Abstract
Chromatin immunoprecipitation followed by sequencing with next-generation technologies (ChIP-Seq) has become the de facto standard for building genome-wide maps of regions bound by a given transcription factor (TF). The regions identified, however, have to be further analyzed to determine the actual DNA-binding sites for the TF, as well as sites for other TFs belonging to the same TF complex or in general co-operating or interacting with it in transcription regulation. PscanChIP is a web server that, starting from a collection of genomic regions derived from a ChIP-Seq experiment, scans them using motif descriptors like JASPAR or TRANSFAC position-specific frequency matrices, or descriptors uploaded by users, and it evaluates both motif enrichment and positional bias within the regions according to different measures and criteria. PscanChIP can successfully identify not only the actual binding sites for the TF investigated by a ChIP-Seq experiment but also secondary motifs corresponding to other TFs that tend to bind the same regions, and, if present, precise positional correlations among their respective sites. The web interface is free for use, and there is no login requirement. It is available at http://www.beaconlab.it/pscan_chip_dev.
INTRODUCTION
The regulation of eukaryotic gene transcription is a complex process, which depends on interactions between transcription factors (TFs) and their binding sites on DNA (TFBSs), as well as on chromatin structure and other epigenetic factors like DNA methylation (1). Chromatin immunoprecipitation (ChIP), followed by the application of next-generation sequencing technologies to the immunoprecipitated DNA (ChIP-Seq), permits to build genome-wide maps of protein–DNA interactions, like TF binding, histone modifications or any other protein involved in the process (2).
The typical output of a ChIP-Seq experiment for a TF (the ChIP’ed TF, hence, on) is a list of thousands of genomic regions with associated enrichment scores and corresponding P-values or false discovery rates. The plot of the enrichment (sequence read density) against the genome shows for these regions a ‘peak’ shape (Supplementary Figure S1), usually with a clear local maximum (‘summit’) point (3). As these regions may range in length from hundreds to thousands of base pairs (bp), further bioinformatic sequence analysis is still required to identify the actual TF-binding sites within them (3), either by de novo motif discovery (4) or by using descriptors (5–8) like position weight matrices [PWMs or motif profiles, (9)] available in databases like TRANSFAC (10) and JASPAR (11). These methods usually assess motif enrichment by comparing motif counts in the regions with background random models, which are essential to obtain reliable significance measures. However, the resolution offered by modern next-generation sequencing technologies is such that binding sites for the ChIP’ed TF are typically located in close proximity of the summit points. Motif discovery and enrichment analyses can take advantage from this fact, confining the analysis to 100–150 bp around them (3) or including positional bias in the measures used to assess motif significance (4). On the other hand, detecting not only the sites bound by the ChIP’ed TF but also secondary motifs for putative co-factors is a point of the utmost importance, as reported by recent large-scale studies (12,13) that unveiled widespread interactions among TFs and precise arrangements of their binding sites.
Indeed, the latest methods using PWMs designed for ChIP-Seq, like CENTDIST (14) and CentriMo (15) altogether bypass the definition of a suitable ‘genome-wide’ background, and they do not evaluate actual motif enrichment in the input regions, but rather ‘maximum central enrichment’. That is, they determine whether putative binding sites described by a given PWM matching it above a given or estimated threshold value tend to cluster toward the center of the regions, and if the clustering is significant assuming a random uniform distribution. However, in this way, actual enrichment of the motifs in the regions with respect to the rest of the genome is not explicitly evaluated, making harder to assess, in case of different motifs presenting the same bias, which one can be considered to be the most likely candidate to describe the binding of the TF studied and/or its most significant co-factors. The latter could in fact be present only in a limited subset of the input regions (and, hence, the positional bias because of a limited number of motif instances), or vice versa, secondary motifs actually enriched might show a much less marked clustering around the peak summit and thus deemed to be less significant.
MATERIALS AND METHODS
To address the issues just discussed, we developed PscanChIP, based on our previous tool for promoter analysis Pscan (5). In PscanChIP we introduce different criteria to assess motif overrepresentation and positional bias in analysis of ChIP-Seq peak regions. Our idea is that motifs actually corresponding to TF binding should be first of all overrepresented in the ChIP-Seq regions with respect to the rest of the genome (that is, the portion of the genome accessible for TF binding), and the same should hold true for motifs corresponding to other TFs interacting with the ChIP’ed one in a significant number of regions. Also, matches to a given PWM should present not only a positional bias, but also yield higher scores in the preferred positions, that is, they should better fit the matrix in the positions where they tend to cluster.
