Abstract
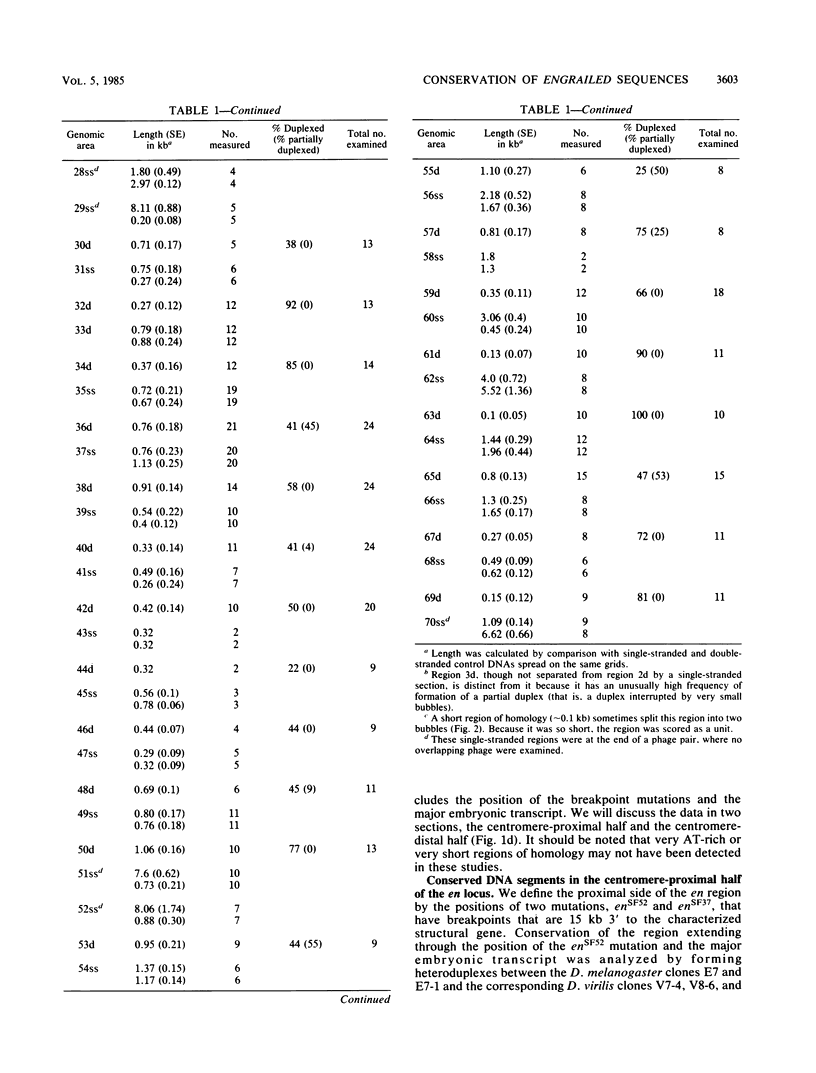
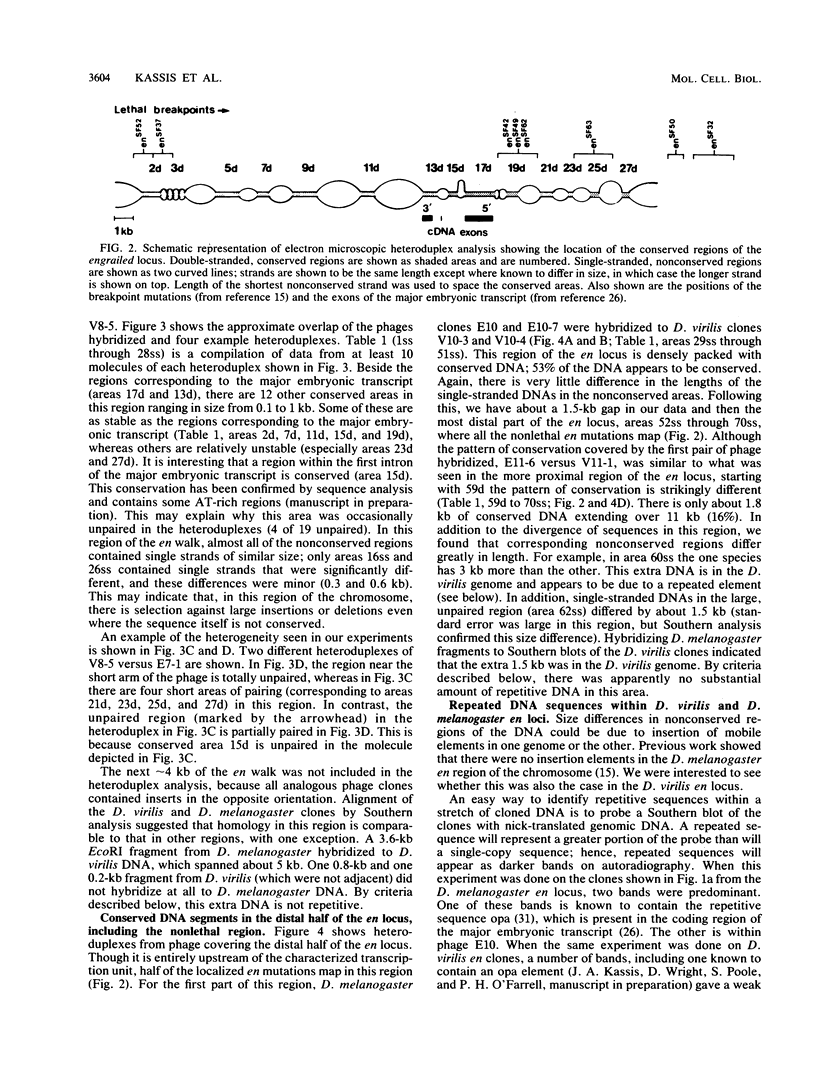
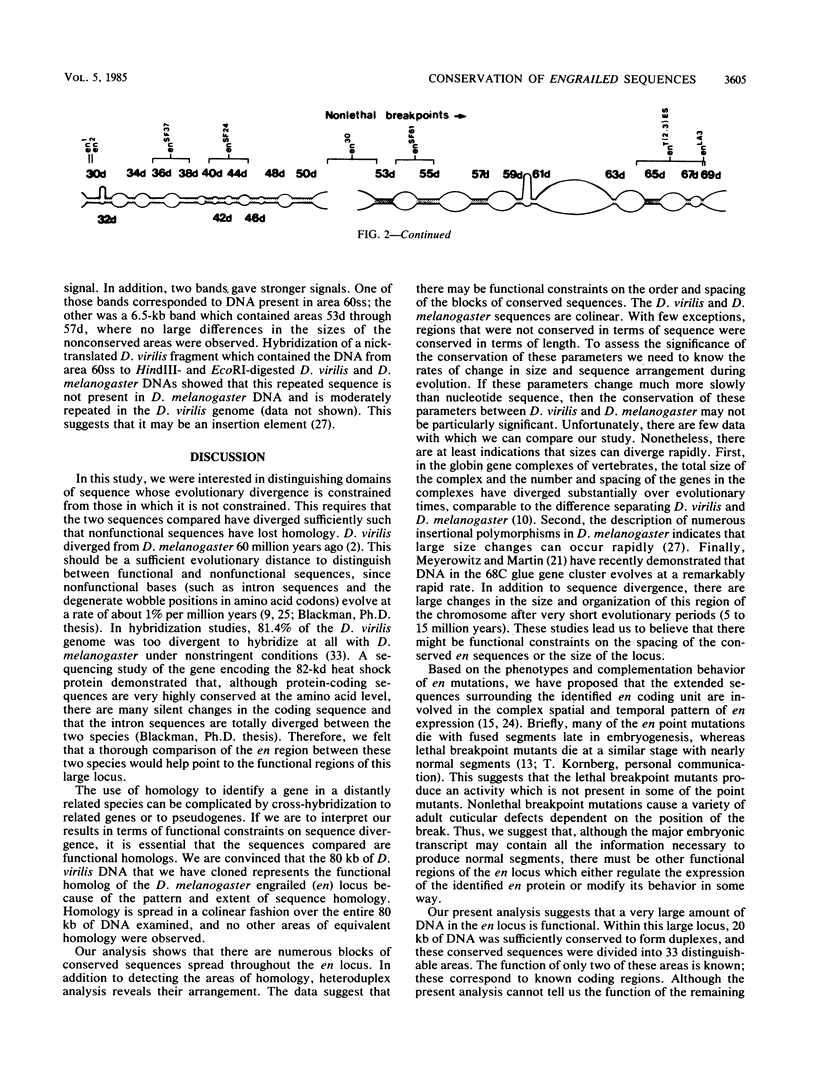
Physical localization of mutations in the engrailed (en) gene suggested that at least 70 kilobases (kb) of genomic sequences contribute to the normal function of this gene. Molecular characterization has suggested that en function is encoded in a small, 4.5-kb primary transcript. To identify functional regions within the 70 kb of the en locus of D. melanogaster, we identified sequences conserved in the D. virilis genome (estimated divergence time, 60 million years). Based on homology to D. melanogaster, we isolated en DNA from a D. virilis genomic library. Electron microscopic heteroduplex analysis indicated that in 70 kb there is 20 kb of conserved DNA in 33 different regions dispersed throughout the en locus, including two which encode parts of the major embryonic transcript. The conserved regions are in the same linear order and are spaced by similar lengths of nonconserved sequences in the D. virilis and D. melanogaster DNAs. What functional constraints have enforced conservation of sequences throughout the entire 70 kb and protected the region from divergence of size and arrangement? Our working hypothesis is that sequences necessary for the complex spatial and temporal pattern of en expression are dispersed throughout the 70-kb en locus and that selection for proper regulation restricts evolutionary divergence.
