Abstract
Platinum complexes related to cisplatin, cis-[PtCl2(NH3)2], are successful anticancer drugs; however, other transition metal complexes offer potential for combating cisplatin resistance, decreasing side effects, and widening the spectrum of activity. Organometallic half-sandwich iridium (IrIII) complexes [Ir(Cpx)(XY)Cl]+/0 (Cpx = biphenyltetramethylcyclopentadienyl and XY = phenanthroline (1), bipyridine (2), or phenylpyridine (3)) all hydrolyze rapidly, forming monofunctional G adducts on DNA with additional intercalation of the phenyl substituents on the Cpx ring. In comparison, highly potent complex 4 (Cpx = phenyltetramethylcyclopentadienyl and XY = N,N-dimethylphenylazopyridine) does not hydrolyze. All show higher potency toward A2780 human ovarian cancer cells compared to cisplatin, with 1, 3, and 4 also demonstrating higher potency in the National Cancer Institute (NCI) NCI-60 cell-line screen. Use of the NCI COMPARE algorithm (which predicts mechanisms of action (MoAs) for emerging anticancer compounds by correlating NCI-60 patterns of sensitivity) shows that the MoA of these IrIII complexes has no correlation to cisplatin (or oxaliplatin), with 3 and 4 emerging as particularly novel compounds. Those findings by COMPARE were experimentally probed by transmission electron microscopy (TEM) of A2780 cells exposed to 1, showing mitochondrial swelling and activation of apoptosis after 24 h. Significant changes in mitochondrial membrane polarization were detected by flow cytometry, and the potency of the complexes was enhanced ca. 5× by co-administration with a low concentration (5 μM) of the γ-glutamyl cysteine synthetase inhibitor L-buthionine sulfoximine (L-BSO). These studies reveal potential polypharmacology of organometallic IrIII complexes, with MoA and cell selectivity governed by structural changes in the chelating ligands.
The PtII metallo-drugs cisplatin, carboplatin, and oxaliplatin are the most widely used anticancer agents, with exemplary success in treating testicular, ovarian, and colorectal cancers.1,2 However, there are several key limitations to their use, including inherent and acquired Pt resistance, which reduces the range of treatable tumors, and severe patient side toxicity.3 Although the mechanism of action (MoA) of cisplatin is not fully understood, activation by hydrolysis and formation of intrastrand DNA cross-links appear to play crucial roles in its cytotoxicity.
Extensive research into other metal-based anticancer complexes strives to produce compounds with higher potency, higher cancer cell selectivity, lower resistance, and reduced side effects.4−13 Metal-based anticancer agents that target mutated biochemical pathways in cancer cells may meet these requirements, especially if more than one pathway is targeted simultaneously. A drug able to cause global cellular effects, perhaps by disrupting the redox balance in cells, may be advantageous, given the heterogeneous nature of solid tumors.14,15
Cytostatic agents have been successfully developed to target one or more biological pathways responsible for tumor growth.16 Often these drugs are used in combination with cytotoxic agents to stabilize cell proliferation before tumor shrinkage. The combination of L-buthionine sulfoximine (L-BSO) with the alkylating agent melphalan is an example of this approach, currently in clinical trials for the treatment of advanced melanomas.17 Anticancer agents with both cytostatic and cytotoxic properties have also been reported, including organometallic RuII compounds.18
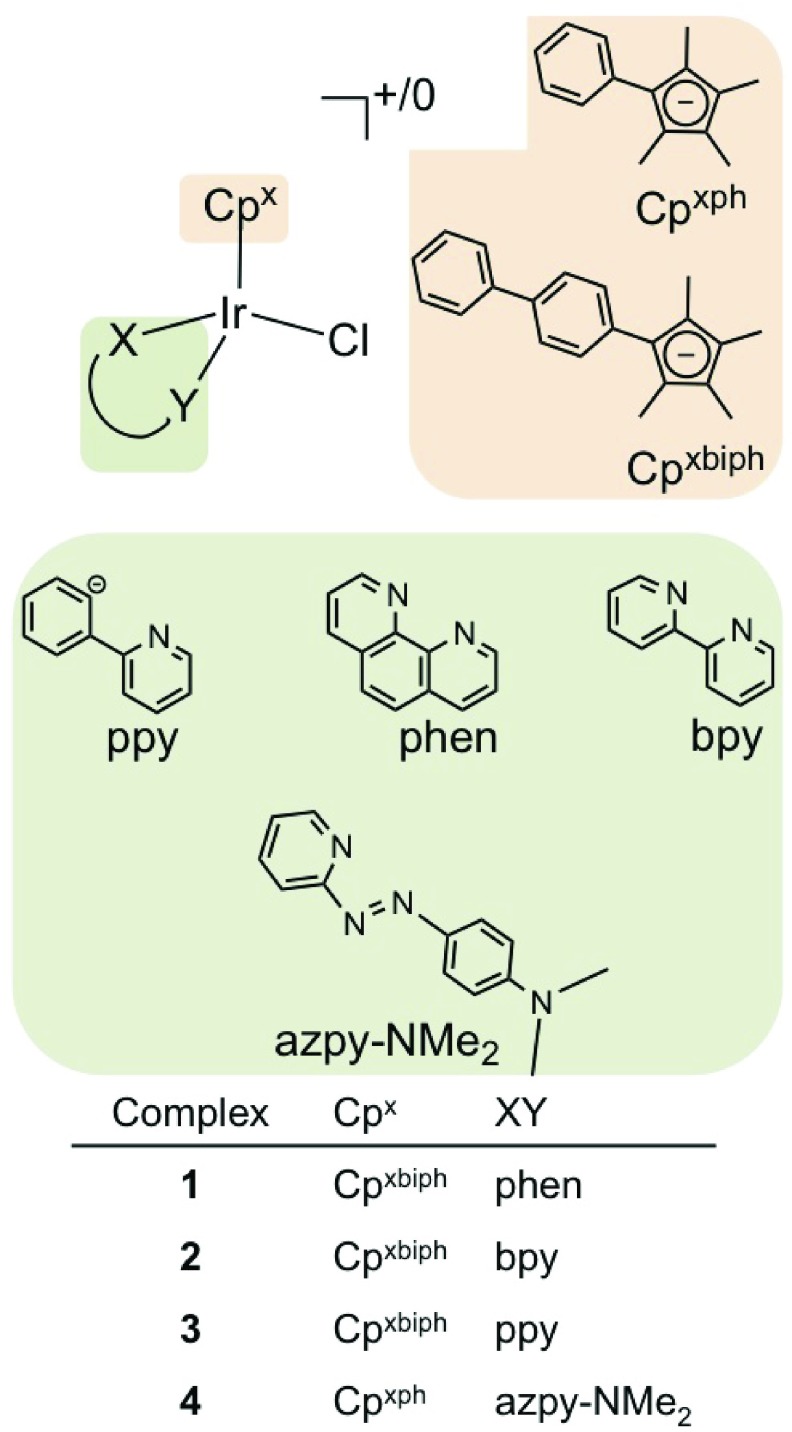
Here, we investigate novel half-sandwich organometallic IrIII cyclopentadienyl complexes as potent cytostatic and cytotoxic anticancer agents. These pseudo-octahedral complexes have carbon-bound cyclopentadienyl ligands that occupy three coordination sites, an N,N- or C,N- chelating ligand that occupies the fourth and fifth sites, and a monodentate Cl ligand at the sixth site. Cancer cell cytotoxicity screening has led to the discovery of a subset of highly potent IrIII complexes (Figure 1 and Table 1).12,13,19
Figure 1.