PscanChIP takes as input a set of genomic coordinates corresponding to ChIP-Seq peaks, assuming that they are centered on their summits and considers the 150 bp genomic regions around their center. The regions are scanned on both strands by PWMs available in the JASPAR and TRANSFAC databases, and optionally with additional PWMs that can be submitted by users. The scan returns for each oligo in the input regions a score between 0 and 1 (Supplementary Methods), and the best (highest scoring) oligo is selected from each input region. As in the original Pscan, we bypass the need to define matching thresholds for PWMs to predict likely TFBS instances, comparing instead for each matrix the mean score of the best-matching oligos in the input regions to expected values defined according to different backgrounds. TFs mostly bind DNA in accessible regions (12), and genome-wide maps of open chromatin can be built through experiments like DNaseI- or FAIRE-Seq (16). The result is a collection of segments of DNA that are accessible to regulatory factors and other DNA interacting molecules. Thus, given a PWM, we compare the matching scores on the input sequences with an expected background value computed by considering the matching scores in a collection of DNA accessible regions of the same length. In particular, we used for this task the ENCODE regions available at the UCSC Genome Browser database (17), which have been identified through DNaseI Digital Genomic Footprinting (18). The regions differ according to the tissue/cell line studied, and thus yield cell-line–specific expected values. Therefore, as input, users have to select also the cell line/tissue on which their ChIP experiment has been performed, or the closest relative in the list of available possibility (e.g. HepG2 for liver cells). But, if no suitable choice is available, we also included a ‘mixed’ background, made by a sample of non-overlapping accessible regions built by taking at random a subset of the regions from each of the cell lines available, and a ‘promoter’ background, to be used if the input comes mostly from promoter regions. Each of the background sequence sets (cell-line specific or mixed) is made of ∼200 000 regions.
Once the matching scores for a PWM are available both in the input and the background sequence sets, global enrichment can be computed, by comparing with a t-test average and standard deviation of the best-match score in the input regions with average and standard deviation of best matches on the background sequence set, yielding enrichment P-values with a two-tailed t distribution. Global enrichment can be used to identify motifs that, in general, are overrepresented in the regions, but with no assumption on their location within the regions or any positional bias with respect to the peak summits. Sites for the ChIP’ed TF should anyway be the most significantly enriched according to this measure, and also PWMs for other ‘co-regulating’ TFs, that is, binding a significant number of the input regions, should yield low P-values.
Also, PscanChIP evaluates local enrichment, comparing with a t-test mean and standard deviation of the score of the best matching oligos in the input regions to mean and standard deviation of the best match in the genomic regions flanking the input ones. Local enrichment can be used to identify motifs with significant preference for binding within the regions, that is, the motif corresponding to the ChIP’ed TF, as well as other TFs likely to interact with it and binding in its neighborhood. It should be noticed that with respect to similar methods, local enrichment not only assesses positional bias but also how well the matches fit the profile used, without establishing a pre-determined matching threshold. Finally, PscanChIP evaluates motif positional bias within the input regions, by splitting them into overlapping sub-regions of 10 bp. Mean and standard deviation for the best match score within each sub-region are compared with mean and standard deviation across all the sub-regions, again with a t-test. Once again, motifs showing a positional bias, but more importantly having their best matches associated with a given sub-region, can be singled out. The bias can be for the center (as usually shown for the sites of the ChIP’ed TF), but it can be identified for any other sub-region. Further details on the calculations performed can be found in the Supplementary Methods.
These different measures can detect different types of overrepresented motifs. The PWM corresponding to the ChIP’ed TF or to the co-factor(s) recruiting it should be the highest ranking (lowest P-values) with respect to all the three different measures. In case of ChIP-Seq experiments with low resolution (e.g. with low quantities of IP’ed DNA or a weakly specific antibody) and without sharp peak summits, the global P-value should be nevertheless low. PWMs corresponding to possible interactors (or competitors) of the ChIP’ed TF (often, but not necessarily always, binding DNA in its neighborhood) should have low global and local P-values, but rank after the ChIP’ed TF. Positional bias could also appear for these—but not as a rule. ‘Co-regulators’ (TFs that often bind the same promoters/enhancers of the ChIP’ed TF) tend to regulate the same genes, but do not have to bind cooperatively with it, and should have low global P-values but not necessarily low local P-values and no positional bias. On the other hand, PWMs with low local P-values only (better if substantiated by positional bias) might correspond to TFs interacting with the ChIP’ed one only in a limited subset of the input.
