Abstract
Marine mammals are often reported to possess reduced variation of major histocompatibility complex (MHC) genes compared with their terrestrial counterparts. We evaluated diversity at two MHC class II B genes, DQB and DRB, in the New Zealand sea lion (Phocarctos hookeri, NZSL) a species that has suffered high mortality owing to bacterial epizootics, using Sanger sequencing and haplotype reconstruction, together with next-generation sequencing. Despite this species' prolonged history of small population size and highly restricted distribution, we demonstrate extensive diversity at MHC DRB with 26 alleles, whereas MHC DQB is dimorphic. We identify four DRB codons, predicted to be involved in antigen binding, that are evolving under adaptive evolution. Our data suggest diversity at DRB may be maintained by balancing selection, consistent with the role of this locus as an antigen-binding region and the species' recent history of mass mortality during a series of bacterial epizootics. Phylogenetic analyses of DQB and DRB sequences from pinnipeds and other carnivores revealed significant allelic diversity, but little phylogenetic depth or structure among pinniped alleles; thus, we could neither confirm nor refute the possibility of trans-species polymorphism in this group. The phylogenetic pattern observed however, suggests some significant evolutionary constraint on these loci in the recent past, with the pattern consistent with that expected following an epizootic event. These data may help further elucidate some of the genetic factors underlying the unusually high susceptibility to bacterial infection of the threatened NZSL, and help us to better understand the extent and pattern of MHC diversity in pinnipeds.
Keywords: MHC, balancing selection, NGS, New Zealand sea lion, Phocarctos hookeri , epizootic
Introduction
The New Zealand sea lion (NZSL, Phocarctos hookeri), of the order Carnivora, suborder Pinnipedia, is the largest native and only seal endemic to New Zealand (Bryden et al., 1998). It is the rarest sea lion in the world, with an estimated population of between 8600 and 11300 individuals (Geschke and Chilvers, 2009). The species has suffered a 40% decline in pup production over the last decade (Chilvers, 2009) attributed largely to adult female mortality in fisheries bycatch (Robertson and Chilvers, 2011). In addition, population decline has been compounded by recurrent infection with bacterial epizootics, that have resulted in high levels of mortality of pups especially, but also adults. Such disease events have occurred three times since the first known event was observed in 1997. This episode resulted in mortality of 58% of pups in the 1997/1998 Austral summer breeding season, along with 70 adult females, and was attributed to infection by a Campylobacter bacterium (Baker, 1999). In 2001/2002 and 2002/2003, further seasons of unusually high mortality took place, with the death of 33% and 21% of pups born in these years, respectively, from infection with the opportunistic bacterium Klebsiella pneumoniae (Wilkinson et al., 2006). No cases of epizootic disease in the NZSL population resulting in mass mortality appear to be recorded before 1997/1998 season, although the isolation of the Auckland Islands may mean that any previous epizootics were undetected.
Before human occupation of New Zealand, NZSLs ranged from the far north of North Island, through Stewart Island and down to the sub-Antarctic, with colonies also present on the Chatham Islands (Childerhouse and Gales, 1998). Sea lions were hunted for food by Maori, and the NZSL was extirpated from the New Zealand mainland by 1826, following European settlement and commercial sealing activities (Childerhouse and Gales, 1998). In consequence, the NZSL range is now restricted mainly to the remote Auckland Islands where 71% of pup production occurs (Robertson and Chilvers, 2011) with a colony also present on Campbell Island (Gales and Fletcher, 1999; Childerhouse et al., 2005). Within the Auckland Islands group, Dundas Island and Enderby Island are the primary breeding sites (Figure 1; Gales, 1995). This low, highly geographically restricted breeding population, coupled with the decline in pup production over the last decade (Castinel et al., 2007; Chilvers, 2009), places the NZSL in jeopardy from stochastic demographic processes, with recruitment seemingly unable to exceed mortality (Wilkinson et al., 2006). Thus, the NZSL is listed as ‘vulnerable' by the IUCN (IUCN, 2008) and as ‘Nationally Critical' by the New Zealand Threat Classification Status (Baker et al., 2010). Vulnerability is exacerbated by on-going fisheries pressures (Robertson and Chilvers, 2011) and potential future epizootic events, which could further accelerate the decline of this species.
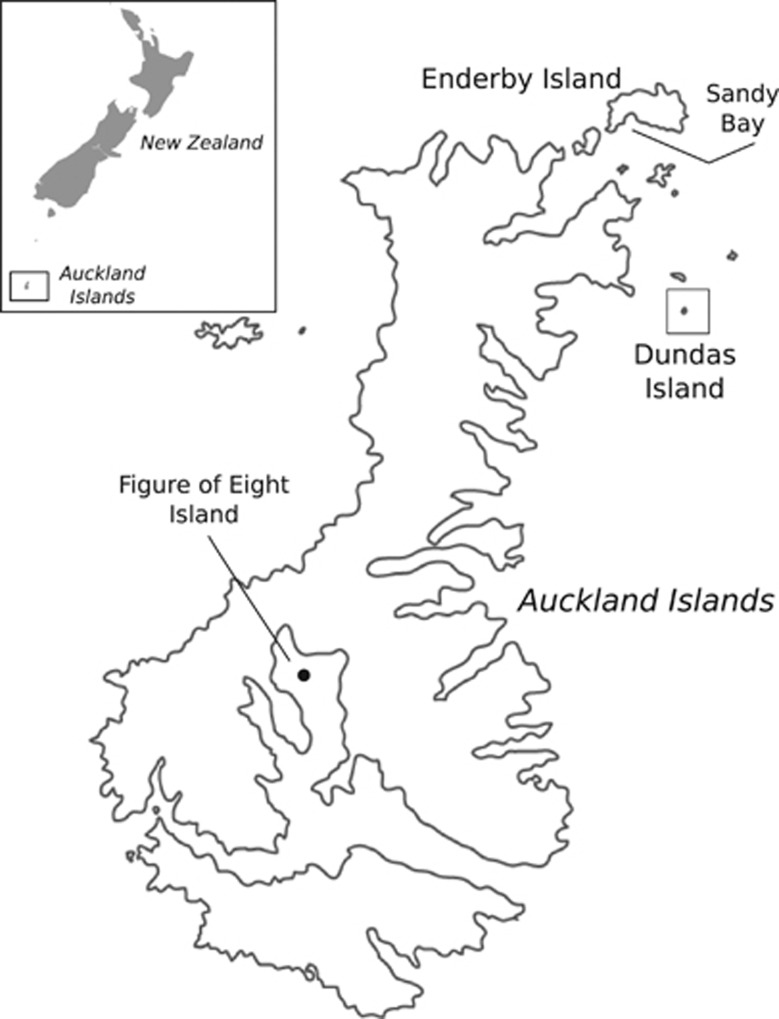
Figure 1.
Breeding distribution of the NZSL. There are three recognised breeding areas: two in the Auckland Islands, one on Campbell Island Motu Ihupuku and also a colony on the Otago Peninsula (Chilvers and Wilkinson, 2008). Sampling locations used in this study are Sandy Bay on Enderby Island, Figure of Eight Island, and Dundas Island.
Genes of the major histocompatibility complex (MHC) have an important role in disease resistance (Hedrick and Kim, 2000) and are among the most polymorphic in the vertebrate genome (O'Brien and Yuhki, 1999; Garrigan and Hedrick, 2003). They encode class I and class II proteins that bind intracellular and extracellular foreign peptides, respectively (Doherty and Zinkernagel, 1975; Jensen, 2007). Class II B loci contain peptide-binding regions encoded in their second exon (Hughes and Nei, 1989), and show high levels of variation in many species (Parham and Ohta, 1996), thought to be related to the region's direct involvement with the interaction and association of foreign peptides, which are then presented to the immune system for processing and disposal (Hughes and Nei, 1988). The level of variation seen at MHC loci generally exceeds that in other regions of the genome (Robinson et al., 2000) and the high ratio of non-synonymous to synonymous substitutions at MHC loci is strongly suggestive of a departure from neutrality (Garrigan and Hedrick, 2003). This comparatively high level of variation is thought to be maintained by balancing selection (Edwards and Hedrick, 1998; Arkush et al., 2002; Harf and Sommer, 2005), and is usually considered beneficial to the generation of a diverse T-cell repertoire (Dyall et al., 2000). This is because maintenance of variation at MHC loci can increase allelic variation and consequently increase the diversity of antigens presented to T cells, which may confer and enhance resistance to infectious diseases (Doherty and Zinkernagel, 1975; Potts and Slev, 1995).
MHC variation over generations is suggested to be driven mainly by pathogenic pressures (Hedrick, 2002) and sexual selection (Sommer et al., 2002). Other mechanisms such as mutation, recombination, gene conversion and drift may also affect the evolution of MHC genes, although their relative contributions to the evolution of these loci are uncertain (Richman et al., 2003; Castro-Prieto et al., 2011). In genetic systems that evolve under balancing selection, such as MHC, long-lasting trans-species polymorphism may occur, in which alleles in one species share their closest ancestry to alleles from another (Klein et al., 1998). Trans-species polymorphism among MHC alleles has been widely documented in mammals, including primates (Go et al., 2002; Huchard et al., 2006), ungulates (Schaschl et al., 2006; Radwan et al., 2007) and carnivores. Within carnivores, trans-species sharing of identical or similar alleles has been found in the families Felidae (Castro-Prieto et al., 2011 and references within), Ursidae (Wan et al., 2006; Goda et al., 2010; Kuduk et al., 2012) and Canidae (Seddon and Ellegren, 2002).
Levels of variation at MHC loci may be lower in endangered species that have experienced a prolonged reduction or ‘bottleneck' in population size, either historically or contemporarily (Hedrick et al., 1999; Bollmer et al., 2007; Eimes et al., 2011). Such a reduction in population size may reduce levels of genetic variability at key loci, including MHC, rendering these species more susceptible to novel pathogens and disease (Hedrick et al., 2001; Siddle et al., 2007). Reduced diversity at MHC genes has been reported for several marine mammal species (Slade, 1992; Murray et al., 1995). Low levels of diversity at MHC loci have been seen to affect the response of endangered species to infection (O'Brien and Evermann, 1988; Siddle et al., 2010); thus, the reduction in genetic variation through bottleneck effects could have additive negative effects on population persistence, potentially leading to an extinction vortex (Soule and Mills, 1998; Frankham, 2005). The population dynamics of the NZSL, which includes a long history of restricted distribution, a static or declining population size (Taylor, 1971; Wilkinson et al., 2006; Robertson and Chilvers, 2011) and a recent history of recurrent bacterial epizootic episodes (Baker, 1999; Wilkinson et al., 2006) leads one to question the possibility that levels of genetic variation may be adversely affecting this population (but see Robertson and Chilvers, 2011).