Full text
PDF


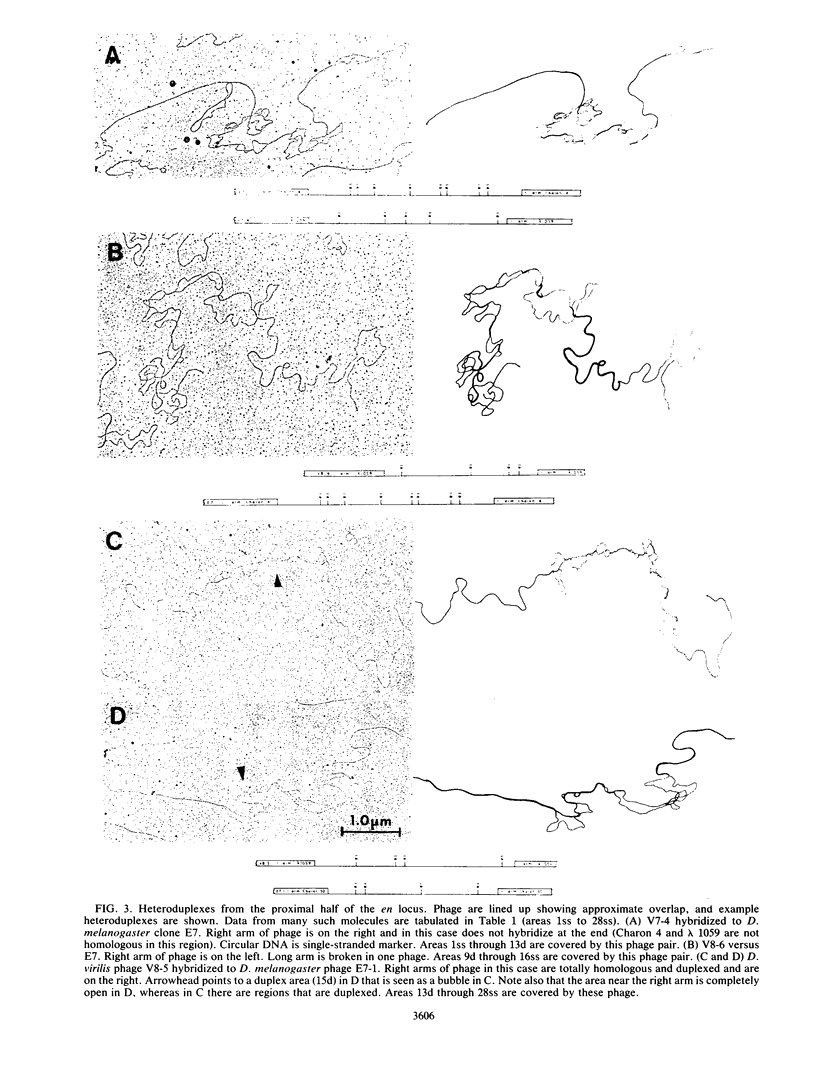
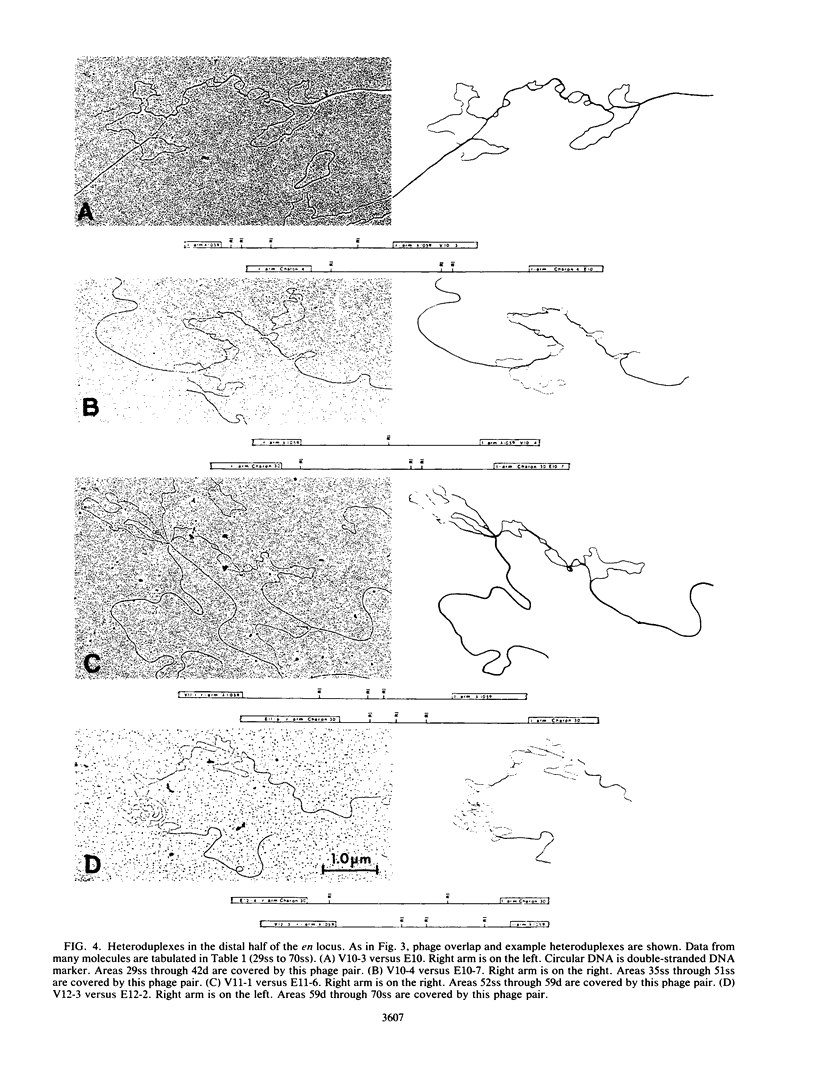


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Carramolino L., Cabrera C. V., Ruíz-Gómez M., Villares R., Boronat A., Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985 Feb;40(2):327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Carrasco A. E., McGinnis W., Gehring W. J., De Robertis E. M. Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes. Cell. 1984 Jun;37(2):409–414. doi: 10.1016/0092-8674(84)90371-4. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Fjose A., McGinnis W. J., Gehring W. J. Isolation of a homoeo box-containing gene from the engrailed region of Drosophila and the spatial distribution of its transcripts. Nature. 1985 Jan 24;313(6000):284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- Hafen E., Kuroiwa A., Gehring W. J. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984 Jul;37(3):833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- Hayashida H., Miyata T. Unusual evolutionary conservation and frequent DNA segment exchange in class I genes of the major histocompatibility complex. Proc Natl Acad Sci U S A. 1983 May;80(9):2671–2675. doi: 10.1073/pnas.80.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller B., Delius H. Intervening sequences in chloroplast genomes. Cell. 1984 Mar;36(3):613–622. doi: 10.1016/0092-8674(84)90341-6. [DOI] [PubMed] [Google Scholar]
- Kornberg T. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T., Sidén I., O'Farrell P., Simon M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell. 1985 Jan;40(1):45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Kuner J. M., Nakanishi M., Ali Z., Drees B., Gustavson E., Theis J., Kauvar L., Kornberg T., O'Farrell P. H. Molecular cloning