IrIII complexes used in this work.
Table 1. IC50 Values for Complexes 1–4 in A2780 Cellsa and Mean GI50 and LC50 Values for the NCI-60 Panel of Cell Lines.
| complex | A2780 IC50 (μM) | mean GI50 (μM) | mean LC50 (μM) |
|---|---|---|---|
| (1) [Ir(η5-Cpxbiph)(phen)Cl]PF6 | 0.72 ± 0.0112 | 2.34 | 31.62 |
| (2) [Ir(η5-Cpxbiph)(bpy)Cl]PF6 | 0.57 ± 0.0912 | 4.17 | 67.61 |
| (3) [Ir(η5-Cpxbiph)(ppy)Cl] | 0.70 ± 0.0413 | 0.71 | 8.13 |
| (4) [Ir(η5-Cpxph)(NMe2-azpy)Cl]PF6 | 0.40 ± 0.03 | 0.59 | 6.61 |
| cisplatin | 1.20 ± 0.20 | 1.49 | 44.00 |
Mean ± standard deviation from two independent experiments, with triplicate measurements in each experiment.
Complex 1 can bind to nucleobases in pSP73KB plasmid DNA, blocking RNA/DNA synthesis.12 However, studies of Ir accumulation in A2780 cells exposed to complex 1 revealed that only 6% of Ir accumulated inside the cells is DNA-bound, suggesting that the antineoplastic activity could result from interactions with other molecular targets.12 The work reported here explores the MoA of these Ir complexes using data from 60-cell-line screening by the National Cancer Institute (NCI-60) together with use of the COMPARE algorithm.20−22 Such an approach has shown success in exploring MoAs of new organic anticancer agents.23−25 In addition, we have employed transmission electron microscopy (TEM) to examine Ir-induced morphological changes in cancer cells, together with determination of changes in mitochondrial membrane polarization. We also assess co-incubation of these complexes with L-buthionine sulfoximine (L-BSO), an agent that depletes cellular levels of glutathione (GSH) and increases the already-elevated levels of oxidative stress inside cancer cells, in order to potentiate the activity of Ir in combination treatment.
Results and Discussion
Anticancer drug discovery is currently dominated by attempts to achieve selective attack on specific biochemical pathways, in particular on key genomic products.26,27 Although this stratified approach is likely to play a key role in the future of cancer research, the process is long and far from evolved. In addition, experience shows that such designed single-target drugs can generate problems of relapse, resistance, and mutations, due to the disease complexity, as well as an ability to avoid drug-affected pathways through use of alternative routes.14,28
Here, we have explored the potential of organometallic IrIII complexes as multitargeting agents. This has involved screening in the wide range of cancer cell lines in the NCI-60 panel and examination of their patterns of activity using the COMPARE algorithm. This has allowed potential mechanisms to be identified and tested experimentally using TEM and flow cytometry. The results highlight our ability to alter the biological activity of these complexes by tailoring their chemical structures.
Iridium Complexes
Complexes 1 [(η5-Cpxbiph)Ir(phen)(Cl)]+, 2 [(η5-Cpxbiph)Ir(bpy)(Cl)]+, and 3 [(η5-Cpxbiph)Ir(ppy)(Cl)] were synthesized as previously described.12,13 In addition we synthesized and characterized the novel complex 4 [(η5-Cpxph)Ir(azpy-NMe2)(Cl)]+ (Figure 1) by IR, UV–vis, 1H NMR spectroscopy, and CHN elemental analysis (Materials and Methods in Supporting Information).
Complexes 1–4 showed promising activity toward A2780 ovarian cancer cells in sulforhodamine B (SRB) assays (IC50 values in Table 1), with all showing higher potency compared to that of cisplatin. They were also submitted to the NCI for screening in the NCI-60 panel and given unique identifier codes: 1 (NSC 756057), 2 (NSC 754721), 3 (NSC 754723), and 4 (NSC 761279).20−22 Table 1 includes the mean GI50 (concentration required to inhibit growth of treated cells by 50% compared to control cells) and LC50 (concentration that kills 50% of treated cells) values, calculated as the average values across each cell line in the NCI-60 panel.
The low GI50 (0.6–4.2 μM) and LC50 (6.6–67.6 μM) values highlight the cytostatic and cytotoxic behavior of the complexes, with 3 and 4 showing potency higher than cisplatin. Dual cytostatic/cytotoxic activity can provide several advantages in cancer treatment.18,29 Given the mechanism of activation of the Pt drugs, we assessed the activity of 1–4 with respect to hydrolysis of the Ir–Cl bond. Complexes 1, 2, and 3 all hydrolyze rapidly, in the order 3 > 2 > 1;12,13 however, complex 4 does not readily undergo aquation (Supplementary Figure 1).
Interestingly, the activity does not appear to depend on hydrolysis, since activity decreases in the order 4 > 2 > 3 > 1 toward A2780 cells and 4 > 3 > 1 > 2 in the NCI-60 screen; the latter correlates with the affinity of binding to guanine nucleobases for the hydrolyzable complexes (3 > 1 > 2). In both tests, the non-hydrolyzable complex 4 is the most active, suggesting that either DNA is not the primary target for this compound or that binding is not achieved through cisplatin-like mechanisms. The bidentate ligand therefore has a significant effect on activity.
Cancer Cell Selectivity
The in vitro growth inhibition patterns in the NCI-60 screen were analyzed for all four compounds using mean graphs (Supplementary Figures 2 and 3). These were constructed by plotting positive and negative values along a vertical line, representing the mean response over all cell lines in the panel (mean GI50). Projections to the right indicate cell lines with susceptibility that exceeds the mean, projections to the left indicate cell lines with lower susceptibility.