Some have alternatively predicted that MHC variation should not be unduly affected by the detrimental changes in genetic variation that occur after a population bottleneck, or those which are the consequence of long-term small population size (Richardson and Westerdahl, 2003; Jarvi et al., 2004). Others believe that genetic drift, which is heightened in small and/or bottlenecked populations, can affect balancing selection and may alter the maintenance of polymorphisms at MHC loci (Miller and Lambert, 2004; van Oosterhout et al., 2006; Mainguy et al., 2007), which can lower the potential for long-term persistence of the species. There is ample evidence for both arguments. MHC variation may not necessarily be disproportionately affected when inbreeding is high (Hedrick et al., 2001); yet conversely, species with reduced genetic diversity in general can also show a reduced MHC diversity (Babik et al., 2005). However, reduced variation at MHC does not necessarily lead to extinction, demonstrated by the long-term survival of the Northern elephant seal (Mirounga angustirostris), which has limited variability at MHC loci (Hoelzel et al., 1999) but its population size has strongly recovered despite a history of exploitation similar to that of the NZSL. Such persistence and recovery may be a consequence of relative environmental stability and the resulting lack of selection pressure on MHC to maintain variability (Bernatchez and Landry, 2003).
It has previously been suggested that MHC variation in marine mammals is generally lower than that seen in terrestrial mammals (Slade, 1992). One hypothesis for this is the expectation of a lower level of pathogen exposure of marine mammals compared with terrestrial mammals, which was speculated to translate to a lack of pathogen selection pressure (Slade, 1992). Studies in numerous marine mammal species have shown that levels of variation at MHC genes are often depressed in marine mammals, relative to terrestrial counterparts (Trowsdale et al., 1989; Hoelzel et al., 1999; Bowen et al., 2002; Weber et al., 2004). However, other data challenge this generalisation; for example, a high level of variation was seen in the Southern elephant seal at MHC DQB exon 2, with the level of variation comparable to that seen in human DQB exon 2 (Hoelzel et al., 1999). Likewise, baleen whales (suborder Mysticeti) showed very high DQB exon 2 diversity, consistent with that observed in related terrestrial mammals (Baker et al., 2006).
Given the known involvement of MHC loci in bacterial infection and in the recognition of extracellular pathogens, such as bacteria and nematodes (Hughes and Yeager, 1998), here we investigate genetic diversity at the MHC class II B region in a broad sample of NZSL pups from the 2003/2004 cohort. The class II B MHC loci, DRB and DQB, are generally the most diverse MHC loci in mammals, and thus the most widely targeted and studied regions in molecular, ecological and evolutionary studies (Baker et al., 2006). As MHC loci are nuclear genes, a typical approach for studying MHC haplotypes would be cloning and sequencing. However, legislative restrictions limiting the development of recombinant organisms containing the DNA of New Zealand native species (Environmental Risk Management Authority, 1998) meant that it was impractical to clone MHC alleles from NZSL. We therefore examined the level of variation at the MHC class II B loci DRB and DQB, and determined MHC alleles in NZSLs using two other methods: (1) Sanger sequencing followed by bioinformatic haplotype reconstruction (Stephens et al., 2001) and (2) high throughput next-generation sequencing (NGS), specifically 454 sequencing on the Roche GS FLX platform (Shirley, NY, USA). Deep amplicon sequencing, such as NGS, is equivalent to cloning amplicons derived from a single DNA molecule in a cell-free system and sequencing a high number of clones (Babik et al., 2009b; Babik, 2010). Moreover, using this technology multiple amplicons can be barcoded/tagged and pooled (Binladen et al., 2007; Babik et al., 2009b) to identify specific genes or amplicons within the pool, allowing sequencing of large number of samples in parallel. Thus, this study provides a thorough investigation of MHC class II B variation using both Sanger- and NGS sequencing methods, providing knowledge about potential selection mechanisms acting on MHC loci in the NZSL. Furthermore, these data allow us to evaluate the application of new NGS methods in comparison with the traditional Sanger sequencing for population MHC allele determination in this non-model species.
Materials and methods
Sampling
All NZSL samples in this study were collected from sites within the Auckland Islands group (50°42′S 166°5′E, Figure 1). The sample set for these analyses consisted of 87 unrelated live pups: 33 samples from Sandy Bay (Enderby Island), 37 samples from Dundas Island and 17 samples from Figure of Eight Island, all of which were sampled in the Austral summer breeding season of 2003/2004 (December–February). These three different breeding sites were included to capture a broad spectrum of MHC alleles present within the cohort.
DNA extraction
DNA was extracted from NZSL skin biopsies using Chelex 100 (Bio-Rad, Hercules, CA, USA) and an adapted protocol (Walsh et al., 1991). Briefly, ∼1 mm2 of tissue was suspended in a digesting solution consisting of 5% Chelex 100, 100 mℳ NaCl, 50 mℳ Tris (pH 8.0), 1% SDS and 10 mℳ EDTA. In all, 10 mg ml−1 each of Proteinase K and RNase was added and samples were digested overnight at 55 °C. Samples were centrifuged at 12 000 g for 1 min to precipitate the debris, and supernatant was transferred to a new tube containing 5% Chelex in TE (10 mℳ Tris pH 8.0, 1 mℳ EDTA). Finally, samples were again centrifuged at 12 000 g for 1 min and stored at −20 °C until used.
Primer development
To amplify the partial second exons, including the putative peptide-binding regions, of DQB and DRB in NZSL DNA by PCR, fusion primers were designed to enable use of the same PCR products for both Sanger sequencing and NGS. For both DQB and DRB loci, we used primer sets previously used in a variety of other mammal species, including pinnipeds. For the DQB locus, we used DQB1 5′-CTGGTAGTTGTGTCTGCACAC-3′ and DQB2 5′-CATGTGCTACTTCACCAACGG-3′, a primer set first reported by Scharf et al. (1988) for human, and then subsequently used in a variety of mammalian species including NZSL (Lento et al., 2003) and Northern elephant seal (Weber et al., 2004). For the DRB locus, we used the primer pair DRB1 5′-AACGGGACGGAGCGGGTGCG-3′ and DRB2 5′-TCGCCGCTGCACCAGGAAGC-3′, previously used in Northern elephant seal (Garza, 1998). Our fusion primers were synthesised to consist of: (1) the locus-specific primers DQB or DRB primers, (2) multiplex identifier sequences (MID, Roche) and (3) the Roche GS FLX 454 sequencing adaptors for Titanium chemistry, as per the 454 Sequencing Technical Bulletin No. 013-2009 (Roche, 2009). The MID sequences (tags/barcodes) for the identification of DQB and DRB in the pool were chosen on the basis of their predicted low propensity for primer-dimer formation and self-dimerisation, which could otherwise inhibit PCR and sequencing (DQB, 5′-CGTGTCTCTA-3′ DRB, 5′-TAGTATCAGC-3′). Note that unlike Sanger sequencing, this NGS design allows the determination of MHC alleles at the level of cohort, but not individual, as our interest for comparison of two sequencing methods is the estimation of variation at population level, not of the individual. Such a tagging and pooling approach at the population level has been used and discussed previously (Druley et al., 2009; Futschik and Schlotterer, 2010; Wang et al., 2011).
Sanger sequencing of MHC loci
Individual PCRs for each of the two loci contained in a 10 μl volume c. 50 ng template DNA, 20 mℳ Tris-HCl, 50 mℳ KCl, 2 nmol each dNTP, 1 pmol each primer, 2.0 mℳ MgCl2, 6% DMSO and 0.1 unit Taq-Ti DNA polymerase (Fisher Biotec, Perth, WA, Australia). Thermal cycling parameters were based on a standard touchdown protocol (Don et al., 1991) and consisted of: 94 °C for 5 min, 10 cycles of: 94 °C for 20 s, 65 °C for 20 s, 72 °C for 30 s, followed by 40 cycles of: 94 °C for 20 s, Tx°C for 20 s (where Tx is a touchdown cycle beginning at 65 °C and reducing by 0.5 °C per cycle to 45 °C), 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. It is important to note that, although this number of PCR cycles may appear non-standard, this method was chosen because of the difficulties associated with cross-species amplification of primers; here we used primers from other mammal species to amplify NZSL DNA. PCR products were separated by agarose gel electrophoresis and visualised under ultraviolet light after staining with ethidium bromide. Amplified individual PCR products were sequenced in both forward- and reverse-primer reactions using an ABI 3730xl DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA) by the Genomic Analysis Service, University of Otago, and analysed using the program Geneious v5.1 (Drummond et al., 2010).
Next-generation sequencing of MHC loci
PCR products obtained as above were purified using SPRI beads, according to the manufacturer's protocol (Agencourt Bioscience Corp., Beverly, MA, USA). Briefly, nucleic acids (PCR products) were immobilised by the addition of SPRI beads, purified using magnetic affinity and contaminants removed by aspiration. The nucleic acids bound to magnetic SPRI beads were washed and then eluted, leaving high-quality nucleic acids. Purified PCR products were quantified using Pico Green (Invitrogen, Carlsbad, CA, USA) and a Victor X3 Multilabel plate reader (Perkin Elmer, Waltham, MA, USA), according to the manufacturer's protocol. Amplicons tagged by locus were pooled in equimolar amounts and submitted for high throughput sequencing using Roche GS FLX Titanium series chemistry, University of Otago High Throughput Sequencing Service. Obtained reads were first evaluated for quality scores and expected length before further analyses.
MHC haplotype reconstruction
MHC DQB and DRB haplotypes were reconstructed from Sanger sequence outputs using the program Phase v2.1 (Stephens et al., 2001), implemented in the Microsoft Windows command line.
Data generated by NGS of PCR amplicons were first screened for artefacts, and allele frequencies were calculated using the program jMHC (Stuglik et al., 2011). We constrained our jMHC analyses by restricting the output to include only those sequences that contained both the full forward primer and reverse primer sequences. On the basis of the number of reads outputted using the above criterion, only those variants present in the data at >40 reads were considered further, to ensure low frequency artefacts were excluded from further analyses (Stuglik et al., 2011). The threshold of 40 reads was calculated based on the number of reads produced when restricting the jMHC analyses as above, in order to capture those alleles present in one copy, the rationale for which is explained fully in the Results section. The alleles resulting from jMHC output were aligned and manually checked using Geneious v5.1 (Drummond et al., 2010) to enable a comparison of both Sanger and NGS methods of MHC haplotype determination and to verify the polymorphic sites identified through Sanger sequencing and subsequent phasing of the data.