of engrailed: a gene involved in the development of pattern in Drosophila melanogaster. Cell. 1985 Aug;42(1):309–316. doi: 10.1016/s0092-8674(85)80126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P. A., Morata G. Compartments in the wing of Drosophila: a study of the engrailed gene. Dev Biol. 1976 Jun;50(2):321–337. doi: 10.1016/0012-1606(76)90155-x. [DOI] [PubMed] [Google Scholar]
- Levine M., Rubin G. M., Tjian R. Human DNA sequences homologous to a protein coding region conserved between homeotic genes of Drosophila. Cell. 1984 Oct;38(3):667–673. doi: 10.1016/0092-8674(84)90261-7. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984 Jun;37(2):403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M., Martin C. H. Adjacent chromosomal regions can evolve at very different rates: evolution of the Drosophila 68C glue gene cluster. J Mol Evol. 1984;20(3-4):251–264. doi: 10.1007/BF02104731. [DOI] [PubMed] [Google Scholar]
- Morata G., Lawrence P. A. Control of compartment development by the engrailed gene in Drosophila. Nature. 1975 Jun 19;255(5510):614–617. doi: 10.1038/255614a0. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., Scalenghe F., Kaufman T. C. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983 Dec;35(3 Pt 2):763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B. T., Kaufman T. C. Analysis of larval segmentation in lethal genotypes associated with the antennapedia gene complex in Drosophila melanogaster. Dev Biol. 1981 Jan 15;81(1):51–64. doi: 10.1016/0012-1606(81)90347-x. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Zwiebel L. J., Cohn V. H., Wright D. R., Moore G. P. Evolution of single-copy DNA and the ADH gene in seven drosophilids. J Mol Evol. 1982;19(1):62–71. doi: 10.1007/BF02100224. [DOI] [PubMed] [Google Scholar]