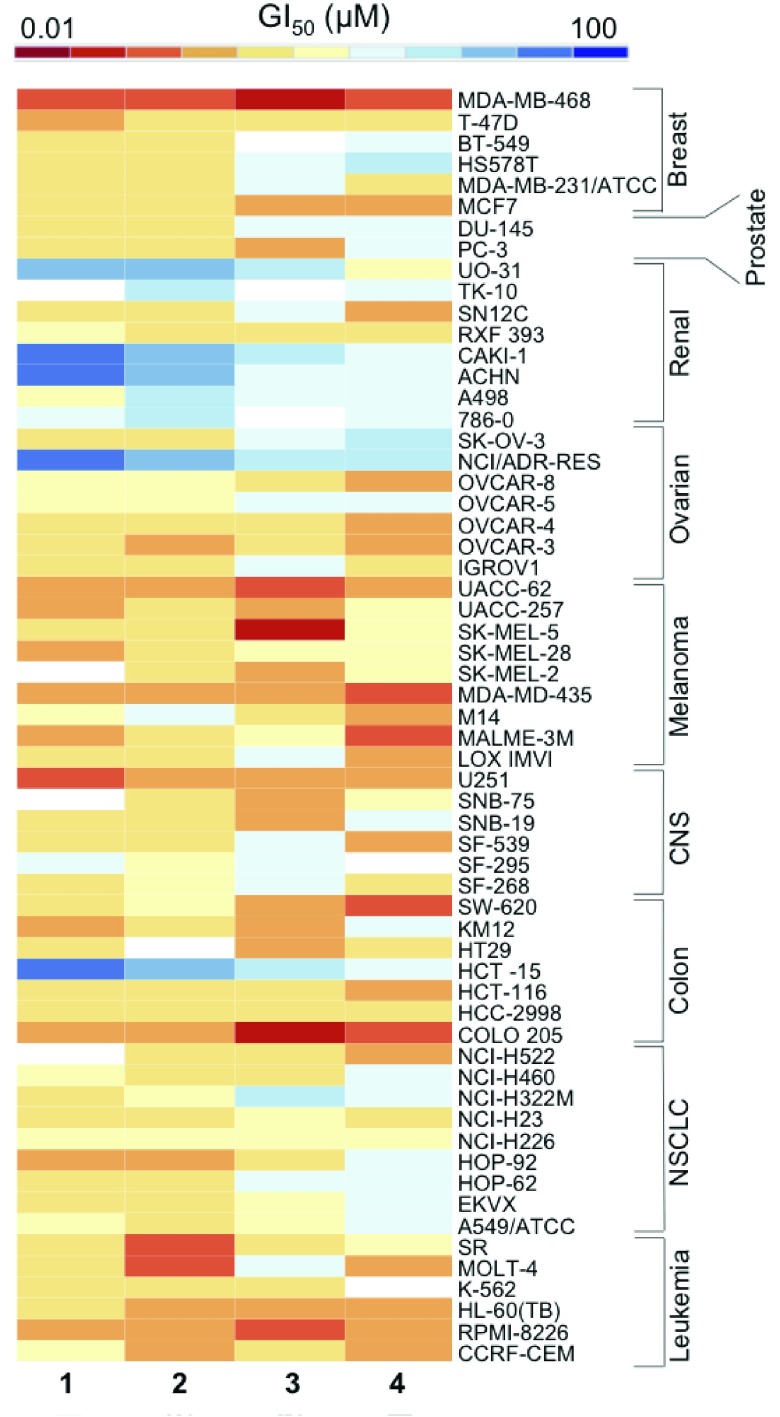
The heat map in Figure 2 summarizes the patterns of cytostatic behavior; high intensity values (red) indicate higher activity (lower GI50), and lower intensity values (blue) indicate lower activity (higher GI50), for each complex. The most striking observation is the level of similarity in the selectivity across the complexes. Most of the renal cell lines are insensitive, shown with light blue boxes, with some completely resistant (GI50 ≥ 100 μM), shown with dark blue boxes. In contrast, the breast cell line MDA-MB-468 (GI50 0.14–0.69 μM), colorectal cell line COLO205 (GI50 0.12–1.39 μM), and most melanoma cell lines (GI50 0.18–17.1 μM), show high sensitivity to each complex.
Figure 2.
Heat map showing the log10 GI50 values for each iridium complex in the NCI-60 screen, where high intensity (red) cells indicate high activity and low intensity (blue) cells indicate low activity.
Renal cell chemoresistance is a common occurrence; these lines often have a high abundance of multi-drug resistant (MDR1) protein expression, related to the levels of P-glycoprotein (Pgp).30,31 This increased expression means that drug efflux is a major problem in the treatment of these cancers.32
COLO205 colorectal cells, which showed high sensitivity, have reduced levels of glutathione-S-transferase P1 (GSTP1), an enzyme that detoxifies drugs by conjugating them to glutathione (GSH) often limiting the efficacy of anticancer agents.33
The breast cell line MDA-MB-468 showed the highest sensitivity in the NCI-60 screen. This cell line has a glucose 6-phosphate dehydrogenase (G6PD) A phenotype, meaning it lacks the proper functioning of this enzyme in the pentose phosphate pathway. G6PD is the rate-limiting enzyme in this pathway; its deficiency blocks the conversion of glucose-6-phosphate to 6-phosphoglucono-δ-lactone and in the process prevents the formation of NADPH from NADP+, a step required in GSH synthesis.34 This suggests that a GSH deficiency plays an important role in potentiating the activity of these Ir complexes, supported by Figure 6.
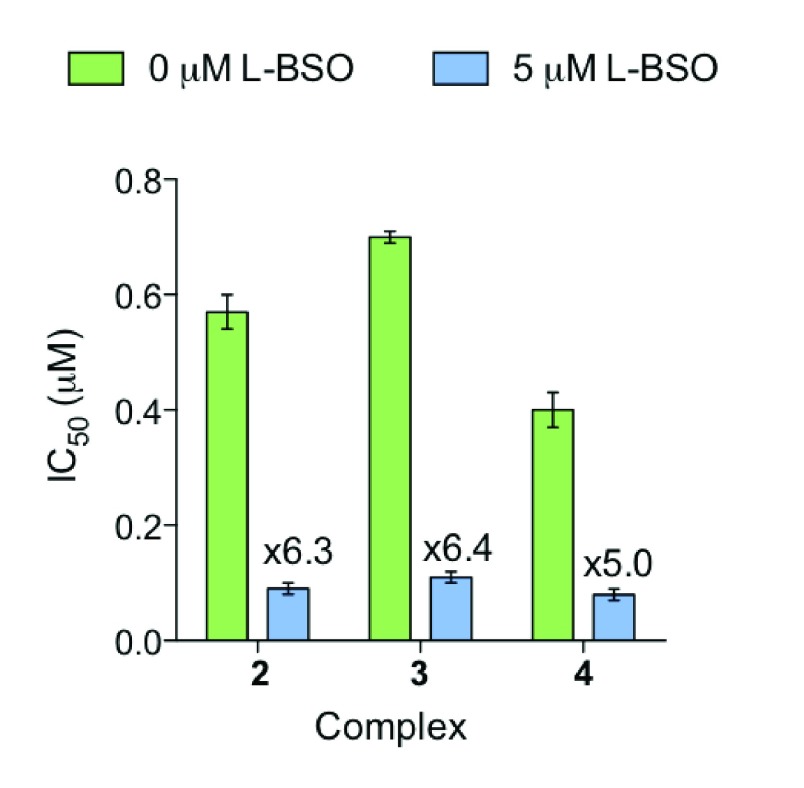
Figure 6.
Enhancement of cytotoxicity by co-incubation of A2780 human ovarian cancer cells with complexes 2–4 and 5 μM L-BSO. Bars show IC50 values with and without L-BSO, with the fold increase in activity above each bar. Each value is a mean of 3 replicates, with error bars for the standard deviation.
Analysis of the selectivity pattern quantitatively, using Pearson’s correlation coefficients (r), between each of the GI50 data sets shows that complexes 1 and 2 share high similarity compared to 3 and 4. The mean graph of 1 correlates to 2 with r = 0.902 and to 3 with r = 0.643 but does not correlate to 4 with a significantly positive coefficient value, i.e., r < 0.5.
The differential behavior of these four complexes highlights the impact of structural alterations, where changing the chelating ligand changes the selectivity toward a wide range of cell lines.
COMPARE Analysis
COMPARE analysis was performed for each Ir complex using the GI50 mean graphs. This algorithm allowed a quantitative comparison of growth inhibition between the Ir complexes and current drugs populating the NCI/NIH Developmental Therapeutics Program (DTP) databases. The Standard Agents Database, which contains ca. 175 agents selected as highly promising by the NCI, and the Synthetics Agents Database, containing >40,000 synthetic compounds and natural products of known structure, were used for this comparison. With each iteration a Pearson’s correlation coefficient (r) is produced, between −1 and 1. All positive correlations with the standard agents are discussed, as well as the top 100 correlations with the synthetic agents (r > 0.5), including only those drugs with established MoAs. The complete list of correlations for each complex is shown in Supplementary Tables 1A–F and 2.