Population differentiation and substructure
In order to test whether identified MHC alleles show population differentiation between sampling locations/breeding beaches used in this study (Figure 1), pairwise FST values (Weir and Cockerham, 1984) from Sanger sequencing data of MHC alleles from each NZSL pup were calculated using Arlequin (Excoffier et al., 2005). In addition, to investigate population substructure and differentiation at neutral loci, the NZSL pups from this study were also genotyped at 21 unlinked pinniped microsatellite loci using standard methodologies (Osborne, 2011; Negro et al., in preparation). FST estimates were calculated and the significance of genetic differentiation between sampling locations was tested with Exact Tests (Raymond and Rousset, 1995; Goudet et al., 1996) using a Markov chain of 10 000 steps. In addition, DST (Jost, 2008) was calculated using SMOGD (Crawford, 2010) with 1 000 bootstrap replicates. Lastly, these microsatellite data were analysed using the program Structure v2.3.3 (Pritchard et al., 2000) to estimate the most likely number of population clusters (K) that derive from these samples. Twenty replicate simulations for K values of 1–10 were performed with a burn-in length of 5 000 followed by 50 000 Markov chain Monte Carlo (MCMC) steps. For these simulations, it was assumed that the allele frequencies between putative population clusters were correlated.
MHC allele frequency differences by sampling location
To determine whether MHC allele frequencies differed between sampling locations, sequential independent two-sample Mann–Whitney U tests were used, followed by construction of a one-way analysis of variance (ANOVA) to assess whether heterozygosity at MHC differed between sampling locations in this study. In addition, to establish whether allele frequencies differed according to the method of allele determination (Sanger vs NGS), a paired Wilcoxon signed-rank test was implemented. All statistical tests described above were undertaken using the program R (R Core Development Team, 2010).
Estimates of mode of MHC allele evolution
To assess the phylogenetic relationships among the MHC DRB alleles identified by Sanger sequencing, an unrooted tree was constructed by maximum likelihood (ML) methods in Mega5 (Tamura et al., 2011). The program PAML 4 (Yang, 1997, 2007) was employed to investigate the potential for species-specific selection on the MHC DRB alleles. The unrooted ML tree was used to evaluate both allelic lineage- and branch-specific models of gene evolution to see which model best fits the DRB data. We tested for lineage-specific-positive selection on DRB using one-ratio (one dN/dS (ω) ratio per tree) and free-ratio (independent dN/dS (ω) ratio per branch of the tree) models, which were evaluated and compared with each other by likelihood ratio test (LRT). The P-values for these comparisons were determined, and the most significantly favoured model was taken forward. We next investigated whether any specific sites (codons) were evolving under adaptive evolution using the models M1a, M2a, M7 and M8 (Yang, 1997, 2007). Likelihood ratio tests favoured M2a over M1a (a model allowing positive selection vs nearly neutral gene evolution) and M8 over M7 (β and ω vs β, essentially positive selection vs neutral evolution). Again, the model that best fitted the data was taken forward and the Bayes Empirical Bayes method was used to calculate the Bayesian probability that each amino-acid site evolved under positive selection (Nielsen and Yang, 1998).
The above methodology was unsuitable to apply to MHC DQB because only two alleles were detected in NZSL. As such, DQB sequence data was investigated for significant departure from neutral evolution using Tajima's D statistic (Tajima, 1989), followed by calculation of 95% confidence intervals (CI) and the critical D value for the estimation of statistical significance by coalescent simulation (10 000 replicates), as implemented in DnaSP v.5 (Librado and Rozas, 2009). For consistency and to enable comparison we applied this methodology also to DRB sequence data.
Tests of trans-species polymorphism
To explore our data for evidence of trans-species polymorphism, we undertook phylogenetic analyses on the consensus alignments of our NZSL DQB and DRB sequences exon 2 sequences, against several hundred sequences from other Carnivora (taxa=287, sequence length=204 bp), spanning the major families: Felidae, Canidae, Ursidae, Phocidae, Otariidae, Procyonidae and Mustelidae available in GenBank (see Supplementary Table S5 for accession numbers). Data were aligned using Clustal Omega (Sievers et al., 2011) and Bayesian phylogenetic inference performed using MrBayes 3.2 (Ronquist et al., 2012) using an ungulate, Sus scrofa, as an out-group to root the trees. We used FindModel (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html) to identify the model of nucleotide substitution that best fit our data using Akaike Information Criterion; selecting the general time-reversible model with gamma-distributed rate variation (GTR+Γ) for both DQB and DRB data sets. Bayesian analysis of our full data set ran for 2 × 107 generations from a random starting tree, with sampling every 500 generations. We checked for convergence by plotting the likelihood scores against generation and discarded the first 25% of the generations as ‘burn-in'. Two separate analyses and four independent chains were then executed to check for the convergence of topology. We also analysed just the pinniped sequences, to explore further the relationship among alleles within this subset of our tree. Here, Bayesian analysis was undertaken as described above, but for only 2 × 106 generations with sampling every 100 generations.
Results
Sanger sequencing and haplotype reconstruction
The number of samples successfully amplified and sequenced using the Sanger method were DQB, n=72; DRB, n=87. All Sanger sequences successfully obtained through PCR were of clear resolution and were derived from a single product that is, one locus. A maximum of two different sequences per individual were detected via both forward- and reverse strand sequencing results that were consistent with the presence of single copies of these genes, allowing for two alleles. BLAST alignment (Altschul et al., 1997) of the consensus sequence for each NZSL allele to the dog genome identified only a single match to the homologous MHC locus (data not shown). This suggests to us that the oligonucleotide primers used in this study are amplifying a single region. Sequence lengths obtained were 231 bp for MHC DQB and 229 bp for MHC DRB.
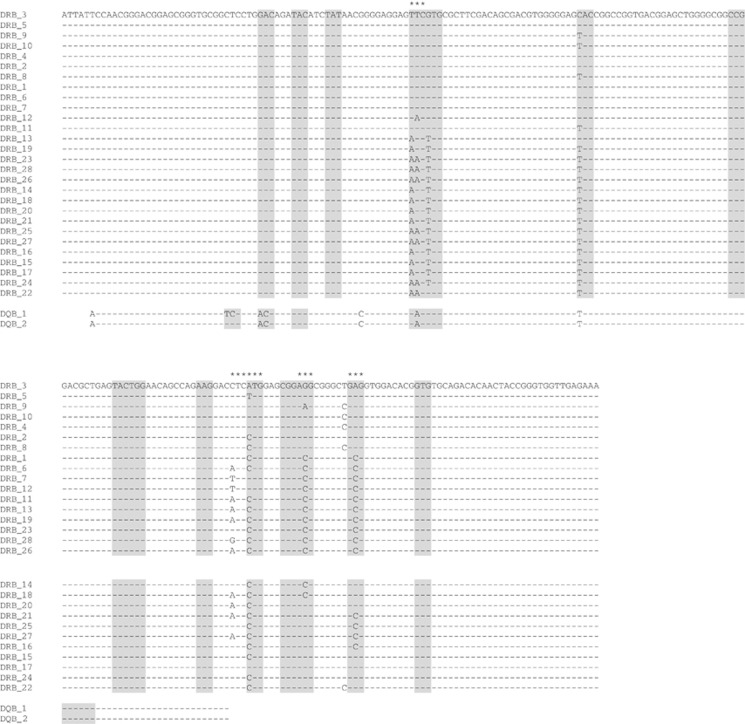
MHC DQB and DRB are highly similar to each other, showing a 92% pairwise identity (Figure 2). Sequence analysis showed two adjacent single-nucleotide polymorphisms in DQB and nine in DRB, including one pair of adjacent polymorphic sites (Figure 2). Haplotype reconstruction showed two alleles by nucleotide sequence for DQB (Table 1) and 28 for DRB (Table 2). Of the nine variable sites of DRB, six showed two possible nucleotides per site, two showed three nucleotides per site and one showed four nucleotides per site.
Figure 2.
ClustalX v2.1 multiple sequence alignment of all 28 MHC DRB (218 bp) and two MHC DQB (147 bp) alleles detected in NZSL. Dashes indicate nucleotides that are identical to the nucleotides of the first sequence (DRB_3) in this alignment. Shaded in grey are codons predicted to be involved in antigen binding in humans (Brown et al., 1993). MHC DRB codons which have ω>1 with a significant Bayes Empirical Bayes probability, and are therefore likely to be under positive selection, are indicated by *. No evidence of positive selection acting at MHC DQB was detected.
Table 1. NZSL MHC DQB alleles and frequency in NZSL live pup population as derived from Sanger sequencing and Phase reconstruction.
| Allele number | Alleles | Count | Frequency | |
|---|---|---|---|---|
|
bp position consensus
|
53
|
54
|
|
|
| 1 | T | C | 117 | 0.812 |
| 2 | C | T | 27 | 0.188 |
Two alleles are present at adjacent nucleotides. Base pair (bp) consensus positions are arbitrarily calculated from the start position of the amplified PCR fragment in NZSL DNA.