THE USER INTERFACE
Input
Users have to submit a list of genomic coordinates in BED format (e.g. chromosome-start-end) corresponding to ChIP-Seq peak regions. PscanChIP assumes that the regions are centered on the point of maximum enrichment within the peak and will automatically retrieve and analyze the 150-bp sequences around that point (Supplementary Methods). There is no limit on the number of regions that can be input. Optionally, if regions are input by users sorted in decreasing order according to region enrichment and/or significance P-value (the most enriched at the top), PscanChIP will also assess the correlation between the enrichment of a motif and the enrichment of the regions in the ChIP-Seq.
Input parameters that have to be set are the organism (human or mouse at the moment) and the genome assembly that was used for the ChIP-Seq analysis. The background field is used to select the genomic background to be used to assess global enrichment of motifs as explained in the previous section. Finally, the ‘Select Descriptors’ option specifies whether the analysis has to be performed with profiles available in the JASPAR or TRANSFAC (public release) databases, and ‘Additional Descriptors’ permits to upload a specific matrix set that will be added to the chosen database (see the online help page for further details on the custom matrix format).
The time required by computations depends on the number of regions and of matrices involved in the analysis, anyway seldom exceeding a couple of minutes on typical TF ChIP-Seq experiments of several thousands of regions.
Output
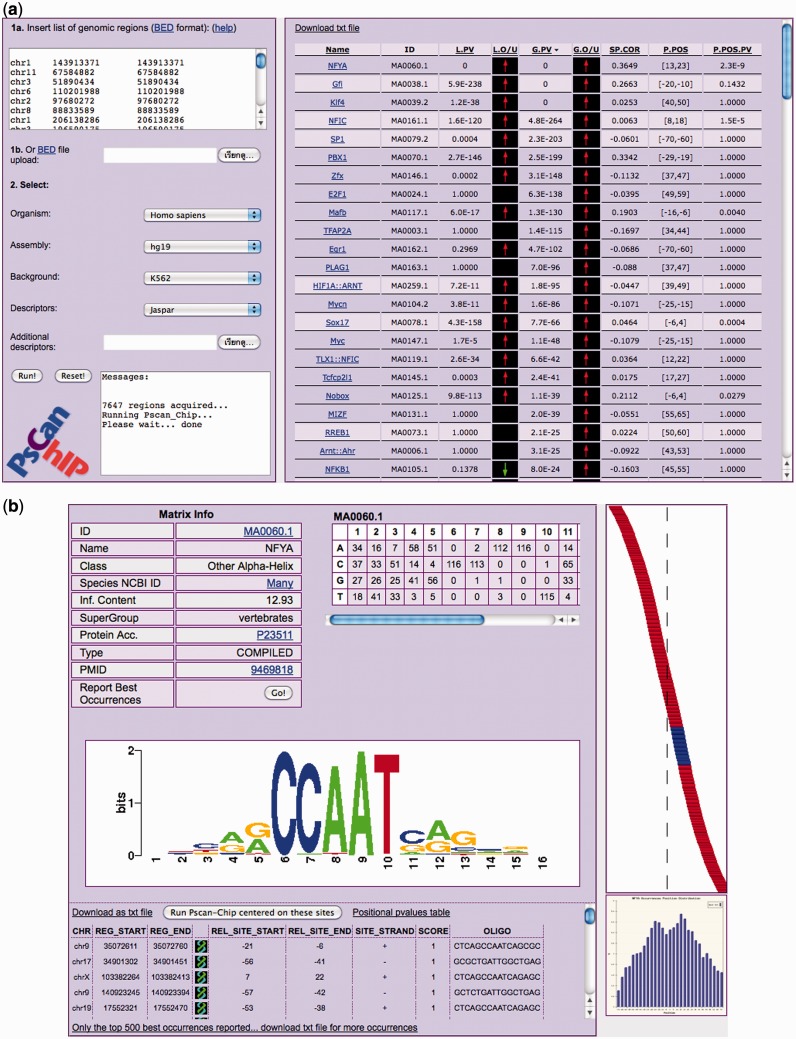
The output will appear in the middle of the page. For each matrix of the selected set and the custom matrices (if any), PscanChIP outputs, as shown in Figure 1a:
Name and ID: the name and the database ID of the matrix.