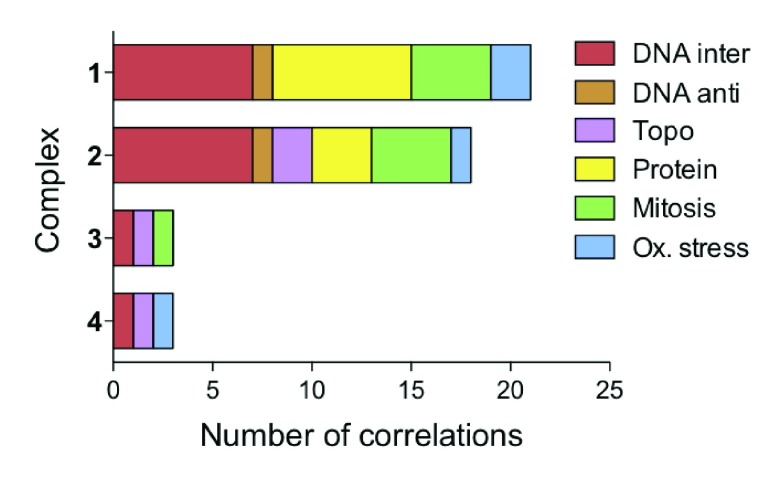
Our COMPARE analysis shows that the Ir complexes have mean graphs distinctly different from those of cisplatin and oxaliplatin, correlating to neither. The algorithm returned positive correlations to drugs with MoAs in six main categories: DNA interactors, DNA antimetabolites, topoisomerase inhibitors, protein synthesis inhibitors, mitosis inhibitors, and redox mediators (Figure 3). Complexes 3 and 4 correlate to only 3 drugs within the Synthetics Agents Database with established MoAs, highlighting their novelty.23 This may stem from the neutral nature of 3 and the novel para-substituted azopyridine ligand of 4. The remaining complexes, 1 and 2, have similar class distinctions, correlating to 18 and 21 drugs, respectively, most commonly to DNA-interacting agents and protein synthesis inhibitors.
Figure 3.
Mechanistic insights derived through use of the COMPARE algorithm. Bar chart summarizing the top 100 correlations, with r > 0.5, and known MoA for each of the four complexes. Correlations were segregated into six classes: oxidative stress, mitosis inhibitors, protein synthesis inhibitors, topoisomerase inhibitors, DNA antimetabolites, and DNA-interacting agents.
The class of DNA interacting agents refers to drugs such as daunorubucin, olivomycin, and chromomycin, all DNA-binding antibiotics.35,36 Our previous work has shown that chlorido IrIII cyclopentadienyl complexes can interact with DNA, binding both directly via Ir coordination to DNA bases (N7 in guanine) and via intercalation of extended (phenylated) cyclopentadienyl ligands.12 As DNA generally exists in nucleosomes (DNA wound around eight core histone proteins), any DNA binding might occur only during DNA replication or protein synthesis. We have previously shown that RNA/DNA replication can be blocked upon binding of IrIII complexes.12
Correlations to protein synthesis inhibitors included phyllanthoside, aurantimycin B, undulatone, and bouvardin, which inhibit translation by a variety of mechanisms.25,37 Often, cancer cells have inherent deficiencies in their protein synthesis machinery, for example, ovarian, thyroid, pancreatic, and colorectal cancer cell lines all have mutations in 12S and 16S rRNA, both of which are required for protein synthesis.39,40 Therefore targeting protein synthesis or the DNA structure inside the nucleus or mitochondria would explain why complexes have high activity toward A2780 ovarian cancer cells.38
The class of mitosis inhibitors includes taxol, vinblastine sulfate, and vincristine sulfate, which disrupt microtubule polymerization through various mechanisms; however, recent work has highlighted their DNA binding potential.41−43 Topoisomerase inhibitors, like doxorubicin, inhibit enzymes that regulate the unwinding of DNA during replication and protein synthesis. Therefore the activity of these enzymes could be indirectly affected through DNA binding. In addition, some of the complexes also correlated to cytotoxic redox mediators such as asiaticoside.44 This type of MoA can be closely linked to mitochondrial effects.
Collectively these Ir complexes show potential polypharmacology, the targeting of more than one biological component or pathway. To investigate further the MoA of these complexes, we address potential redox involvement, as proposed by COMPARE. We have obtained ultrastructural information and assessed activity when cellular levels of the ROS scavenger GSH were depleted.
Mitochondrial Swelling
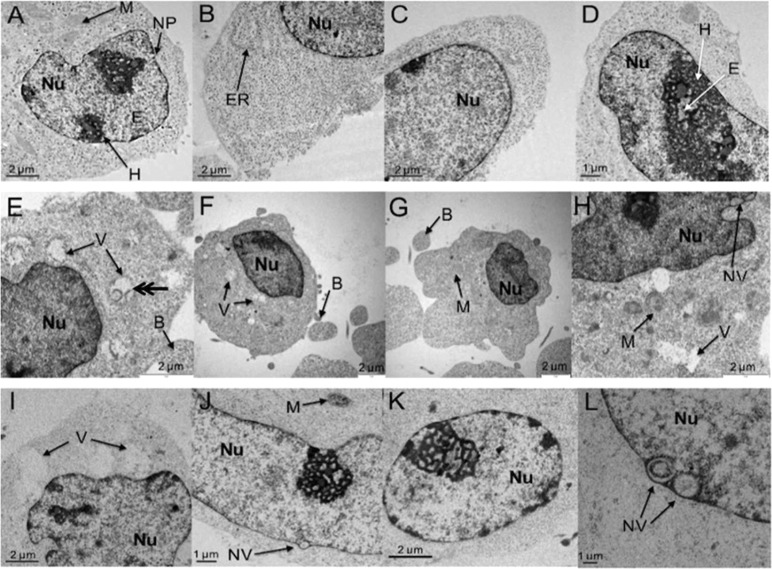
Human ovarian cancer cells (A2780) were exposed to complex 1 at 1 μM (1.4 × IC50) and 5 μM (6.9 × IC50) for 24 h, fixed, stained with uranyl acetate and lead citrate, embedded, sectioned, and analyzed by TEM. Images in Figure 4 are representative subsets of the total images recorded: control (total 48 images), 1 μM exposure (total 23 images), and 5 μM exposure (total 51 images). Figures 4(A–D) show control (untreated) cells, with Figure 4A highlighting the abundant mitochondria (M), 4B the granular endoplasmic reticulum (ER), 4C the nuclear membrane with membrane pores (NP), and 4A and 4D heterochromatin (H) with euchromatin (E) channels.
Figure 4.
Detection of apoptosis in A2780 ovarian cancer cells after treatment with 1. (A–D) Control cells showing typical heterochromatin (H) and euchromatin (E) distributions, mitochondria (M), nuclear pores (NP), and rough endoplasmic reticulum (ER). (E–H) Cells exposed to 1 μM 1 for 24 h, showing swollen mitochondria (V), nuclear vacuoles (NV), and membrane blebbing (B). (I–L) Cells exposed to 5 μM 1 for 24 h, showing abnormal chromatin distributions and further vacuole formations.