Table 2. NZSL MHC DRB alleles and frequency in NZSL live pup population as derived from Sanger sequencing and Phase reconstruction.
| Allele number | Alleles—bp position consensus | Count | Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
58
|
59
|
61
|
88
|
148
|
151
|
161
|
168
|
170
|
|
|
| 1 | T | T | G | C | C | C | C | T | C | 1 | 0.57 |
| 2/5 | T | T | G | C | C | C | G | T | A | 1 | 0.57 |
| 3/4 | T | T | G | C | C | A | G | T | A | 43 | 24.71 |
| 6 | T | T | G | C | A | C | C | T | C | 2 | 1.15 |
| 7 | T | T | G | C | T | A | C | T | C | 4 | 2.3 |
| 8 | T | T | G | T | C | C | G | C | A | 3 | 1.72 |
| 9 | T | T | G | T | C | A | A | C | A | 1 | 0.57 |
| 10 | T | T | G | T | C | A | G | C | A | 21 | 12.07 |
| 11 | T | T | G | T | A | C | C | T | C | 1 | 0.57 |
| 12 | T | A | G | C | T | A | C | T | C | 3 | 1.72 |
| 13 | A | T | T | C | A | C | C | T | C | 1 | 0.57 |
| 14 | A | T | T | T | C | C | C | T | A | 2 | 1.15 |
| 15 | A | T | T | T | C | C | G | T | A | 6 | 3.45 |
| 16 | A | T | T | T | C | C | G | T | C | 1 | 0.57 |
| 17 | A | T | T | T | C | A | G | T | A | 1 | 0.57 |
| 18 | A | T | T | T | A | C | C | T | A | 1 | 0.57 |
| 19 | A | T | T | T | A | C | C | T | C | 2 | 1.15 |
| 20 | A | T | T | T | A | C | G | T | A | 2 | 1.15 |
| 21 | A | T | T | T | A | C | G | T | C | 1 | 0.57 |
| 22 | A | A | G | T | C | C | G | C | A | 1 | 0.57 |
| 23 | A | A | T | T | C | C | C | T | C | 2 | 1.15 |
| 24 | A | A | T | T | C | C | G | T | A | 1 | 0.57 |
| 25 | A | A | T | T | C | C | G | T | C | 4 | 2.3 |
| 26 | A | A | T | T | A | C | C | T | C | 67 | 38.51 |
| 27 | A | A | T | T | A | C | G | T | C | 1 | 0.57 |
| 28 | A | A | T | T | G | C | C | T | C | 1 | 0.57 |
The most frequent alleles and those used in further analyses are shaded grey. Base pair (bp) consensus positions are arbitrarily calculated from the start position of the amplified PCR fragment in NZSL DNA. Alleles 2 and 5, and 3 and 4, were synonymous at the amino-acid level, forming two distinct alleles by protein sequence. Here, frequencies are combined, and the most common nucleotide sequence is listed.
To ensure that undetected population substructure did not affect downstream analyses, MHC DRB allele frequencies at each of the three sampling sites were evaluated. No significant difference in allele frequency was found between sampling sites (Supplementary Table S1), as determined by sequential independent two-sample Mann–Whitney U tests: Dundas vs Figure of Eight, W=366.5, P=0.08; Enderby vs Figure of Eight, W=305.5, P=0.70; Enderby vs Dundas, W=231, P=0.21. There was no significant difference in mean heterozygosity of MHC alleles between sampling locations (one-way ANOVA; F2,84=0.5628; P=0.5718). Pairwise FST on these data ranged from 0.04 to 0.19, and an Exact test of significance indicated that all pairwise comparisons were non-significant (P>0.05).
Further tests to examine the strength of demographic influences in our study using genotypic data from 21 microsatellite loci for each of our NZSL pups suggest such influences are weak in our sample set. FST estimates ranged from 0.006 (Enderby:Dundas and Enderby:Figure of Eight) to 0.009 (Dundas:Figure of Eight), and no significant differentiation was observed among any of our study populations (Exact tests of population differentiation, all P>0.05). Likewise, harmonic means of DST that were estimated for each population pair showed no population differentiation (Supplementary Table S2). Lastly, structure analysis using the mean likelihood of each value of K from 1 to 10 indicated that K=1 was the most likely (Supplementary Table S3), suggesting that there is no evident substructuring.
Effects of nucleotide variants on MHC peptide sequences
DQB variants
To determine whether both alleles identified here had the potential to be expressed as different peptide sequences, nucleotide sequences were entered into ExPASy (Gasteiger et al., 2003) accounting for the correct reading frame as determined by Lento et al. (2003). It was found that the two nucleotide variants were in the same codon and were non-synonymous, and that these changes altered the amino-acid sequence: the codon containing the TC variant codes for serine; the CT variant codes for leucine. Serine has a hydroxyl group, which means this amino acid is able to form hydrogen bonds with a variety of substrates, while leucine is hydrophobic and therefore is buried in protein hydrophobic cores (Betts and Russell, 2003). Despite this, the amino-acid substitution was not predicted to affect protein structure per se (Adzhubei et al., 2010), although comparison with sequences defined by Brown et al. (1993) suggested that these polymorphisms are components of a codon important in peptide binding.
DRB variants
To reduce the complexity of the analyses, alleles were grouped according to their translated amino-acid sequence. Although they differed in nucleotide sequence, alleles 2 and 5, and alleles 3 and 4, were synonymous at the amino-acid level. As with DQB, all substitutions were predicted to have no effect on protein structure (Adzhubei et al., 2010); however, seven of the nine polymorphic nucleotides identified here are situated in codons, which are predicted to be important in peptide binding (Brown et al., 1993, Figure 2).
NGS and allele frequency determination
MHC amplicons from live NZSL pups provided 20 077 reads for DQB and 28 571 reads for DRB. The modal fragment lengths for DQB were 223 and 236 bp for DRB, consistent with the read lengths predicted from Sanger sequencing. Reads that were excessively larger or smaller than the expected product size were removed from the alignments, leaving 13 695 reads (∼190 reads per amplicon) for DQB (220–226 bp) and 11 933 reads (∼137 reads per amplicon) for DRB (230–240 bp). Mean Phred quality scores for both DQB and DRB in the required size range were between 30 and 40, with approximately 60% of sites having scores of 40 (that is, base call accuracy of 99.99%). On the basis of above outputs, reads were screened for low frequency artefacts in jMHC (Stuglik et al., 2011), using a conservative cut-off of 40 reads per variant, leaving a total of 7439 reads for DQB and 5425 reads for DRB. Assuming a maximum of two variants for each amplicon, as identified in Sanger sequencing data, this conservative cut-off of 40 reads per variant was chosen so that even those variants present in one copy should be detected, if these data yield ∼137 reads per amplicon at the lowest level; that is, ∼68 reads per variant. These screened data were then used to calculate frequencies of variants (alleles), and their relative proportions within the NZSL population were calculated and compared with Sanger sequencing and bioinformatic haplotype reconstruction via Phase (Stephens et al., 2001).
After filtering as described above, NGS identified two alleles of MHC DQB identical to alleles 1 and 2 identified through Sanger sequencing and Phase reconstruction. The frequencies of each DQB allele are comparable between the two approaches (Supplementary Table S4a).
Analysis of DRB allele frequencies derived from Sanger sequencing and Phase reconstruction determined that three alleles (3, 10 and 26) were common in the NZSL population (frequencies from Sanger 24.7%, 12.07% and 38.51%, respectively), with the remainder much less frequent (<3.45%, Supplementary Table S4b). Frequencies for alleles 3, 10 and 26 were not identical in both Sanger and NGS, but were represented in similar proportions within the data derived using each approach; that is, the most common vs much rarer alleles. Two alleles (6 and 12) that were identified in very low frequency in Sanger sequencing were present in fractionally higher frequencies in NGS data. Allele 7 was detected through Sanger sequencing at a frequency of 2.3% but it was not detected through NGS. Small differences in frequency are likely due to the conservative cut-off of 40 reads; the pooling of PCR products for NGS depends on equimolar distribution of each PCR product to ensure accurate frequencies, and either handling error or an error in the NGS reaction can lead to loss of very low copy alleles (that is, those present only once in a given sample). Increasing the coverage of NGS would likely solve this problem. Nevertheless, there was no detectable difference between allele frequencies of the most common alleles in the two methods employed here and shown in Supplementary Table S4b (paired Wilcoxon signed-rank test, P=0.8438). Nucleotides present at each polymorphic site were identical between Sanger and NGS. Empirical confirmation of allele presence and frequency using NGS serves to validate the outputs of Sanger sequencing and haplotype reconstruction by the program Phase.
Prediction of mode of MHC gene evolution
An unrooted tree of MHC DRB alleles was constructed by ML methods (Supplementary Figure S1) using Mega5 (Tamura et al., 2011), and this tree was used to evaluate both lineage- and branch-specific models of gene evolution to see which model best fit the DRB data. The one-ratio model was compared with the free-ratios model using a LRT to determine whether an equal dN/dS (ω) ratio for each branch fit the data better than a scenario wherein each branch is allowed an independent dN/dS ratio (ω). In these analyses, the free-ratio model was not favoured over the one-ratio model (χ2 12.71, d.f. 52, P>0.9), suggesting the one-ratio model was a better fit to the data. Under the favoured one-ratio model, the dN/dS ratio (ω) was 1.70, which is suggestive of positive selection at the DRB locus.
Next, we asked whether there was any evidence of site-specific selection in the NZSL MHC DRB, because of the large number of polymorphic sites situated in codons thought to be directly involved in antigen binding (Brown et al., 1993). These analyses allow the ω ratio to vary among sites (Nielsen and Yang, 1998; Yang, 2000) and the data can be used to help identify codons that are subject to adaptive evolution. The models chosen to use here were M1a vs M2a (nearly neutral model vs positive selection) and M7 vs M8 (β vs β and ω). The comparison of M1a vs M2a showed that the positive selection model (M2a) was significantly favoured at P<0.001 (χ2 56.77, d.f. 2). Likewise the M7 vs M8 comparison strongly favoured the positive selection model (M8) at P<0.001 (χ2 33.24, d.f. 2).
The Bayes Empirical Bayes method calculates the Bayesian probability for each amino-acid site that ω >1. If this probability is high, the sites are likely to be under positive selection (Nielsen and Yang, 1998). Our analyses show that the strongly favoured positive selection models (M2a and M8) identify five amino-acid sites (out of seven variable amino-acid sites) with a high probability that ω>1 (Figure 2), with their Bayesian probabilities ranging from 0.997 to 1.000. Four of these five sites reside in codons that are predicted to be involved in antigen binding in humans (Figure 2; Brown et al., 1993).
MHC DQB showed a segregating site of two adjacent base pairs contained within the same codon (perhaps a consequence of one mutational event), which resulted in an amino-acid change. The average number of nucleotide differences between sequences was 0.62. In addition to the number of nucleotide differences, the nucleotide diversity was calculated, which is the mean number of differences per site between two randomly chosen sequences from the sample set; nucleotide diversity at MHC DQB was 0.004. Tajima's D was 1.02 (95% CI: −1.69, 1.67) and was not significantly different from zero. For consistency and to enable comparison, we applied this methodology also to DRB. There were nine segregating sites detected in MHC DRB, with 13 total mutations. The average number of nucleotide differences between sequences was estimated at 4.23 and the nucleotide diversity was 0.02. The measure of Tajima's D for MHC DRB was 2.53. This lies outside of the 95% CI of neutral expectations, which was determined by coalescence simulation (95% CI: −1.73, 1.63), and is therefore significantly different from zero at P<0.05 and suggestive of balancing selection. Tajima's D did not vary significantly from this level, according to breeding beach/sampling location (data not shown).