Local enrichment P-value (L.PV): describing whether the motif is over- or underrepresented in the 150 bp input regions with respect to the genomic regions flanking them, with an arrow (L.O/U) indicating whether the motif is over- (red upward) or underrepresented (green downward).
Global enrichment P-value (G.PV): describing whether the motif is over- or underrepresented in the input regions with respect to the global background used, with the respective arrow (G.O/U). Notice that for computation reasons, this value is not calculated for user-submitted matrices.
Spearman correlation coefficient (SP.COR): if the input regions were ranked according to their enrichment, this value represents the Spearman rank correlation coefficient between the ranking of the input regions and their ranking with respect to the motif. Positive values indicate that the more enriched regions are the best matches they contain for the motif, and vice versa.
Preferred position (P.POS) and position bias P-value (P.POS.PV): this indicates the position (10 bp sub-region) within the input regions where oligos tend to be found with best matches to the matrix. Coordinates are relative to the center of the regions, which has coordinate 0. The associated P-value indicates the significance of the positional bias of the motif.
Figure 1.
The overall output of PscanChIP (a), and detailed NFYA PWM output (b) for the ENCODE ChIP-Seq of NF-YA in K562 cells.
The output table can be sorted according to any column value by clicking on the corresponding header.
Clicking on a matrix name (NFYA in Figure 1a), opens a detailed output page for it (Figure 1b). This page permits to retrieve, for each region submitted, its genomic coordinates and the best-matching oligo, its score, and its position and strand, with respect to the region itself (0 is the center of the region). It also includes a simple graphical representation of the location of the best matching oligos within the regions, as well as the corresponding histogram. Notice that the mostly enriched sub-region according to the P-value does not have to correspond to the local maximum sub-region of the histogram. Rather, disagreement between those two measures is an indicator of low-quality sites clustering in the histogram.
Performing a motif-centered analysis
The ‘Run Pscan-Chip centered on these sites’ button associated with the occurrences list of any PWM permits to start automatically a motif-centered analysis on the PWM itself. For all the regions containing a best match with score higher than the global background average, PscanChiP will process the 150 bp genomic regions around the matching oligo.
Similar computations are performed, e.g. by the SpaMo tool (19), but in our implementation, positional correlations between PWMs are incorporated and integrated with other types of enrichment analysis, overall providing a more complete picture.
The computations of the motif-centered analysis and its output are identical to the default one, with the sole difference that for the matrix chosen (whose best matches fall exactly in the middle of the regions analyzed) PscanChIP will instead report in the detailed output page the position of the second best match, to detect whether there is preferential arrangements of sites for the same TF.
RESULTS
We ran PscanChIP on several different data sets retrieved from literature and compared the results with other similar tools. The full results are available in Supplementary Material and can be reproduced by running PscanChIP on the samples included in the web interface. All the parameters required will be set automatically.