After exposure to 1 μM complex 1, the cells developed small vacuoles throughout the cytoplasm (Figure 4E, F, and H), representing swollen mitochondria, identified by their double-layer membrane (double arrow) in Figure 4E.45 The mitochondria appear devoid of structure inside the inner membrane, although the outer membrane appears to stay intact in some cases. After exposure to 5 μM 1 (Figure 4I), these vacuoles are larger and the mitochondrial membrane no longer visible.
Swelling of mitochondria, which could distort the visualization of organelle structure by TEM, is often associated with calcium imbalance inside the cells, caused by drug-induced oxidative stress.46−48 However, substituted triptycenes, which exert anticancer activity through DNA-dependent mechanisms such as topoisomerase inhibition, can also disrupt calcium influx into the mitochondria of leukemia cells.47 This causes changes in transmembrane potentials by altering membrane permeability through the mitochondrial permeability transition pore (mPTP), causing mitochondrial swelling. This mechanism has also been shown in cardiac toxicity studies for doxorubicin, a DNA topoisomerase inhibitor that correlates to several of the Ir complexes in the COMPARE analysis (see Supplementary Tables 1A–F and 2). Doxorubicin can cause oxidative stress inside cells as a secondary mechanism and in turn activates calcium-dependent mitochondrial swelling.49
Polarization of the Mitochondrial Membrane
The mitochondria are semi-autonomous organelles, with their own genome and protein synthesis machinery.39 They play important roles in metabolism, free radical generation, and cell death. However, mitochondria in cancer cells differ from those found in normal cells, highlighting them as potential therapeutic targets.50
Anaerobic glycolysis plays a significant role in the metabolism of cancer cells; this adaptation allows them to survive under hypoxic conditions.45 The membrane potential of the mitochondria in cancer cells is higher (more negative), which can allow accumulation of cationic lipophilic compounds down an electrochemical gradient.45,51 Complex 1, which appeared to have an effect on mitochondria by TEM, has a Log P value of 1.11 ± 0.17, making it hydrophobic, and therefore could be readily taken up by mitochondria, an example of passive drug delivery.12
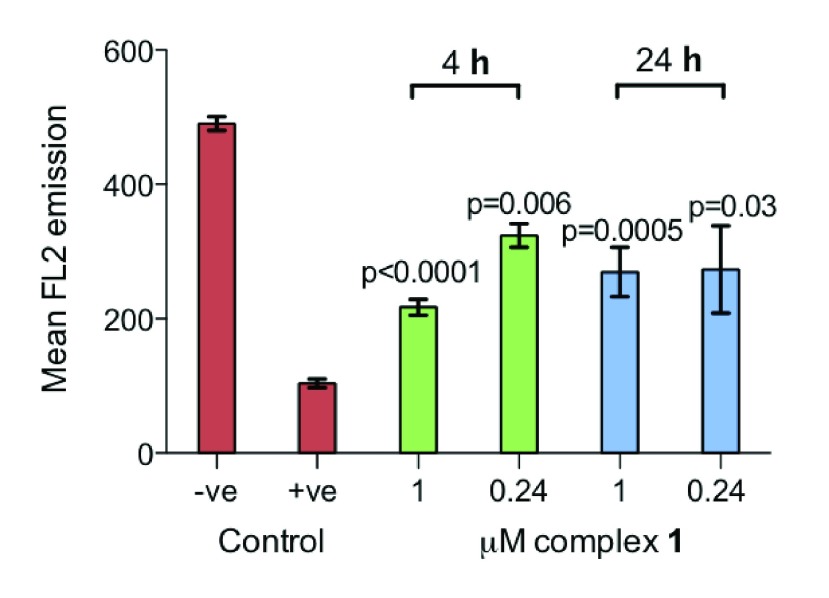
We investigated the integrity of the mitochondrial membrane by measuring the membrane potential by observing the fluorescence of JC-10, a cationic and lipophilic dye, using flow cytometry. In normal mitochondria, JC-10 aggregates and emits red fluorescence; however, upon mitochondrial membrane polarization, aggregation of JC-10 reduces, together with red emission. The level of membrane polarization after cells were exposed to complex 1 for 4 and 24 h (Figure 5) is approximately midway between the negative and positive controls. The p-values of each treatment relative to the control are all significant at 5% after a Welch t test.
Figure 5.
Bar graph showing the mitochondrial membrane polarization, determined by a reduction in red (FL2) fluorescence, by complexes 1–4 against that of a negative and positive control (5 μM carbonyl cyanide m-chlorophenylhydrazone). p-values were calculated after a Welch t test with the negative control data. Each value is the mean taken from 3 replicates, with error bars for the standard deviation.
The proposed effect on the mitochondria could in turn cause ATP depletion and increased permeability of the outer membrane, allowing transfer of pro-apoptotic proteins and the activation of intrinsic apoptosis cascades. This release of proteins is sometimes accompanied by membrane rupturing, as shown in Figure 4E (labeled V).50
Inducing Apoptosis
Apoptosis is the process by which cells die in a programmed fashion, as a result of particular stimuli. The initiation of apoptosis by pro-apoptotic stimuli is followed by an effector phase where the machinery required for cell death is activated.52 After this, the cell enters into the degradation phase, at which point apoptosis becomes visible.53 There are a plethora of routes by which anticancer agents or radiotherapy can activate apoptosis, normally characterized by activation of intrinsic or extrinsic pathways. Caspase enzymes (cysteine proteases) are effector molecules activated during either extrinsic (receptor) or intrinsic (mitochondrial) pathways.54 Caspases further activate each other through cleavage next to aspartate residues, amplifying caspase activity down a cascade, accounting for many of the morphological changes associated with apoptosis.54
The onset of apoptosis, characterized by condensation of nuclear chromatin into delineated masses that localize on the nuclear membrane, is evident in Figure 4J and K.52,55 This nuclear destabilization results from caspase-activated DNase (CAD) enzymes, which cleave DNA at internucleosomal linker sites.56 Also visible are small vacuoles formed in the nucleus and nuclear membrane (Figure 4J and L). These morphological developments can result from abnormalities in chromatid packing and destabilization of the genetic material.56 We have shown that Ir complexes such as [Ir(η5-Cpxph)(phen)(H2O)]2+ can react with coenzyme NADH to generate the Ir hydride and H2 catalytically.57 It is also possible that some of the nuclear vacuoles seen in Figure 4J and L arise from H2 release.57
Apoptosis causes the outline of the cell to become convoluted and form extensions, termed ‘budding’. These extensions separate from the cell and form so-called ‘apoptotic bodies’. This is shown in Figure 4E–G.52,55 Ordinarily cellular organelles, such as mitochondria, would remain preserved until the apoptotic bodies are phagocytozed or are degraded by secondary necrosis.58 However, the mitochondria inside the apoptotic cells in Figure 4E, F, H, and I are not preserved, suggesting that something other than the normal processes of cell death may be occurring.
Cancer cells can acquire resistance to anticancer compounds designed to activate apoptosis. They do this by up-regulating pro-survival factors, including inhibitors of apoptosis proteins (IAPs).59 IAPs block caspase cascades required to activate apoptosis. However, if the Ir complexes affect mitochondria as proposed, then energy deficiency is a key consequence. If IAPs block the caspase cascade successfully, cell growth can still be halted through ATP deficiency, giving rise to cytostasis.