Trans-species polymorphism
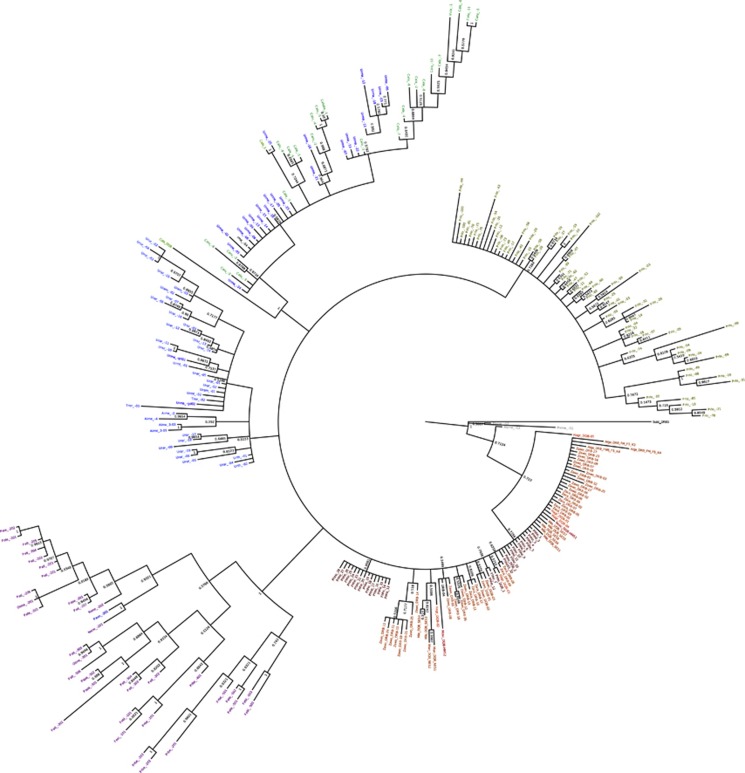
Our prior analysis suggests that MHC diversity at the NZSL DRB locus is maintained by balancing selection. Under such a scenario, long-lasting trans-species polymorphism may occur (Klein et al., 1998); thus, we used Bayesian phylogenetic inference to explore our data for evidence of such a possibility. Phylogenetic reconstruction (Figure 3) using our NZSL MHC DQB and DRB sequences, together with those derived from other carnivore DQB and DRB sequences, shows pinniped DQB and DRB alleles are clearly divergent from those of felids, canids and ursids. Within the felids, ursids and canids there is a strong evidence for trans-species polymorphism as reported previously by multiple authors (Seddon and Ellegren, 2002; Wan et al., 2006; Goda et al., 2010; Castro-Prieto et al., 2011; Kuduk et al., 2012). For example, we see evidence of allele sharing among cat species, bear species, and even among bears and dogs (Figure 3). However, the relationships among alleles within the pinnipeds resolve poorly, with the pinniped sequences forming a polytomy that makes interpretation of the relationships within this group of carnivores difficult. We do not believe this is a consequence of the short length of the sequence, as the relations among other alleles resolve quite strongly; rather we suggest this polytomy is suggestive of a recent common ancestry among pinniped alleles and relatively recent diversification of MHC alleles.
Figure 3.
Bayesian 50% majority-rule consensus phylogenetic tree illustrating the relationships among MHC DQB/DRB from NZSL, and other representative carnivores: felids (purple), canids (green), ursids (blue), mustelids (grey), procyonids (light brown) and pinnipeds (red). The tree is rooted using a DRB sequence from the ungulate Sus scrofa. Details of the sequences used and their accession numbers are shown in Supplementary Table S5. Bayesian posterior probabilities for each node are shown.
Further phylogenetic analysis of pinniped DQB and DRB alleles independent of the other carnivore sequences (Supplementary Figure S2) did not provide much greater resolution, with the majority of branches very weakly supported, based on posterior probabilities. As expected, DQB alleles form a separate clade, albeit with weak posterior probability, from all but two DRB alleles (Arga-FM-FS-K4 and Arga-FM6-S-K4) that may have been misannotated. Across the tree there are multiple instances of allelic similarity among distantly related pinniped species for DQB and DRB, but the support for these is universally weak. For example, alleles from Zalophus wollebaeki (Zawo DRB 14 and 25) form a clade with those from NZSL (Phho 1–28); however, the posterior probability on every node supporting this clade is <0.1. Similar relations are observed among DRB alleles from the close sister taxa Z. californianus and Z. wollebaeki (Zaca DRB 1–7 and Zawo DRB 5 and 14), as well as among more distant taxa, such as elephant, monk and grey seals for DQB alleles (Mile, Monsi and Hagr). Although tempting, it is clearly fraught to interpret specific relationships among alleles when the support for these is so low, and thus we cannot confirm or refute trans-species polymorphism within pinnipeds.
Discussion
Patterns of MHC diversity
The MHC DQB sequences of NZSL in our study showed two segregating sites consistent with two alleles and peptides. This study confirms previous work with MHC DQB in the NZSL that showed the same, very low variation present at this locus that would otherwise be expected to show high levels of variability (Lento et al., 2003) considering its function as a peptide-binding site (Hughes and Nei, 1989). The observed MHC DQB variability in NZSL is comparable to the two alleles detected in Northern elephant seal, a species that like NZSL has undergone a significant bottleneck (Hoelzel et al., 1999; Weber et al., 2004). A similar pattern of low variation in DQ loci was also detected in California sea lion, Z. californianus (Bowen et al., 2004). In contrast, the opposite pattern has also been observed in, for example, Crabeater seals (Lobodon carcinophaga) and Baiji (Chinese river dolphin, Lipotes vexillifer), where variation at MHC DQ loci were higher than expected and more akin to the variation seen in land-based mammals (Lehman et al., 2004; Yang et al., 2005).
The detection of 28 nucleotide sequences that corresponded to 26 DRB peptides in 87 NZSL pups, three peptides of which account for the majority of observed alleles, is consistent with observations in terrestrial mammalian species (Schaschl et al., 2004; Becker et al., 2009; Srithayakumar et al., 2011). In fact, the detection of extensive variation at MHC DRB in mammalian species that are not of conservation concern is common; primates especially are highly variable at DRB (Otting et al., 2000; Schwensow et al., 2007). However, it should be noted that the above species, apart from the European mink (Mustela lutreola, Becker et al., 2009), are of low conservation concern and have an expectation of overall moderate-to-high genetic diversity, which would extend to genes of the MHC. Thus, given the recent population history of the NZSL, along with its restricted area of occupancy and declining population size, the level of MHC DRB diversity observed here was higher than expected.
We have considered the possibility that this unexpectedly high level of variation is a consequence of amplification of more than one DRB locus; however, we do not think this is the case in this species, for either DRB or DQB, for the following reasons. First, MHC DQB and DRB do not appear to be duplicated in NZSLs (DQB, Lento et al., 2003) or in pinnipeds (Slade, 1992) although this is the first study to assess DRB in the NZSL. Second, reciprocal BLAST alignment of the NZSL DQB and DRB consensus sequences to the dog genome provided only one hit to the canine DRB gene, and based on the observed synteny between species of the Carnivora (Osborne et al., 2011), these data suggest that the DRB gene is not duplicated in the NZSL. Third, if more than one DRB (or DQB) locus was being amplified, sequencing outputs would likely include polymorphic loci with more than two variants within an individual, and this is not detected here. Last, if more than one locus were being amplified, then Phase reconstruction of the alleles amplified by Sanger sequencing would not result in the detection of the same alleles to those identified by NGS, as shown by these data.
Examining diversity of two MHC class II B loci in our study produced two interesting results. First, we observed contrasting level of variability between the two loci, with DQB locus showing much lower variability than DRB locus at both nucleotide and amino-acid levels. Although the observed variability at NZSL DQB locus is very low, the same overall pattern of differential variability between these two MHC loci was also observed in, for example, Northern elephant seals (Weber et al., 2004). Second, this study demonstrates extensive variation at MHC DRB in the NZSL despite a history of a population bottleneck and an observed limited level of allelic diversity at microsatellite loci in the NZSLs from the sampling sites used in this study (expected heterozygosity 0.66, average number of alleles per locus 7.29 (Osborne, 2011) compared with Table 1 in Robertson and Chilvers (2011)). A more extreme example of this pattern of variability is the San Nicholas Island fox (Urocyon littoralis dickeyi), a critically endangered species that possesses high MHC DRB heterozygosity yet appears monomorphic at microsatellite loci, having in the past been subject to a severe population bottleneck (Aguilar et al., 2004; Hedrick, 2004). Likewise, bighorn sheep (Ovis aries), a species with a population history of bottlenecks and epizootic disease, like NZSL, possesses 21 MHC DRB alleles with extensive nucleotide and amino-acid divergence, but relatively modest levels of variability at microsatellite loci (Gutierrez-Espeleta et al., 2001). In stark contrast, the brown trout (Salmo trutta), a widespread species of no particular conservation concern, showed comparable levels of diversity at both MHC class II B and four microsatellite loci (Campos et al., 2006). Although there are undoubtedly exceptions, these studies and others suggest that a high variability at MHC DRB coupled with low microsatellite diversity appears to be a relatively frequent occurrence in natural populations that are the subject of conservation concern because of small population size. Why this might be so has been the subject of considerable discussion and debate (Aguilar et al., 2004; van Oosterhout et al., 2006; Babik et al., 2009a).
Use of NGS for MHC allele detection
Implementation of a computational approach (Stephens et al., 2001) coupled with the use of NGS on PCR amplicons, provides a high throughput method for MHC haplotyping (Babik, 2010). The key advantage of this approach is that it is not reliant on slow and costly cloning steps (Abdelkrim et al., 2009; Babik, 2010), and may ultimately be less error prone than the more traditional approaches (Longeri et al., 2002; Harrigan et al., 2008). Nonetheless, the generation of artefacts during PCR and sequencing are a known problem, so outputs of NGS were screened to discern between true sequences and artefacts, the latter of which should be relatively rare compared with true sequences (Stuglik et al., 2011). We chose a conservative cut-off of 40 reads per allele in order to accommodate true sequences only; however, this stringency, coupled with low coverage in the NGS reaction, may lead to the non-detection of rarer alleles; for example, those only present in the sample population in one copy. It is important to note that the haplotypes presented will need to be validated further by, for example, investigation of RNA to check for splice variants and to eliminate pseudogenes (Klein and Figueroa, 1986; Klein et al., 1993), and results should be interpreted with this in mind.