‘Core’ TFs in mouse embryonic stem cells
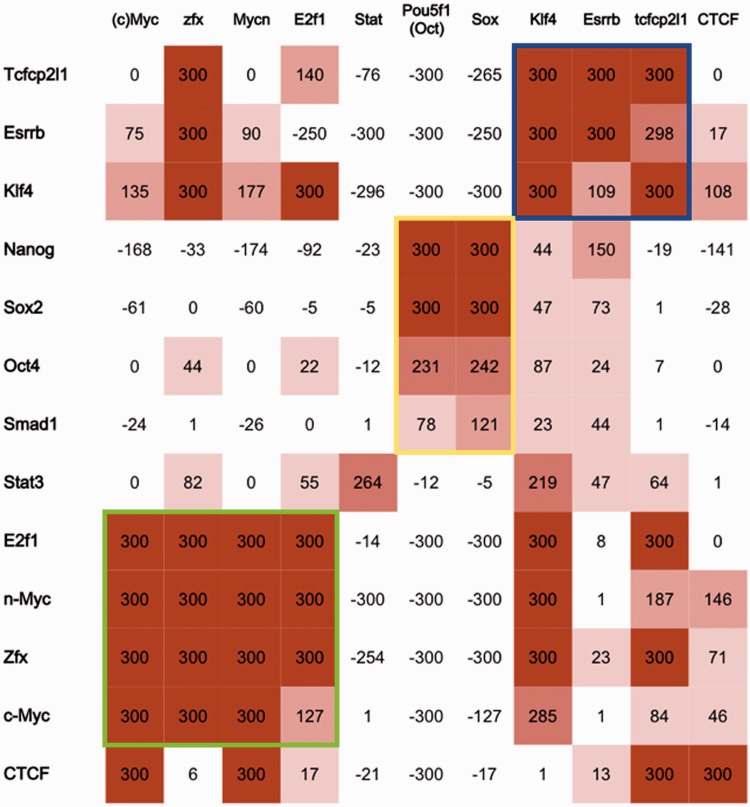
A seminal work showing the full potential of ChIP-Seq is the analysis of 13 different ‘core’ TFs in mouse embryonic stem cells [Nanog, Oct4, STAT3, Smad1, Sox2, Zfx, c-Myc, n-Myc, Klf4, Esrrb, Tcfcp2l1, E2F1 and CTCF (20)]. Once the binding regions on the genome for each TF had been determined, further processing identified binding correlations among the different TFs, forming two main ‘multi transcription factor loci (MTL)’, the first always containing c-Myc and/or n-Myc-binding sites, but not Nanog, Oct4 and Sox2, and vice versa the second.
This data set represents a good benchmark to assess whether the global enrichment measure used by PscanChIP is able to retrieve the same correlations obtained in the original study. The matrix for the main TF investigated was singled out in each experiment as the most enriched both locally and globally, with a marked positional bias for the summit location, showing the reliability of the enrichment measures we used (see Supplementary Table S1 for the full results), whereas for Nanog and Smad1, a PWM for the corresponding TF is not available either in JASPAR or in the free version of TRANSFAC included in the interface. Moreover, although we cannot expect the results to be symmetrical, that is, the enrichment for motif X in the ChIP for factor Y will be different from the enrichment of motif Y in the ChIP of X, matrices corresponding to TFs co-binding in a significant number of loci can be anyway singled out consistently in the ChIP of their partners, with global enrichment P-values reproducing the correlations reported in the study [see Figure 2 and compare with Figure 4A in (20)]. The two MTLs can be clearly identified, one comprising n-Myc, c-Myc, Zfx and E2F1 and the other Oct4, Sox2, Nanog and Smad1 (in the latter two the Oct4 and Sox2 matrices are highly enriched), and also the high correlations among Esrrb, Klf4 and Tcfcp2l1. Also, the Myc PWMs are underrepresented in the Oct/Sox MTL regions, and vice versa. The STAT3 motif, on the other hand, shows little enrichment in all the data sets, confirming its lower correlation to the other experiments shown in the study. The sole exception is the E2F1 regions, in which, as also previously observed (15), E2F-like matrices lack marked central enrichment and positional bias. On the same data set, CENTDIST (run on the best 10 000 regions included in the web interface, which is the maximum number of regions it can process) ranks E2F the second place after SP1, but using a weakly conserved SP1-like non-canonical motif (SSGCSS), whereas for PscanChIP also the canonical JASPAR E2F1 site (TTTGGCGC) has very low global and local P-values, but no relevant positional bias. In this data set, E-box–like motifs (c-Myc and n-Myc) are among the highest ranking ones for PscanChIP (global P-value = 0), with also positional bias toward the center of the regions, whereas in CENTDIST, they are ranked at the seventh place. On the other hand, these ChIP-Seqs show a significant overlap between E2F1 and/or n-Myc- and c-Myc-bound sites in ESC cells (35–45% of Myc sites falling within 100 bp from an E2F1 site). To further substantiate the hypothesis that an E-box motif could be required for E2F1 binding, at least in some contexts, we applied PscanChIP to the ENCODE E2F1-binding sites in HeLa cells retrieved from the UCSC Genome Browser database (17). Once again, the two highest ranking motifs are E2F1 (canonical) and the E-box (n-Myc or c-Myc), with the lowest global and local P-values simultaneously, and both showing a positional bias toward the summit (Supplementary Table S1). CENTDIST again reports SP1 as the most enriched motif, ranking the canonical E2F1 motif at the fourth place, and E-boxes around the 10th. YY1, identified by CentriMo as the more likely candidate for the recruitment of E2F1 (15), presents only marginal local enrichment. All in all, our results hint to possible recruitment of E2F1 by one of the E-box–binding factors, as indeed has already been demonstrated for c-Myc (21). Finally, in the CTCF sample (>30 000 regions), we identified again the E-box as a possible co-binding motif, confirming its preference for c/n-Myc MTLs and recovering already known possible associations between CTCF and MYC (22). The same test could not be replicated with CENTDIST because of the limit of 10 000 regions allowed as input.
