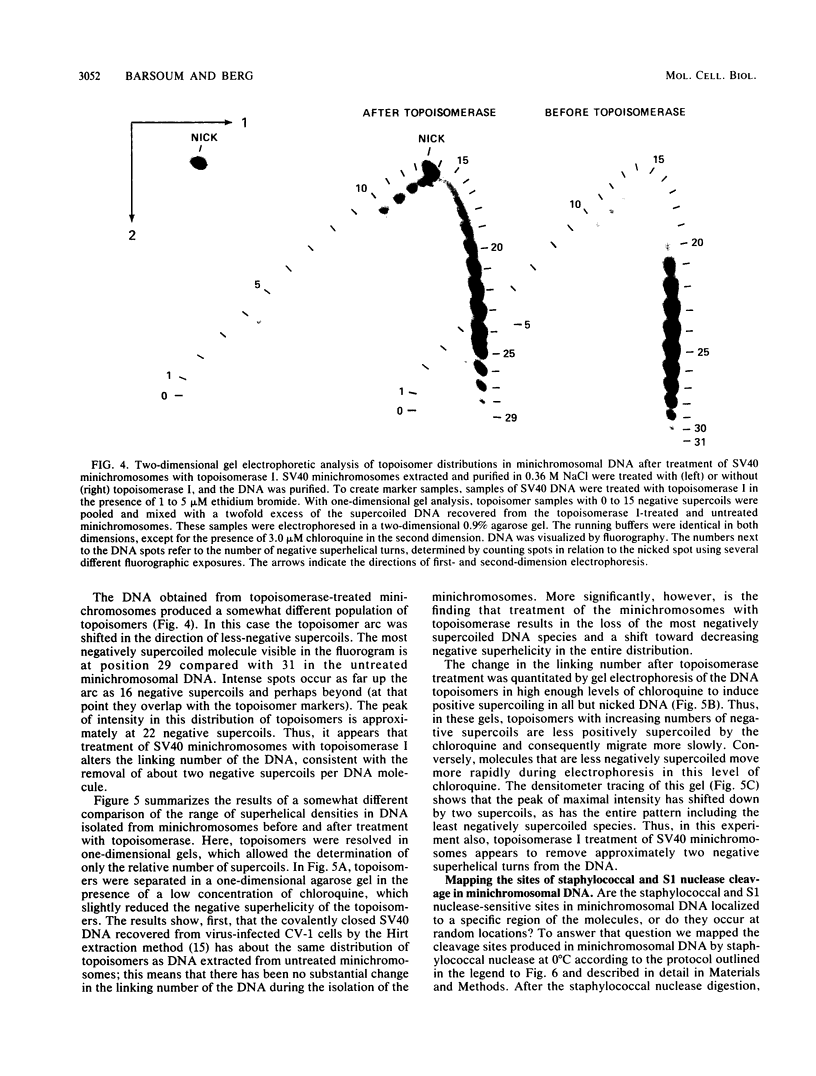
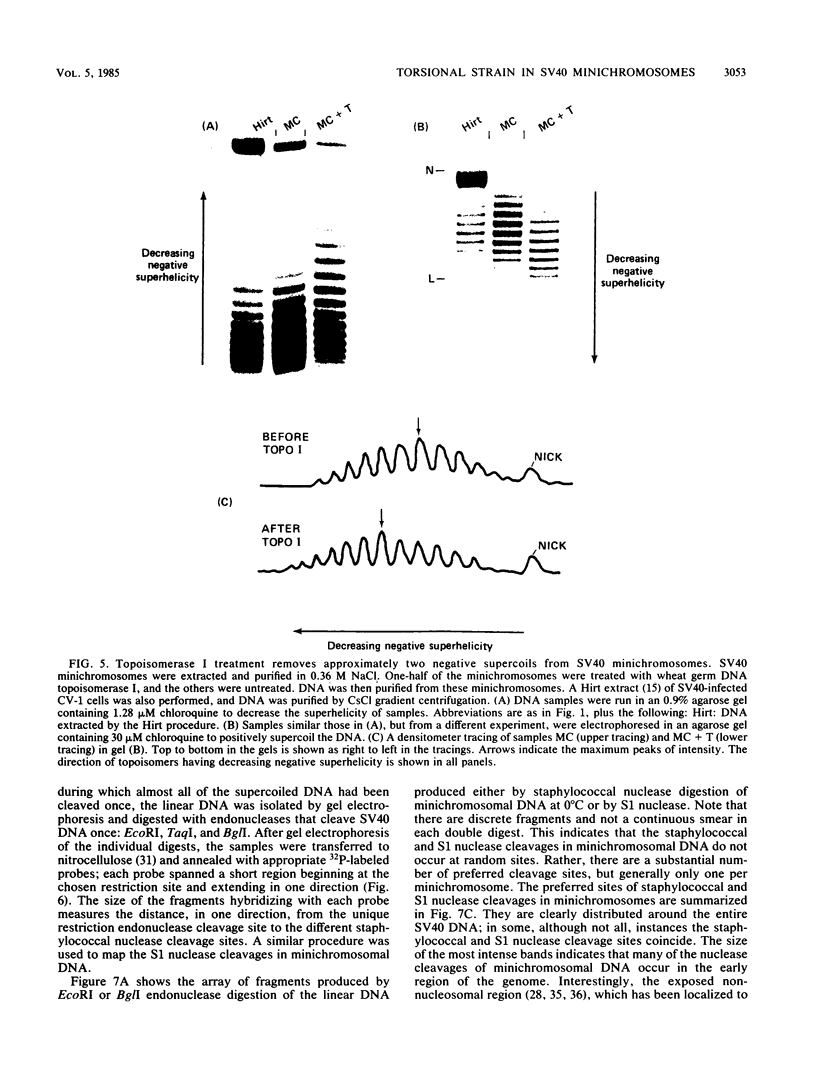
Sundin and Varshavsky (J. Mol. Biol. 132:535-546, 1979) found that nearly two-thirds of simian virus 40 (SV40) minichromosomes obtained from nuclei of SV40-infected cells become singly nicked or cleaved across both strands after digestion with staphylococcal nuclease at 0 degrees C. The same treatment of SV40 DNA causes complete digestion rather than the limited cleavages produced in minichromosomal DNA. We have explored this novel behavior of the minichromosome and found that the nuclease sensitivity is dependent upon the topology of the DNA. Thus, if minichromosomes are pretreated with wheat germ DNA topoisomerase I, the minichromosomal DNA is completely resistant to subsequent digestion with staphylococcal nuclease at 0 degrees C. If the minichromosome-associated topoisomerase is removed, virtually all of the minichromosomes are cleaved to nicked or linear structures by the nuclease treatment. The cleavage sites are nonrandomly located; instead they occur at discrete loci throughout the SV40 genome. SV40 minichromosomal DNA is also cleaved to nicked circles and full-length linear fragments after treatment with the single strand-specific endonuclease S1; this cleavage is also inhibited by pretreatment with topoisomerase I. Thus, it may be that the nuclease sensitivity of minichromosomes is due to the transient or permanent unwinding of discrete regions of their DNA. Direct comparisons of the extent of negative supercoiling of native and topoisomerase-treated SV40 minichromosomes revealed that approximately two superhelical turns were removed by the topoisomerase treatment. The loss of these extra negative supercoils from the DNA probably accounts for the resistance of the topoisomerase-treated minichromosomes to the staphylococcal and S1 nucleases. These findings suggest that the DNA in SV40 intranuclear minichromosomes is torsionally strained. The functional significance of this finding is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Boyce F. M., Sundin O., Barsoum J., Varshavsky A. New way to isolate simian virus 40 nucleoprotein complexes from infected cells: use of a thiol-specific reagent. J Virol. 1982 Apr;42(1):292–296. doi: 10.1128/jvi.42.1.292-296.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Gariglio P., Llopis R., Oudet P., Chambon P. The template of the isolated native simian virus 40 transcriptional complexes is a minichromosome. J Mol Biol. 1979 Jun 15;131(1):75–105. doi: 10.1016/0022-2836(79)90302-4. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Menzel R., Mizuuchi K., O'Dea M. H., Friedman D. I. Regulation of DNA supercoiling in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):763–767. doi: 10.1101/sqb.1983.047.01.087. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Gourlie B. B., Pigiet V., Breaux C. B., Krauss M. R., King C. R., Benbow R. M. Polyoma virus minichromosomes: associated enzyme activities. J Virol. 1981 Jun;38(3):826–832. doi: 10.1128/jvi.38.3.826-832.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. H., Brooks T. L. Isolation of two forms of SV40 nucleoprotein containing RNA polymerase from infected monkey cells. Virology. 1976 Jul 1;72(1):110–120. doi: 10.1016/0042-6822(76)90316-0. [DOI] [PubMed] [Google Scholar]
- Hamelin C., Yaniv M. Nicking-closing enzyme is associated with SV40 DNA in vivo as a sodium dodecyl sulfate-resistant complex. Nucleic Acids Res. 1979 Oct 10;7(3):679–687. doi: 10.1093/nar/7.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Liu L. F. Association of eukaryotic DNA topoisomerase I with nucleosomes and chromosomal proteins. Nucleic Acids Res. 1983 Jan 25;11(2):461–472. doi: 10.1093/nar/11.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M. R., Gourlie B. B., Bayne M. L., Benbow R. M. Polyomavirus minichromosomes: associated DNA topoisomerase II and DNA ligase activities. J Virol. 1984 Feb;49(2):333–342. doi: 10.1128/jvi.49.2.333-342.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopis R., Perrin F., Bellard F., Gariglio P. Quantitation of transcribing native simian virus 40 minichromosomes extracted from CV1 cells late in infection. J Virol. 1981 Apr;38(1):82–90. doi: 10.1128/jvi.38.1.82-90.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Carlson J. O., Pettijohn D. E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980 Oct;21(3):773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Staphylococcal nuclease makes a single non-random cut in the simian virus 40 viral minichromosome. J Mol Biol. 1979 Aug 15;132(3):535–546. doi: 10.1016/0022-2836(79)90274-2. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Nedospasov S. A., Georgiev G. P. On the structure of eukaryotic, prokaryotic, and viral chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):457–473. doi: 10.1101/sqb.1978.042.01.049. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O. H., Bohn M. J. SV40 viral minichromosome: preferential exposure of the origin of replication as probed by restriction endonucleases. Nucleic Acids Res. 1978 Oct;5(10):3469–3477. doi: 10.1093/nar/5.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldeck W., Zentgraf H., Sauer G. In vitro formation of multimeric DNA structures mediated by purified simian virus 40 chromatin. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1027–1031. doi: 10.1073/pnas.79.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Young L. S., Champoux J. J. Interaction of the DNA untwisting enzyme with the SV40 nucleoprotein complex. Nucleic Acids Res. 1978 Feb;5(2):623–635. doi: 10.1093/nar/5.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]