Selection at MHC loci and departure from neutrality
Fitness-related genes are often under selection (either directional or balancing), and therefore may also be more likely to show higher levels of variation than neutral genomic regions regardless of loss of variation elsewhere in the genome, and of population size (Maruyama and Nei, 1981; Nevo et al., 1997). Indeed, variation and diversity at MHC loci has been associated with fitness and individual outcome upon infection (Oliver et al., 2009; Radwan et al., 2010; Spurgin and Richardson, 2010), and pathogen-mediated selection is predicted to alter the frequencies of MHC alleles within a population (Meyer and Thomson, 2001; Hedrick, 2002). Polymorphism at exon 2 of MHC DQB and DRB is generally accepted to have a considerable role in the development of an adequate immune response (Hedrick and Kim, 2000), and variation is thought to be driven by its function as a peptide-binding region, which may confer resistance to a wider range of pathogens (Doherty and Zinkernagel, 1975). This is also thought to be reflective of the fact that balancing selection may be maintaining diversity at MHC (Meyer and Thomson, 2001; Aguilar et al., 2004; van Oosterhout et al., 2006).
Balancing selection takes two forms: heterozygote advantage (also termed overdominant selection) and frequency-dependent selection (or negative frequency-dependent selection). In the first, heterozygosity is favoured because some alleles will be advantageous when heterozygous, and disadvantageous when homozygous (Gemmell and Slate, 2006). Negative frequency-dependent selection (also termed rare allele advantage) can be driven by the selective advantage conferred by new or rare alleles in the population. Rare alleles may offer a greater protection against common pathogens than that of common alleles, because the fitness of an allele can decrease as it becomes more common and pathogen defences co-evolve (Takahata and Nei, 1990). Such mechanisms allow variation at MHC to be maintained after bottleneck despite potential loss of variation elsewhere in the genome; for example, loss of variation at microsatellite loci (Aguilar et al., 2004; van Oosterhout et al., 2006; Mona et al., 2008). Balancing selection is therefore important in maintaining high variability at MHC loci (Hedrick, 1998; Bernatchez and Landry, 2003).
Through the analyses in this study, we have shown evidence of selection acting on the MHC DRB gene of the NZSL that does not appear to be influenced by undetected population substructure. The free-ratio model was not favoured over the one-ratio model, suggesting that the selective pressure may be relatively constant, rather than acting on specific alleles of DRB. For the strongly favoured one-ratio model, the dN/dS ratio (ω) was 1.70, which suggests DRB is under positive selection. Further, analysis using site-specific models, to examine whether any particular codons might be evolving under positive selection, significantly favoured the two positive selection models, M2a and M8 (Yang, 1997, 2007) over the nearly neutral models of gene evolution (M1a and M7) for DRB. Specifically, out of seven variable amino-acid sites in the NZSL, five had ω>1 and four of these sites reside in codons predicted to be involved in antigen binding in humans (Brown et al., 1993). Thus, we can conclude it is likely that DRB loci/alleles are evolving under positive selection and that these four particular codons might be subject to adaptive evolution.
We were unable to implement the methodology applied above to estimate departure from neutral expectations for the NZSL MHC DQB gene, because only two alleles were detected, and thus construction of an unrooted tree and downstream analyses that were applied to DRB were not possible. Further, the absence of singleton mutations (those nucleotide differences observed only once in the sample) in our DQB data makes the use of many other common tests of selection, such as Fu and Li's D, D* (Fu and Li, 1993) inappropriate in this instance. Consequently, we chose to use Tajima's D statistic, which tests the hypothesis that all mutations are selectively neutral.
We did not detect any evidence of positive selection acting at MHC DQB in the NZSL. However, consistent with the output from PAML, we see a strongly positive value of Tajima's D at MHC DRB, which suggests that balancing selection may be operating at MHC DRB in the NZSL population. However, it is important to note that currently there are no approaches that allow accurate dissection of the different mechanisms of selection in wild populations, and this is because of the non-exclusivity of such mechanisms (Spurgin and Richardson, 2010). In addition, although there was no evidence of population substructure among pups from sites sampled in this study based on both MHC DRB allele sequences and microsatellite analyses, there remains the slight possibility that much larger numbers of markers or individuals may lead to the detection of subtle population structure in the wider population.
Trans-species polymorphism
Phylogenetic analysis of MHC DQB and DRB sequences from pinnipeds and a wide representation of other carnivores could not confirm or refute the possibility of trans-species polymorphism within pinnipeds. We readily confirmed previously described instances of trans-species polymorphism in canids, felids and ursids (Seddon and Ellegren, 2002, Castro-Prieto et al., 2011; Kuduk et al., 2012), but were unable to resolve the relationships among DQB and DRB alleles from NZSLs and other pinnipeds. Our pinniped sequences form an extended polytomy (Figure 3 and Supplementary Figure S2), a pattern previously described for DQB by Hoelzel et al. (1999), albeit for a smaller number of taxa. The lack of resolution of DQB and DRB loci in pinnipeds is unlikely to be a consequence of the short sequence length used (consensus alignment=204 bp), because the relationships among other alleles in the tree resolve quite strongly. Rather, we believe this polytomy is suggestive of a relatively recent common ancestry among pinniped alleles and subsequent diversification of MHC alleles. As in other parts of the carnivore DQB/DRB tree we see a large number of unique DRB alleles within the pinnipeds. However, the pinniped clade shows quite shallow branches in comparison with other parts of the tree. Such a pattern is typical of a recent selective sweep. Although further work examining the age of DRB alleles is needed, one possibility worthy of further examination is that pinnipeds have experienced a relatively recent, common, epizootic event that resulted in selection of just a few ancestral alleles, from which the modern diversity has emerged. A further possibility is that allelic variation for DQB and DRB in pinnipeds is heavily constrained, and the variations we observe are simply minor deviations from a sequence that is potentially adapted strongly for optimal response to just a few common pathogens.
Conclusion
The NZSL has maintained a large amount of variability at MHC DRB despite a history of population size contraction, and the documentation of relatively limited allelic diversity at neutral loci among individuals from the sampling sites studied (Osborne, 2011). Further, MHC DRB appears to be displaying balancing selection, consistent with its role as an antigen-binding region. Four of five codons where dN/dS>1 are demonstrated to be in codons predicted to be important in antigen binding, which may have important ramifications for understanding the maintenance of the immune repertoire, and thus immunological response of this species to pathogens. This is in contrast to MHC DQB, which appears to be dimorphic. We show that Sanger sequencing followed by bioinformatic haplotype reconstruction provides an accurate measure of the level of allelic diversity at MHC, demonstrated by equivalence with results from NGS, an empirical measure of allele type and frequency. That we have identified such extensive variation at MHC DRB may be of great use in the further investigation and elucidation of the genetic factors underlying susceptibility to bacterial infection of the threatened NZSL and other pinniped species. The significant diversity seen in pinniped MHC DRB alleles, together with the limited phylogenetic depth of this diversity, suggests some significant evolutionary constraint on this locus in the recent past.
Data archiving
Details of the data archiving can be found in the Supplementary Table S5. All sequences were deposited in GenBank with the exception of the Phho_DQB_1 and Phho_DQB_2 sequences that were deposited in the Dryad repository: doi:10.5061/dryad.2kt7s.
Acknowledgments
We are grateful to Rudiger Brauning (AgResearch, Invermay) for help with statistical analysis of NGS data and Catherine Grueber (University of Otago) and anonymous reviewers for their comments and advice on an earlier version of this manuscript. We also thank Ian Wilkinson, Padraig Duignan, Simon Childerhouse, Jacinda Amey, Wally Hockly and all other field workers for NZSL sample collection and field data. We acknowledge the following funding bodies: University of Otago; subcontract to NJG from Landcare Research's FRST OBI ‘Sustaining and Restoring Biodiversity' contract no. C09X0503; University of Otago Department of Anatomy PhD Scholarship to AJO.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abdelkrim J, Robertson BC, Stanton JAL, Gemmell NJ. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. Biotechniques. 2009;46:185. doi: 10.2144/000113084. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar A, Roemer G, Debenham S, Binns M, Garcelon D, Wayne RK. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc Natl Acad Sci USA. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkush KD, Giese AR, Mendonca HL, Mcbirde AM, Marty GD, Hedrick PW. Resistance to three pathogens in the endangered winter-run chinook salomon (Oncorhynchus tshawytscha): effects of inbreeding and major histocompatiblity complex genotypes. Can J Fish Aqu Sci. 2002;59:966–975. [Google Scholar]