Figure 2.
Heatmap of PscanChIP global P-values associated with the JASPAR PWMs of core TFs in mouse ESC peak regions (positive and negative values denote over- and underrepresented motifs, respectively). Rows correspond to ChIP-Seq experiments, columns to motifs. MTLs reported previously (20) are highlighted in green (c-Myc/n-Myc) and yellow (Nanog/Oct4/Sox2).
STAT3 binding in four different cell lines
In a recent work (23), binding regions of STAT3 have been determined through ChIP-Seq in four different mouse cell lines [embryonic stem cells (ESC), CD4+ T cells, macrophages and AtT-20 cells (PEC)]. Analysis of the regions through motif discovery, protein–protein interaction data and overlap with other ChIP-Seq experiments revealed distinct transcription regulatory modules, with different co-factors singled out to be essential for cell-line–specific binding of STAT3. PscanChIP was able to identify STAT motifs as the most enriched with respect to every measure used (globally, locally and with positional bias) in the ESC and AtT-20 data sets (Supplementary Table S2). In CD4+, STAT motifs are the most enriched globally, have a local P-value near 0 and are the only ones presenting positional bias, whereas CENTDIST ranks them at the second place. In this cell line, IRF and GATA motifs are the most likely candidates to represent main interactors of STAT3 [confirming the findings of (23)], and indeed, the analysis motif centered on STAT3 shows their preferential arrangement immediately upstream or downstream (Supplementary Figure S2). On the other hand, on the ESC data set, PscanChIP confirmed Esrrb as a possible interactor, whereas OCT4 and Sox2, identified in the study as essential co-factors, did not show any significant global or local motif enrichment and a moderate enrichment for CENTDIST with a low number of estimated regions containing the motif. The fact that PWMs for other ESC core TFs like Klf4 and Tcfcp2l1 are more enriched both globally and locally (the latter confirmed also by CENTDIST) leads us to conjecture that other factors could be involved in the recruitment of STAT3, or that the respective binding motifs are not essential for the function of Oct4 and Sox2. As described previously (23), on the data set comprised by regions in any cell line shared with other ChIP-Seq experiments (‘any three’), PscanChIP is able to retrieve as significantly enriched also E-box motifs (c-Myc or n-Myc).
Notably, in the PEC macrophage data set, composed of 1300 regions, PscanChIP could not find any ‘dominating motif’ (i.e. ranking first for all the measures used as in the previous examples). However, the only motifs with significantly low P-value (<10−5) both for global and local enrichment were those corresponding to STAT matrices, even if with no clear positional bias toward the region summits. We think that the result might be due to experimental issues in the ChIP-Seq experiment or in the peak calling pipeline, because in the other cell lines STAT binding could be clearly determined, and most importantly in these regions no other motif could be detected showing both global and local enrichment, differently from virtually all the other published summit-centered data sets we analyzed with PscanChIP. The fact that the study reports that >70% of these regions do not contain any variant of the annotated STAT3 motifs seems to further consolidate our hypothesis. On these regions, CENTDIST ranked STAT matrices at the third place, with a P-value of 0.002, because of the moderate bias of STAT sites toward the center of the regions. This example shows how using different measures of enrichment can help solve cases in which there is no clear signal emerging from the data, and to point out possible issues either in the ChIP or in the downstream bioinformatic analysis. Finally, a ‘core’ set of 32 binding regions was determined, conserved across the four cell lines, which co-regulates a set of key genes important for STAT3 function in multiple cell types. These 400-bp regions were not summit centered, as they were derived from the intersection of four different experiments with different summits. On this small data set, PscanChIP reveals again STAT profiles as the most locally and globally enriched, with, among others, E2F1 (shown to be needed to recruit STAT3 to these genomic loci), MYC and KLF4 as putative co-regulators. This example shows the robustness of the enrichment measures we used even in small and not summit-centered data sets. On this example, CENTDIST fails to find any enriched matrix, all the profiles output having P-values of 0.5.