Combination with L-BSO Increases Potency
L-Buthionine sulfoximine (L-BSO) depletes the levels of glutathione (GSH) in cells by targeting γ-glutamyl cysteine synthetase. GSH is an antioxidant that protects the cells from reactive oxygen species (ROS). We co-incubated A2780 cells with a non-toxic dose of L-BSO (5 μM) and complexes 2–4 (1/3 IC50) for 24 h and compared their IC50 values with and without L-BSO (Figure 6). Each experiment used two negative controls: one untreated and one treated with 5 μM L-BSO. After 24 h, cell viability was still 98.5–99.5% after L-BSO treatment, ≤ 5% difference from the untreated control. There is a significant decrease in IC50 for all three complexes by factors of 5–6.4 down to the nanomolar range (Figure 6): 90 nM for complex 2 (6.3× increase in activity), 110 nM for complex 3 (6.4× increase), and 80 nM for complex 4 (5× increase) (Table 1). Cells with lower levels of GSH are more vulnerable to oxidative stress, suggesting that this mechanism may be involved either directly or indirectly in the MoA of these Ir complexes. This would explain the high activity toward MDA-MB-468 cells in the NCI-60 screen (Figure 2).
Cancer cells are, in general, more active than normal cells with regard to ROS production, which is directly related to the behavior of mitochondria.60 COMPARE correlations to agents that cause oxidative stress, as well as the promising data with L-BSO (Figure 6), suggest further the involvement of these complexes in ROS production.
Redox mediation by metal complexes is a well-documented mechanism of anticancer activity, either through GSH binding and depletion, or through direct production of ROS.61 The variation in activity is dependent on both the metal center and the chelating ligands.
Conclusions
We have studied four organoiridium complexes with potency higher than that of the PtII drugs in the clinic and with different MoAs. We have demonstrated an ability to vary the reactivity and selectivity of these Ir complexes by changing the chelating ligand. Using the tools of the NCI, we propose polypharmacology for these complexes, potentially increasing ROS, damaging DNA, and disrupting protein synthesis. Future work will further investigate the polypharmacology of these complexes, where perhaps the primary MoA is redox mediation, leading to secondary and tertiary MoAs involving DNA damage and protein synthesis inhibition by ROS. It is apparent, from this study, that DNA interference is not always through cisplatin-like binding, given that complex 4 is non-hydrolyzable. We highlighted two complexes (neutral complex 3 and azopyridine complex 4) that show highly novel patterns of growth inhibition. Complex 4 showed high levels of activity in renal cell lines in the NCI-60 screen, in particular SN12C (GI50 0.22 μM), suggesting that the azpy-NMe2 chelating ligand plays a role in increasing Ir-based drug sensitivity.
TEM and biochemical assays supported involvement in ROS production, with mitochondrial targeting and apoptosis occurring within a 24 h exposure period. These observations support our COMPARE-based predictive findings, where DNA interaction, protein synthesis disruption, and redox mediation can all be linked to the mitochondria.
Methods
Synthesis and Characterization of the Complexes
Complexes 1–3 were synthesized from their corresponding dimers and characterized as previously reported.12,13 Synthesis and characterization of novel complex 4 can be found in the Supporting Information.
Cell Culture
The A2780 ovarian cell line was obtained from ECACC (European Collection of Animal Cell Culture), and grown in RPMI-1640 medium supplemented with 10% (v/v) fetal calf serum, 1% (v/v) 2 mM l-glutamine, and 1% (v/v) penicillin (10 k units/mL)/streptomycin (10 mg/mL), at 310 K in an humidified atmosphere containing 5% CO2; maintenance passages were done at ca. 80% confluency using trypsin.
IC50 Determinations
Briefly, A2780 cells were seeded in 96-well plates with ca. 5000 cells per well. The plates were incubated at 310 K for 48 h. Drug solutions were added, and the cells were left to incubate for 24 h. Drug-containing medium was replaced with fresh medium, and cells were left to recover for 72 h. The sulforhodamine B assay was used to determine cell survival. See Supporting Information for full details.
TEM Sample Preparation
Briefly, A2780 cells were seeded at a density of 5 × 106 cells/100 mm Petri dish, left to incubate for 24 h at 310 K, and exposed to the drug for 24 h more. Cells were fixed with 2% glutaraldehyde and dehydrated with graded levels of ethanol (20–100% ethanol), before embedding with Embed 812 resin and curing. Blocks were trimmed and sectioned on a Leica Ultracut E ultramicrotome (Leica Microsystems) and imaged on a JEOL 1200EXII TEM with a Gatan 1 k × 1 k CCD camera. See Supporting Information for full details.
Mitochondrial Polarization Assay
Briefly, cells were seeded at 1 × 106 cells per well in 6-well plates and left to incubate for 24 h. Drug solutions were added, and the cells were left to incubate for a further 4 or 24 h. Cells were collected, and to each sample was added 500 μL of a solution of JC-10 dye (Abcam JC-10 Mitochondrial Membrane Potential Assay kit). Samples were immediately analyzed on a Beckton Dickinson FACScan with fluorescence detection. See Supporting Information for full details.
NCI-60 Screening
Complexes were submitted to the National Cancer Institute for screening. The protocols used by the NCI have been described previously.21,22,62 Briefly, cells in the NCI-60 screen were exposed to compound and incubated for 48 h at 310 K in a humidified atmosphere containing 5% CO2 before measurement of the GI50 (concentration of compound which inhibits 50% cell growth), TGI (concentration of compound which inhibits 100% of cell growth), and LC50 (concentration of compound which kills 50% of cell population).
Acknowledgments
We thank the National Cancer Institute for NCI-60 screening and COST Action CM1105 for stimulating discussions. We also thank A. Pizarro and A. Habtemariam for assistance with cell culture and chemical synthesis, respectively. This research was supported by ERC (grant no. 247450), Science City (AWM/ERDF), BBSRC (University of Warwick, Scholarship for J.M.H.), IAS (University of Warwick, U.K.; Fellowship for I.R.-C.).