- Babik W. Methods for MHC genotyping in non-model vertebrates. Mol Ecol Resour. 2010;10:237–251. doi: 10.1111/j.1755-0998.2009.02788.x. [DOI] [PubMed] [Google Scholar]
- Babik W, Durka W, Radwan J. Sequence diversity of the MHC DRB gene in the Eurasian beaver Castor fiber. Mol Ecol. 2005;14:4249–4257. doi: 10.1111/j.1365-294X.2005.02751.x. [DOI] [PubMed] [Google Scholar]
- Babik W, Pabijan M, Arntzen JW, Cogalniceanu D, Durka W, Radwan J. Long-term survival of a urodele amphibian despite depleted major histocompatibility complex variation. Mol Ecol. 2009a;18:769–781. doi: 10.1111/j.1365-294X.2008.04057.x. [DOI] [PubMed] [Google Scholar]
- Babik W, Taberlet P, Ejsmond MJ, Radwan J. New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol Ecol Resour. 2009b;9:713–719. doi: 10.1111/j.1755-0998.2009.02622.x. [DOI] [PubMed] [Google Scholar]
- Baker A. Unusual mortality of the New Zealand sea lion, Phocarctos hookeri, Auckland Islands, January-February 1998. Department of Conservation; 1999. [Google Scholar]
- Baker CS, Chilvers BL, Constantine R, Dufresne S, Mattlin RH, van Helden A, et al. Conservation status of New Zealand marine mammals (suborders Cetacea and Pinnipedia), N Zeal J Mar Fresh Res. 2010;2009;44:101–115. [Google Scholar]
- Baker CS, Vant MD, Dalebout ML, Lento GM, O'Brien SJ, Yuhki N. Diversity and duplication of DQB and DRB-like genes of the MHC in baleen whales (suborder: Mysticeti) Immunogenetics. 2006;58:283–296. doi: 10.1007/s00251-006-0080-y. [DOI] [PubMed] [Google Scholar]
- Becker L, Nieberg C, Jahreis K, Peters E. MHC class II variation in the endangered European mink Mustela lutreola (L. 1761)-consequences for species conservation. Immunogenetics. 2009;61:281–288. doi: 10.1007/s00251-009-0362-2. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years. J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. Amino acid properties and consequences of substituitons. Wiley; 2003. [Google Scholar]
- Binladen J, Gilbert MTP, Bollback JP, Panitz F, Bendixen C, Nielsen R, et al. The use of coded PCR Primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. Plos One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmer JL, Vargas FH, Parker PG. Low MHC variation in the endangered Galapagos penguin (Spheniscus mendiculus) Immunogenetics. 2007;59:593–602. doi: 10.1007/s00251-007-0221-y. [DOI] [PubMed] [Google Scholar]
- Bowen L, Aldridge B, Gulland F, Woo J, Van Bonn W, Delong R, et al. Molecular characterization of expressed DQA and DQB genes in the California sea lion (Zalophus californianus) Immunogenetics. 2002;54:332–347. doi: 10.1007/s00251-002-0472-6. [DOI] [PubMed] [Google Scholar]
- Bowen L, Aldridge BM, Stott JL, Gulland F, Woo J. Class II multiformity generated by variable MHC-DRB region configurations in the California sea lion (Zalophus californianus) Immunogenetics. 2004;56:12–27. doi: 10.1007/s00251-004-0655-4. [DOI] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Bryden M, Marsh H, Shaughnessy P. Dugongs, whales, dolphins and seals: a guide to the sea mammals of Australasia. Allen & Unwin: St Leonards, Australia; 1998. [Google Scholar]
- Campos JL, Posada D, Morán P. Genetic variation at MHC, mitochondrial and microsatellite loci in isolated populations of brown trout (Salmo trutta) Conserv Genet. 2006;7:515–530. [Google Scholar]
- Castinel A, Duignan PJ, Pomroy WE, Lopez-Villalobos N, Gibbs NJ, Chilvers BL, et al. Neonatal mortality in New Zealand sea lions (Phocarctos hookeri) at Sandy Bay, Enderby Island, Auckland Islands from 1998 to 2005. J Wildlife Dis. 2007;43:461–474. doi: 10.7589/0090-3558-43.3.461. [DOI] [PubMed] [Google Scholar]
- Castro-Prieto A, Wachter B, Sommer S. Cheetah paradigm revisited: MHC diversity in the world's largest free-ranging population. Mol Biol Evol. 2011;28:1455–1468. doi: 10.1093/molbev/msq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childerhouse S, Gales N. Historical and modern distribution and abundance of the New Zealand sea lion Phocarctos hookeri. N Zeal J Zool. 1998;25:1–16. [Google Scholar]
- Childerhouse S, Gibbs N, Mcalister G, Mcconkey S, Mcconnell H, Mcnally N, et al. Distribution, abundance and growth of New Zealand sea lion Phocarctos hookeri pups on Campbell Island. N Zeal J Mar Fresh Res. 2005;39:889–898. [Google Scholar]
- Chilvers BL. Demographic parameters and at-sea distribution of New Zealand sea lions breeding on the Auckland Islands (POP2007/01) Department of Conservation; 2009. [Google Scholar]
- Chilvers BL, Wilkinson IS. Philopatry and site fidelity of New Zealand sea lions (Phocarctos hookeri) Wildlife Res. 2008;35:463–470. [Google Scholar]
- Crawford NG. smogd: software for the measurement of genetic diversity. Mol Ecol Res. 2010;10:556–557. doi: 10.1111/j.1755-0998.2009.02801.x. [DOI] [PubMed] [Google Scholar]
- DoC . New Zealand sea lion species management plan: 2009–2014. Department of Conservation; 2009. [Google Scholar]
- Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druley TE, Vallania FLM, Wegner DJ, Varley KE, Knowles OL, Bonds JA, et al. Quantification of rare allelic variants from pooled genomic DNA. Nat Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, et al. 2010. Geneious v5.1 Available from http://www.geneious.com .
- Dyall R, Messaoudi I, Janetzki S. MHC polymorphism can enrich the T Cell repertoire of the species by shifts in intrathymic selection. J Immunol. 2000;164:1695–1698. doi: 10.4049/jimmunol.164.4.1695. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Hedrick PW. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol Evol. 1998;13:305–311. doi: 10.1016/s0169-5347(98)01416-5. [DOI] [PubMed] [Google Scholar]
- Eimes JA, Bollmer JL, Whittingham LA, Johnson JA, Van Oosterhaut C, Dunn PO. Rapid loss of MHC class II variation in a bottlenecked population is explained by drift and loss of copy number variation. J Evol Biol. 2011;24:1847–1856. doi: 10.1111/j.1420-9101.2011.02311.x. [DOI] [PubMed] [Google Scholar]
- Environmental Risk Management Authority 1998Annotated Methodology for the Consideration of Applications for Hazardous Substances and New Organisms Under the HSNO Act 1996ERMA New Zealand: Wellington, New Zealand.
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik A, Schlotterer C. The next generation of molecular markers from massively parallel sequencing of pooled dna samples. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gales NJ. Threatened Species Recovery Plan Series no. 17. Department of Conservation: Wellington, New Zealand; 1995. Hooker's sea lions (Phocarctos hookeri) recovery plan. [Google Scholar]
- Gales NJ, Fletcher DJ. Abundance, distribution and status of the New Zealand sea lion, Phocarctos hookeri. Wildlife Res. 1999;26:35–52. [Google Scholar]
- Garrigan D, Hedrick PW. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- Garza JC. Population genetics of the Northern elephant seal. University of California: Berkeley; 1998. [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ, Slate J. Heterozygote advantage for fecundity. PLoS ONE. 2006;1:e125. doi: 10.1371/journal.pone.0000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschke K, Chilvers BL. Managing big boys: a case study on remote anaesthesia and satellite tracking of adult male New Zealand sea lions (Phocarctos hookeri) Wildlife Res. 2009;36:666–674. [Google Scholar]
- Go Y, Satta Y, Kawamoto Y, Rakotoarisoa G, Randrianjafy A, Koyama N, et al. MHC-DRB genes evolution in lemurs. Immunogenetics. 2002;54:403–417. doi: 10.1007/s00251-002-0480-6. [DOI] [PubMed] [Google Scholar]
- Goda N, Mano T, Kosintsev P, Vorobiev A, Masuda R. Allelic diversity of the MHC class II DRB genes in brown bears (Ursus arctos) and a comparison of DRB sequences within the family Ursidae. Tissue Antigens. 2010;76:404–410. doi: 10.1111/j.1399-0039.2010.01528.x. [DOI] [PubMed] [Google Scholar]
- Goudet J, Raymond M, Demeeus T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Espeleta GA, Hedrick PW, Kalinowski ST, Garrigan D, Boyce WM. Is the decline of desert bighorn sheep from infectious disease the result of low MHC variation. Heredity. 2001;86:439–450. doi: 10.1046/j.1365-2540.2001.00853.x. [DOI] [PubMed] [Google Scholar]
- Harf R, Sommer S. Association between MHC Class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the Southern Kalahari. Mol Ecol. 2005;14:85–91. doi: 10.1111/j.1365-294X.2004.02402.x. [DOI] [PubMed] [Google Scholar]
- Harrigan RJ, Mazza ME, Sorenson MD. Computation vs. cloning: evaluation of two methods for haplotype determination. Mol Ecol Res. 2008;8:1239–1248. doi: 10.1111/j.1755-0998.2008.02241.x. [DOI] [PubMed] [Google Scholar]
- Hedrick P. Balancing selection and MHC. Genetica. 1998;104:207–214. doi: 10.1023/a:1026494212540. [DOI] [PubMed] [Google Scholar]
- Hedrick P. Foxy MHC selection story. Heredity. 2004;93:237–238. doi: 10.1038/sj.hdy.6800539. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Kim KJ.2000Genetics of complex polymorphisms: parasites and maintenance of the major histocompatibility complex variationIn Singh RS, Krimbas CB, (eds)Evolutionary Genetics: from molecules to morphology Cambridge University Press: Cambridge [Google Scholar]
- Hedrick PW, Kim TJ, Parker KM. Parasite resistance and genetic variation in the endangered Gila topminnow. Anim Conserv. 2001;4:103–109. [Google Scholar]
- Hedrick PW, Parker KM, Miller EL, Miller PS. Major histocompatibility complex variation in the endangered Przewalski's horse. Genetics. 1999;152:1701–1710. doi: 10.1093/genetics/152.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel AR, Stephens JC, O'Brien SJ. Molecular genetic diversity and evolution at the MHC DQB locus in four species of pinnipeds. Mol Biol Evol. 1999;16:611–618. doi: 10.1093/oxfordjournals.molbev.a026143. [DOI] [PubMed] [Google Scholar]
- Huchard E, Cowlishaw G, Raymond M, Weill M, Knapp LA. Molecular study of MHC-DRB in wild chacma baboons reveals high variability and evidence for trans-species inheritance. Immunogenetics. 2006;58:805–816. doi: 10.1007/s00251-006-0156-8. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:948–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–434. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- IUCN 2008. 2008 IUCN Red List of Threatened Species.