NF-Y binding in human K562 cells
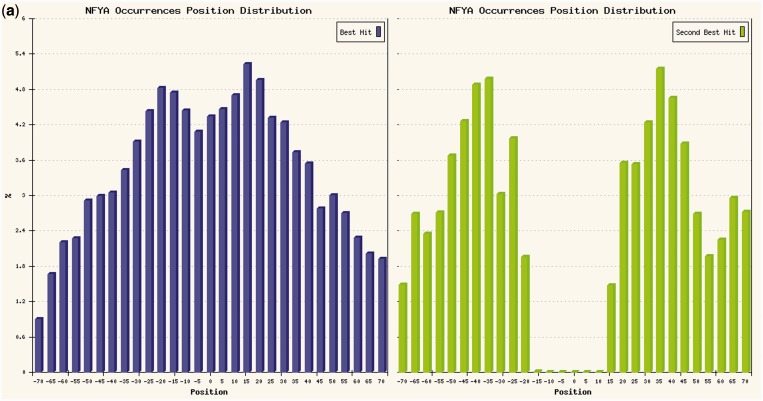
In this experiment, we analyzed the list of ENCODE peaks for NF-YA in K562 cells retrieved from the UCSC Genome Browser database (17). The output, also shown in Figure 1a, lists several JASPAR matrices with low P-values, and expectedly the one associated with NF-YA (NFYA, CCAAT-box) is the highest ranking one for all the measures. Moreover, as the input was ranked according to enrichment, it shows a positive Spearman correlation (0.36) with the enrichment in the ChIP. In the detailed output page for NFYA (Figure 1b), the histogram in the bottom right corner shows a clear preference for the center of the regions, but with a bimodal distribution (Figure 3a, left), mostly because of the fact that NF-Y often binds DNA in two consecutive CCAAT boxes at a preferred distance of ∼33 bp.
Figure 3.
(a) Histogram of the best motif occurrences of the NFYA PWM in ChIP-Seq regions for NF-YA in K562 cells (left), and of the second-best occurrences in regions centered on the NFYA motif (right). (b) Relative positioning of other motifs in regions for NF-YA centered on the NFYA motif.
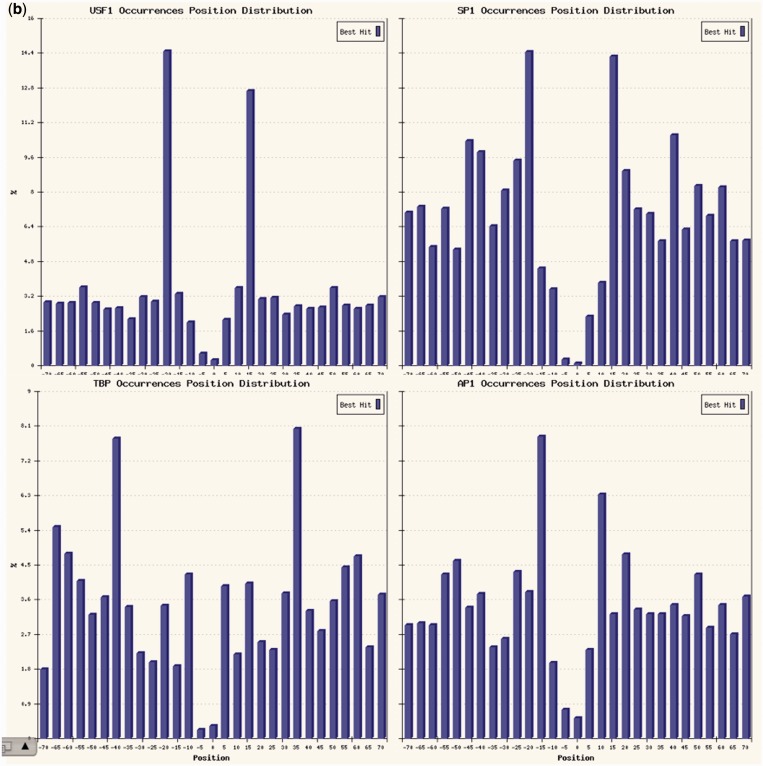
The fact that there are several other matrices with low local and global P-values is an indicator that there might be other TFs binding cooperatively, each in a significant subset of the regions. Among these, AP1 (bound by the FOS-JUN complex) shows only a low local P-value but no global enrichment, whereas for CENTDIST it does not have any significant enrichment. Indeed (Supplementary Table S3 and histograms in Figure 3) results of the analysis centered on the NFYA motif show that there are other matrices presenting—other than local or global enrichment—also marked positional bias for sub-regions not corresponding to the center (which is occupied by NF-Y itself), like E-boxes (Myc, MAX, USF and so on), TBP, SP1 and also AP1 PWMs (Figure 3b). Also, the second-best NF-Y motif occurrences have a clear preference for a 30–35 bp distance with respect to the central CCAAT box (Figure 3a, right). Indeed, sites predicted for AP1 could be further ‘validated’ by regions bound by FOS in K562 cells in the respective ChIP-Seq experiment, showing a correlation between sites for the two factors in LTR repeated regions [see (24), also for further discussion on the other motif arrangements found with respect to the CCAAT box]. The AP1-NFY relationship is an example of a correlation resulting from a limited number of secondary sites that are not enough to reach a significant global but only a marginal local enrichment, but when present have a precise positional correlation highlighted by the corresponding P-values. This latter case, however, should be thoroughly checked, to determine whether the lack of global enrichment is indeed because of the presence of just a low number of ‘high-quality’ sites (which can be confirmed by the positional P-value as in this example), or instead to a high number of ‘low-quality sites’, unlikely to be bound by the corresponding TF.
CONCLUSIONS
PscanChIP is a web server that given a set of ChIP-Seq enriched regions finds overrepresented TFBS motifs described by PWMs. Overrepresentation is evaluated according to different criteria, by adapting the statistical framework we introduced in our previous tool Pscan.