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): P.J.S. and Z.L. are named inventors on a patent application relating to the iridium complexes used in this work filed by the University of Warwick.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Monneret C. (2011) Platinum anticancer drugs. From serendipity to rational design. Ann. Pharm. Fr. 69, 286–295. [DOI] [PubMed] [Google Scholar]
- Kelland L. (2007) The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 7, 573–584. [DOI] [PubMed] [Google Scholar]
- Siddik Z. (2003) Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279. [DOI] [PubMed] [Google Scholar]
- Garbutcheon-Singh K. B.; Grant M. P.; Harper B. W.; Krause-Heuer A. M.; Manohar M.; Orkey N.; Aldrich-Wright J. R. (2011) Transition metal based anticancer drugs. Curr. Top. Med. Chem. 11, 521–542. [DOI] [PubMed] [Google Scholar]
- Hannon M. J. (2007) Metal-based anticancer drugs: From a past anchored in platinum chemistry to a post-genomic future of diverse chemistry and biology. Pure Appl. Chem. 79, 2243–2261. [Google Scholar]
- Fu Y.; Habtemariam A.; Pizarro A. M.; Van Rijt S. H.; Healey D. J.; Cooper P. A.; Shnyder S. D.; Clarkson G. J.; Sadler P. J. (2010) Organometallic osmium arene complexes with potent cancer cell cytotoxicity. J. Med. Chem. 53, 8192–8196. [DOI] [PubMed] [Google Scholar]
- Fu Y.; Habtemariam A.; Basri A. M. B. H.; Braddick D.; Clarkson G. J.; Sadler P. J. (2011) Structure-activity relationships for organometallic osmium arene phenylazopyridine complexes with potent anticancer activity. Dalton Trans. 40, 10553–10562. [DOI] [PubMed] [Google Scholar]
- Park G. Y.; Wilson J. J.; Song Y.; Lippard S. J. (2012) Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc. Natl. Acad. Sci. U.S.A. 109, 11987–11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharova J.; Zerzankova L.; Stepankova J.; Novakova O.; Farrer N. J.; Sadler P. J.; Brabec V.; Kasparkova J. (2012) Interactions of DNA with a new platinum(IV) azide dipyridine complex activated by UVA and visible light: relationship to toxicity in tumor cells. Chem. Res. Toxicol. 25, 1099–1111. [DOI] [PubMed] [Google Scholar]
- Castonguay A.; Doucet C.; Juhas M.; Maysinger D. (2012) New ruthenium(II)-letrozole complexes as anticancer therapeutics. J. Med. Chem. 55, 8799–8806. [DOI] [PubMed] [Google Scholar]
- Hartinger C. G.; Phillips A. D.; Nazarov A. A. (2011) Polynuclear ruthenium, osmium and gold complexes. The quest for innovative anticancer chemotherapeutics. Curr. Top. Med. Chem. 11, 2688–2702. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Habtemariam A.; Pizarro A. M.; Fletcher S. A.; Kisova A.; Vrana O.; Salassa L.; Bruijnincx P. C. A.; Clarkson G. J.; Brabec V.; Sadler P. J. (2011) Organometallic half-sandwich iridium anticancer complexes. J. Med. Chem. 54, 3011–3026. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Habtemariam A.; Pizarro A. M.; Clarkson G. J.; Sadler P. J. (2011) Organometallic iridium(III) cyclopentadienyl anticancer complexes containing C,N-chelating ligands. Organometallics 30, 4702–4710. [Google Scholar]
- Petrelli A.; Giordano S. (2008) From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr. Med. Chem. 15, 422–432. [DOI] [PubMed] [Google Scholar]
- Bozic I.; Allen B.; Nowak M. A. (2012) Dynamics of targeted cancer therapy. Trends Mol. Med. 18, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. W.; Lynch K. P. (2003) Rethinking clinical trials for cytostatic drugs. Nat. Rev. Cancer 3, 540–545. [DOI] [PubMed] [Google Scholar]
- Bailey H. H.; Mulcahy R. T.; Tutsch K. D.; Arzoomanian R. Z.; Alberti D.; Tombes M. B.; Wilding G.; Pomplun M.; Spriggs D. R. (1994) Phase I clinical trial of intravenous L-buthionine sulfoximine and melphalan: an attempt at modulation of glutathione. J. Clin. Oncol. 12, 194–205. [DOI] [PubMed] [Google Scholar]
- Gaiddon C.; Jeannequin P.; Bischoff P.; Pfeffer M.; Sirlin C.; Loeffler J. P. (2005) Ruthenium (II)-derived organometallic compounds induce cytostatic and cytotoxic effects on mammalian cancer cell lines through p53-dependent and p53-independent mechanism. J. Pharmacol. Exp. Ther. 315, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Salassa L.; Habtemariam A.; Pizarro A. M.; Clarkson G. J.; Sadler P. J. (2011) Contrasting reactivity and cancer cell cytotoxicity of isoelectronic organometallic iridium(III) complexes. Inorg. Chem. 50, 5777–5783. [DOI] [PubMed] [Google Scholar]
- Paull K.; Shoemaker R.; Hodes L.; Monks A.; Scudiero D. A.; Rubinstein L.; Plowman J.; Boyd M. R. (1989) Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 81, 1088–1092. [DOI] [PubMed] [Google Scholar]
- Holbeck S.; Collins J. M.; Doroshow J. H. (2010) Analysis of Food and Drug Administration-approved anti-cancer agents in the NCI-60 panel of human tumor cell lines. Mol. Cancer. Ther. 9, 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R. H. (2006) The NCI-60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 22, 7265–7279. [DOI] [PubMed] [Google Scholar]
- Doroshow J. H.; Juhasz A.; Ge Y.; Holbeck S.; Lu J.; Antony S.; Wu Y.; Jiang G.; Roy K. (2012) Antiproliferative mechanisms of action of the flavin dehydrogenase inhibitors diphenylene iodonium and di-2-thienyliodonium based on molecular profiling of the NCI-60 human tumor cell panel. Biochem. Pharmacol. 83, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan V.; Bonham S.; Carty M. P.; Saenz-Méndez P.; Eriksson L. A.; Aldabbagh F. (2012) COMPARE analysis of the toxicity of an iminoquinone derivative of the imidazo[5,4-f]benzimidazoles with NAD(P)H:quinone oxidoreductase 1 (NQO1) activity and computational docking of quinones as NQO1 substrates. Bioorg. Med. Chem. 20, 3223–3232. [DOI] [PubMed] [Google Scholar]
- Chan J.; Khan S.; Harvey I.; Merrick W.; Pelletier J. (2004) Eukaryotic protein synthesis inhibitors identified by comparison of cytotoxicity profiles. RNA 10, 528–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrero A.; Garuti A.; Cirmena G.; Rocco I.; Palermo C.; Nencioni A.; Scabini S.; Zoppoli G.; Parodi S.; Patrone F. (2012) Patient-tailored treatments with anti-EGFR monoclonal antibodies in advanced colorectal cancer: KRAS and beyond. Curr. Cancer Drug Targets 12, 316–328. [DOI] [PubMed] [Google Scholar]
- Zhu Z.; Shen Z.; Xu C. (2012) Targeted therapy for advanced urothelial cancer of the bladder: Where do we stand?. Anticancer Agents Med. Chem. 12, 1081–1087. [DOI] [PubMed] [Google Scholar]
- Lu J.-J.; Pan W.; Hu Y.-J.; Wang Y.-T. (2012) Multi-target drugs: the trend of drug research and development. PLoS One 7, e40262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixe O.; Fojo T. (2007) Is cell death a critical end point for anticancer therapies or is cytostasis sufficient?. Clin. Cancer Res. 13, 7280–7287. [DOI] [PubMed] [Google Scholar]