- Jarvi SI, Tarr CL, Mcintosh CE, Atkinson CT, Fleischer RC. Natural selection of the major histocompatibility complex (MHC) in Hawaiian honeycreepers (Drepanidinae) Mol Ecol. 2004;13:2157–2168. doi: 10.1111/j.1365-294X.2004.02228.x. [DOI] [PubMed] [Google Scholar]
- Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- Jost L. G(ST) and its relatives do not measure differentiation. Mol Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Klein J, Figueroa F. Evolution of the major histocompatibility complex. Crit Rev Immunol. 1986;6:295–386. [PubMed] [Google Scholar]
- Klein J, Sato A, O'hUigin C. Evolution by gene duplication in the major histocompatibility complex. Cytogenet Cell Genet. 1998;80:123–127. doi: 10.1159/000014967. [DOI] [PubMed] [Google Scholar]
- Klein J, Satta Y, Ohuigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- Kuduk K, Johanet A, Allainé D, Cohas A, Radwan J. Contrasting patterns of selection acting on MHC class I and class II DRB genes in the Alpine marmot (Marmota marmota) J Evol Biol. 2012;25:1686–1693. doi: 10.1111/j.1420-9101.2012.02537.x. [DOI] [PubMed] [Google Scholar]
- Lehman N, Decker DJ, Stewart BS. Divergent patterns of variation in major histocompatibility complex class II alleles among Antarctic phocid pinnipeds. J Mammol. 2004;85:1215. [Google Scholar]
- Lento GM, Baker CS, David V, Yuhki N, Gales NJ, O'Brien SJ. Automated single-strand conformation polymorphism reveals low diversity of a major histocompatibility complex class ii gene in the threatened New Zealand sea lion. Mol Ecol Notes. 2003;3:346–349. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Longeri M, Zanotti M, Damiani G. Recombinant DRB sequences produced by mismatch repair of heteroduplexes during cloning in Escherichia coli. Eur J Immun. 2002;29:517–523. doi: 10.1046/j.1365-2370.2002.00356.x. [DOI] [PubMed] [Google Scholar]
- Mainguy J, Worley K, Côté S, Coltman D. Low MHC DRB class II diversity in the mountain goat: past bottlenecks and possible role of pathogens and parasites. Conserv Genet. 2007;8:885–891. [Google Scholar]
- Maruyama T, Nei M. Genetic variability maintained by mutation and over-dominant selection in finite populations. Genetics. 1981;98:441–459. doi: 10.1093/genetics/98.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Thomson G. How selection shapes variation of the human major histocompatibility complex: a review. Ann Hum Genet. 2001;65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. [DOI] [PubMed] [Google Scholar]
- Miller HC, Lambert DM. Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae) Mol Ecol. 2004;13:3709–3721. doi: 10.1111/j.1365-294X.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- Mona S, Crestanello B, Bankhead-Dronnet S, Pecchioli E, Ingrosso S, D'Amelio S, et al. Disentangling the effects of recombination, selection, and demography on the genetic variation at a major histocompatibility complex class II gene in the alpine chamois. Mol Ecol. 2008;17:4053–4067. doi: 10.1111/j.1365-294x.2008.03892.x. [DOI] [PubMed] [Google Scholar]
- Murray BW, Malik S, White BN. Sequence variation at the major histocompatibility complex locus DQ beta in beluga whales (Delphinapterus leucas) Mol Biol Evol. 1995;12:582–593. doi: 10.1093/oxfordjournals.molbev.a040238. [DOI] [PubMed] [Google Scholar]
- Negro SS, et al. The impacts of epizootics on genetic diversity and genotype-fitness association in the threatened New Zealand sea lion, Phocarctos hookeri (in preparation).
- Nevo E, Kirzher V, Beiles A, Karol A. Selection versus random drift: Long-term polymorphism persistence in small populations (evidence and modelling) Phil Trans R Soc B - Biol Sci. 1997;352:381–389. [Google Scholar]
- Nielsen R, Yang ZH. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Evermann JF. Interactive influence of infectious disease and genetic diversity in natural populations. Trends Ecol Evol. 1988;3:254–259. doi: 10.1016/0169-5347(88)90058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Yuhki N. Comparative genome organisation of the major histocompatibility complex: lessons from the Felidae. Immunol Rev. 1999;167:133–144. doi: 10.1111/j.1600-065X.1999.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver MK, Telfer S, Piertney SB. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris) Proc R Soc B-Biol Sci. 2009;276:1119–1128. doi: 10.1098/rspb.2008.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AJ. Assessment of genetic variation in the threatened New Zealand sea lion, Phocarctos hookeri, and its association with fitness. Department of Anatomy and Department of Pathology. Dunedin and Christchurch, University of Otago; 2011. [Google Scholar]
- Osborne AJ, Brauning R, Schultz JK, Kennedy MA, Slate J, Gemmell NJ. Development of a predicted physical map of microsatellite locus positions for pinnipeds, with wider applicability to the Carnivora. Mol Ecol Res. 2011;11:503–513. doi: 10.1111/j.1755-0998.2010.02962.x. [DOI] [PubMed] [Google Scholar]
- Otting N, de Groot NG, Noort MC, Doxiadis GGM, Bontrop RE. Allelic diversity of MHC-DRB alleles in rhesus macaques. Tissue Antigens. 2000;56:58–68. doi: 10.1034/j.1399-0039.2000.560108.x. [DOI] [PubMed] [Google Scholar]
- Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- Potts WK, Slev PR. Pathogen-based models favouring MHC genetic diversity. Immunol Rev. 1995;143:181–197. doi: 10.1111/j.1600-065x.1995.tb00675.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team 2010. R: A language and environment for statistical computing Available at http://www.R-project.org/R Foundation for Statistical Computing: Vienna, Austria [Google Scholar]
- Radwan J, Biedrzycka A, Babik W. Does reduced MHC diversity decrease viability of vertebrate populations. Biol Conserv. 2010;143:537–544. doi: 10.1016/j.biocon.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan J, Kawalko A, Wojcik JM, Babik W. MHC-DRB3 variation in a free-living population of the European bison, Bison bonasus. Mol Ecol. 2007;16:531–540. doi: 10.1111/j.1365-294X.2006.03179.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Westerdahl H. MHC diversity in two Acrocephalus species: the outbred great reed warbler and the inbred Seychelles warbler. Mol Ecol. 2003;12:3523–3529. doi: 10.1046/j.1365-294x.2003.02005.x. [DOI] [PubMed] [Google Scholar]
- Richman AD, Herrera LG, Nash D, Schierup MH. Relative roles of mutation and recombination in generating allelic polymorphism at an MHC class II locus in Peromyscus maniculatus. Genet Res. 2003;82:89–99. doi: 10.1017/s0016672303006347. [DOI] [PubMed] [Google Scholar]
- Robertson BR, Chilvers BL. The population decline of the New Zealand sea lion Phocarctos hookeri: a review of possible causes. Mamm Rev. 2011;41:253–275. [Google Scholar]
- Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA Database - a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- Roche 2009. 454 Sequencing Technical Bulletin No. 013-2009. Amplicon fusion primer design guidelines for GS FLX Titanium Series Lib-A Chemistry.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic Inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf SJ, Long CM, Erlich HA. Sequence analysis of the HLA-DRsz and HLA-DQsz loci from three Pemphigus vulgaris patients. Hum Immunol. 1988;22:61–69. doi: 10.1016/0198-8859(88)90052-3. [DOI] [PubMed] [Google Scholar]
- Schaschl H, Goodman SJ, Suchentrunk F. Sequence analysis of the MHC class II DRB alleles in Alpine chamois (Rupicapra r. rupicapra) Devel Comp Immunol. 2004;28:265–277. doi: 10.1016/j.dci.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Schaschl H, Wandeler P, Suchentrunk F, Obexer-Ruff G, Goodman SJ. Selection and recombination drive the evolution of MHC class II DRB diversity in ungulates. Heredity. 2006;97:427–437. doi: 10.1038/sj.hdy.6800892. [DOI] [PubMed] [Google Scholar]
- Schwensow N, Fietz J, Dausmann KH, Sommer S. Neutral versus adaptive genetic variation in parasite resistance: importance of major histocompatibility complex supertypes in a free-ranging primate. Heredity. 2007;99:265–277. doi: 10.1038/sj.hdy.6800993. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Ellegren H. A Temporal Analysis Shows Major Histocompatibility Complex Loci in the Scandinavian Wolf Population Are Consistent with Neutral Evolution. Proc R Soc Lond B-Biol Sci. 2002;271:2283–2291. doi: 10.1098/rspb.2004.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle HV, Kreiss A, Eldridge MDB, Noonan E, Clarke CJ, Pyecroft S, et al. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc Nl Acad Sci. 2007;104:16221–16226. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle HV, Marzec J, Cheng YY, Jones M, Belov K. MHC gene copy number variation in Tasmanian devils: implications for the spread of a contagious cancer. Proc R Soc B-Biol Sci. 2010;277:2001–2006. doi: 10.1098/rspb.2009.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade RW. Limited MHC polymorphism in the Southern elephant seal: Implications for MHC evolution and marine mammal population biology. Proc R Soc B-Biol Sci. 1992;249:163–171. doi: 10.1098/rspb.1992.0099. [DOI] [PubMed] [Google Scholar]
- Sommer S, Schwab D, Ganzhorn JU. MHC diversity of endemic Malagasy rodents in relation to range contraction and social system. Behav Ecol Sociobiol. 2002;51:214–221. [Google Scholar]
- Soule ME, Mills LS. Population genetics: no need to isolate genetics. Science. 1998;282:1658–1659. [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc R Soc Lond B-Biol Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srithayakumar V, Castillo S, Rosatte RC, Kyle CJ. MHC class II DRB diversity in raccoons (Procyon lotor) reveals associations with raccoon rabies virus (Lyssavirus) Immunogenetics. 2011;63:103–113. doi: 10.1007/s00251-010-0485-5. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuglik MT, Radwan J, Babik W. jMHC: software assistant for multilocus genotyping of gene families using next-generation amplicon sequencing. Mol Ecol Res. 2011;11:739–742. doi: 10.1111/j.1755-0998.2011.02997.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RH. Influence of man on vegetation and wildlife on Enderby and Rose Island, Auckland Islands. N Zeal J Bot. 1971;9:225–268. [Google Scholar]
- Trowsdale J, Groves V, Arnason A. Limited MHC polymorphism in whales. Immunogenetics. 1989;41:19–24. doi: 10.1007/BF02341609. [DOI] [PubMed] [Google Scholar]
- van Oosterhout C, Joyce DA, Cummings SM, Blais J, Barson NJ, Ramnarine IW, et al. Balancing selection, random genetic drift, and genetic variation at the major histocompatibility complex in two wild populations of guppies (Poecilia reticulata) Evolution. 2006;60:2562–2574. [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex-100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Wan Q-H, Zhu L, Wu HUA, Fang S-G. Major histocompatibility complex class II variation in the giant panda (Ailuropoda melanoleuca) Mol Ecol. 2006;15:2441–2450. doi: 10.1111/j.1365-294X.2006.02966.x. [DOI] [PubMed] [Google Scholar]
- Wang T, Pradhan K, Ye K, Wong L-J, Rohan TE.2011Estimating Allele Frequency from Next-Generation Sequencing of Pooled Mitochondrial DNA Samples Front Genet 2Article 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DS, Stewart BS, Schienman J, Lehman N. Major histocompatibility complex variation at three class II loci in the northern elephant seal. Mol Ecol. 2004;13:711–718. doi: 10.1111/j.1365-294x.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson IS, Duignan P, Castinel A, Grinberg A, Chilvers BL, Robertson BC. Sea Lions of the World - Conservation and Research in the 21st Century. 22nd Wakefield Fisheries Symposium: Alaska; 2006. Klebsiella pneumoniae epidemics: Possible impact on New Zealand sea lion recruitment. [Google Scholar]
- Yang G, Yan J, Zhou K, Wei F. Sequence variation and gene duplication at MHC DQB Loci of Baiji (Lipotes vexillifer), a Chinese river dolphin. J Hered. 2005;96:310–317. doi: 10.1093/jhered/esi055. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang ZH. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J Mol Evol. 2000;51:423–432. doi: 10.1007/s002390010105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.