As other similar tools, PscanChIP can be used to identify the correct descriptor for the binding specificity of TFs investigated by ChIP-Seq experiments, but as we have shown in the examples can correctly single out additional motifs corresponding to other TFs cooperating with the ChIP’ed one, with or without taking advantage of positional arrangements of their respective binding sites within the regions. Indeed, the three different measures of enrichment can solve dubious cases, or help in presence of noisy data, and their combination can be used to identify general co-regulators (i.e. usually binding the same promoters or enhancers), or to discover precise positional correlations hinting a cooperative binding even in a limited number of the ChIP’ed regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–2 and Supplementary Methods.
FUNDING
Funding for open access charge: Italian Ministry of University and Research Fondo Italiano per la Ricerca di Base (FIRB) project ‘Laboratorio Internazionale di Bioinformatica’ (LIBI); Consiglio Nazionale delle Ricerche (CNR) flagship project EPIGEN.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the anonymous reviewers for their thorough comments, criticisms and suggestions, which helped us to improve the manuscript both in its substance and its form.
REFERENCES
- 1.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J. Cell. Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 2.Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zambelli F, Pesole G, Pavesi G. Motif discovery and transcription factor binding sites before and after the next-generation sequencing era. Brief. Bioinform. 2013;14:225–237. doi: 10.1093/bib/bbs016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hestand MS, van Galen M, Villerius MP, van Ommen GJ, den Dunnen JT, Hoen PA. CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes. BMC Bioinformatics. 2008;9:495. doi: 10.1186/1471-2105-9-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 10.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Chang CW, Goh WL, Sung WK, Cheung E. CENTDIST: discovery of co-associated factors by motif distribution. Nucleic Acids Res. 2011;39:W391–W399. doi: 10.1093/nar/gkr387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey TL, Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128. doi: 10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabo PJ, Hawrylycz M, Wallace JC, Humbert R, Yu M, Shafer A, Kawamoto J, Hall R, Mack J, Dorschner MO, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl Acad. Sci. USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitington T, Frith MC, Johnson J, Bailey TL. Inferring transcription factor complexes from ChIP-seq data. Nucleic Acids Res. 2011;39:e98. doi: 10.1093/nar/gkr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 21.Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008;27:4172–4179. doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]
- 22.Lee BK, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012;22:9–24. doi: 10.1101/gr.127597.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, Miranda-Saavedra D. Distinct transcriptional regulatory modules underlie STAT3's cell type-independent and cell type-specific functions. Nucleic Acids Res. 2013;41:2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y co-associates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013 doi: 10.1101/gr.148080.112. gr.148080.112 [pii] doi:10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]