- Türk D.; Hall M. D.; Chu B. F.; Ludwig J. A.; Fales H. M.; Gottesman M. M.; Szakács G. (2009) Identification of compounds selectively killing multidrug-resistant cancer cells. Cancer Res. 69, 8293–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy W. T. (1996) P-glycoproteins and multidrug resistance. Annu. Rev. Pharmacol. Toxicol. 36, 161–183. [DOI] [PubMed] [Google Scholar]
- Huo H.; Margro P. G.; Pietsch E. C.; Patel B. B.; W S. K. (2010) Histone methyltransferase MLL1 regulates MDR1 transcription and chemoresistance. Cancer Res. 70, 8726–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler D.; Brockmann A.; Mueller J.; Nachbur U.; Corazza N.; Renzulli P.; Hemphill A.; Brunner T. (2012) Thiazolide-induced apoptosis in colorectal cancer cells is mediated via the Jun kinase-Bim axis and reveals glutathione-S-transferase P1 as Achilles’ heel. Oncogene 31, 4095–4106. [DOI] [PubMed] [Google Scholar]
- Frank J. E. (2005) Diagnosis and management of G6PD deficiency. Am. Fam. Physician 72, 1277–1282. [PubMed] [Google Scholar]
- Dyke M. W. Van; Dervan P. B. (1983) Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry 22, 2373–2377. [DOI] [PubMed] [Google Scholar]
- Purewal M.; Liehr J. G. (1993) Covalent modification of DNA by daunorubicin. Cancer Chemother. Pharmacol. 33, 239–244. [DOI] [PubMed] [Google Scholar]
- Zalacaín M.; Zaera E.; Vázquez D.; Jiménez A. (1982) The mode of action of the antitumor drug bouvardin, an inhibitor of protein synthesis in eukaryotic cells. FEBS Lett. 148, 95–97. [DOI] [PubMed] [Google Scholar]
- Carew J. S.; Huang P. (2002) Mitochondrial defects in cancer. Mol. Cancer 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S.; Hassinen I. E.; Majamaa K. (2001) Phylogenetic analysis of mitochondrial DNA in patients with an occipital stroke. Evaluation of mutations by using sequence data on the entire coding region. Mutat. Res. 458, 31–39. [DOI] [PubMed] [Google Scholar]
- Van den Bogert C.; Van Kernebeek G.; De Leij L.; Kroon A. M. (1986) Inhibition of mitochondrial protein synthesis leads to proliferation arrest in the G1-phase of the cell cycle. Cancer Lett. 32, 41–51. [DOI] [PubMed] [Google Scholar]
- Jordan M. A.; Toso R. J.; Thrower D.; Wilson L. (1993) Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc. Natl. Acad. Sci. U.S.A. 90, 9552–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai S. S.; Wolff J. (1996) Localization of the vinblastine-binding site on beta-tubulin. J. Biol. Chem. 271, 14707–14711. [DOI] [PubMed] [Google Scholar]
- Tsutsui T.; Suzuki N.; Maizumi H.; Barrett J. C. (1986) Vincristine sulfate-induced cell transformation, mitotic inhibition and aneuploidy in cultured Syrian hamster embryo cells. Carcinogenesis 7, 131–135. [DOI] [PubMed] [Google Scholar]
- Pittella F.; Dutra R. C.; Junior D. D.; Lopes M. T. P.; Barbosa N. R. (2009) Antioxidant and cytotoxic activities of Centella asiatica (L) Urb. Int. J. Mol. Sci. 10, 3713–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindo D.; Dahm F.; Szulc Z.; Bielawska A.; Obeid L. M.; Hannun Y. A.; Graf R.; Clavien P.-A. (2006) Cationic long-chain ceramide LCL-30 induces cell death by mitochondrial targeting in SW403 cells. Mol. Cancer Ther. 5, 1520–1529. [DOI] [PubMed] [Google Scholar]
- Zhou D.-F.; Chen Q.-Y.; Qi Y.; Fu H.-J.; Li Z.; Zhao K.-D.; Gao J. (2011) Anticancer activity, attenuation on the absorption of calcium in mitochondria, and catalase activity for manganese complexes of N-substituted di(picolyl)amine. Inorg. Chem. 50, 6929–6937. [DOI] [PubMed] [Google Scholar]
- Perchellet E. M.; Wang Y.; Lou K.; Zhao H.; Battina S. K.; Hua D. U. Y. H.; Perchellet J.-P. H. (2007) Antitumor triptycene analogs directly interact with isolated mitochondria to rapidly trigger markers of permeability transition. Anticancer Res. 27, 3259–3272. [PubMed] [Google Scholar]
- McKeage M. J.; Maharaj L.; Berners-Price S. J. (2002) Mechanisms of cytotoxicity and antitumor activity of gold(I) phosphine complexes: the possible role of mitochondria. Coord. Chem. Rev. 232, 127–135. [Google Scholar]
- Zhang Y.-W.; Shi J.; Li Y.-J.; Wei L. (2009) Cardiomyocyte death in doxorubicin-induced cardiotoxicity. Arch. Immunol. Ther. Exp. 57, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantini P.; Jacotot E.; Decaudin D.; Kroemer G. (2000) Mitochondrion as a novel target of anticancer chemotherapy. J. Natl. Cancer Inst. 92, 1042–1053. [DOI] [PubMed] [Google Scholar]
- Porteous C. M.; Logan A.; Evans C.; Ledgerwood E. C.; Menon D. K.; Aigbirhio F.; Smith R. A. J.; Murphy M. P. (2010) Rapid uptake of lipophilic triphenylphosphonium cations by mitochondria in vivo following intravenous injection: implications for mitochondria-specific therapies and probes. Biochim. Biophys. Acta 1800, 1009–1017. [DOI] [PubMed] [Google Scholar]
- Kroemer G.; El-Deiry W. S.; Golstein P.; Peter M. E.; Vaux D.; Vandenabeele P.; Zhivotovsky B.; Blagosklonny M. V; Malorni W.; Knight R. A.; Piacentini M.; Nagata S.; Melino G.; Cell Death N. C. (2005) Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 12(Suppl 2), 1463–1467. [DOI] [PubMed] [Google Scholar]
- Lazebnik Y. A.; Cole S.; Cooke C. A.; Nelson W. G.; Earnshaw W. C. (1993) Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J. Cell. Biol. 123, 7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A.; Boyce M.; Yuan J. (2003) A decade of caspases. Oncogene 22, 8543–8567. [DOI] [PubMed] [Google Scholar]
- Saraste A.; Pulkki K. (2000) Morphologic and biochemical hallmarks of apoptosis. Cardiovasc. Res. 45, 528–537. [DOI] [PubMed] [Google Scholar]
- Nagata S.; Nagase H.; Kawane K.; Mukae N.; Fukuyama H. (2003) Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 10, 108–116. [DOI] [PubMed] [Google Scholar]
- Betanzos-Lara S.; Liu Z.; Habtemariam A.; Pizarro A. M.; Qamar B.; Sadler P. J. (2012) Organometallic ruthenium and iridium transfer-hydrogenation catalysts using coenzyme NADH as a cofactor. Angew. Chem., Int. Ed. 51, 3897–3900. [DOI] [PubMed] [Google Scholar]
- Kerr J. F.; Winterford C. M.; Harmon B. V. (1994) Apoptosis. Its significance in cancer and cancer therapy. Cancer 73, 2013–2026. [DOI] [PubMed] [Google Scholar]
- Dai Y.; Lawrence T. S.; Xu L. (2009) Overcoming cancer therapy resistance by targeting inhibitors of apoptosis proteins and nuclear factor-kappa B. Am. J. Transl. Res. 1, 1–15. [PMC free article] [PubMed] [Google Scholar]
- Hileman E. A.; Achanta G.; Huang P. (2001) Superoxide dismutase: an emerging target for cancer therapeutics. Expert Opin. Ther. Targets 5, 697–710. [DOI] [PubMed] [Google Scholar]
- Jungwirth U.; Kowol C. R.; Keppler B. K.; Hartinger C. G.; Berger W.; Heffeter P. (2011) Anticancer activity of metal complexes: involvement of redox processes. Antioxid. Redox Signaling 15, 1085–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R. H.; Monks A.; Alley M. C.; Scudiero D. A.; Fine D. L.; McLemore T. L.; Abbott B. J.; Paull K. D.; Mayo J. G.; Boyd M. R. (1988) Development of human tumor cell line panels for use in disease-oriented drug screening. Prog. Clin. Biol. Res. 276, 265–286. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.