Abstract
Many important functions have been attributed to the high-risk human papillomavirus (HPV) E6 and E7 proteins, including binding and degradation of p53 as well as interacting with Rb proteins. In contrast, the physiological roles of the low-risk E6 and E7 proteins remain unclear. Previous studies demonstrated that the high-risk E6 and E7 proteins also play roles in the productive life cycle by facilitating the maintenance of viral episomes (J. T. Thomas, W. G. Hubert, M. N. Ruesch, and L. A. Laimins, Proc. Natl. Acad. Sci. USA 96:8449-8454, 1999). In order to determine whether low-risk E6 or E7 is similarly necessary for the stable maintenance of episomes, HPV type 11 (HPV-11) genomes that contained translation termination mutations in E6 or E7 were constructed. Upon transfection into normal human keratinocytes, genomes in which E6 function was abolished were unable to be maintained episomally. Transfection of genomes containing substitution mutations in amino acids conserved in high- and low-risk HPV types suggested that multiple protein domains are involved in this process. Examination of cells transfected with HPV-11 genomes in which E7 function was inhibited were found to exhibit a more complex phenotype. At the second passage following transfection, mutant genomes were maintained as episomes but at significantly reduced levels than in cells transfected with the wild-type HPV-11 genome. Upon further passage in culture, however, the episomal forms of these E7 mutant genomes quickly disappeared. These findings identify important new functions for the low-risk E6 and E7 proteins in the episomal maintenance of low-risk HPV-11 genomes and suggest that they may act in a manner similar to that observed for the high-risk proteins.
Human papillomaviruses (HPVs) are small, double-stranded DNA viruses that induce hyperproliferative lesions in epithelial tissues (16, 56). More than 100 types of HPV have been identified, with each differing by at least 10% in the coding sequence of the major capsid gene, L1 (28, 35, 55). High-risk HPV types, including HPV type 16 (HPV-16), HPV-18, HPV-31, HPV-33, and HPV-42, induce lesions in the genital tract that can progress to malignancy. In contrast, low-risk HPV types, such as HPV-6 and HPV-11, which also infect genital epithelia, primarily induce benign lesions (16, 28, 55).
In the high-risk HPV types, E6 and E7 have been shown to function as oncoproteins (18, 25). E7 acts by binding to members of the Rb tumor suppressor protein family and inhibiting their ability to modulate the function of E2F transcription factors (7, 34). The low-risk E7 proteins have also been shown to bind to Rb but with a 10-fold-lower affinity (15). One of the primary activities of the high-risk forms of E6 is to target p53 for degradation (32, 43, 53), which is mediated by complex formation with the cellular ubiquitin ligase E6-AP (19, 20, 28, 41, 42). Although degradation of p53 by E6 is specific to high-risk HPV types, some evidence suggests that low-risk forms of E6 may be able to bind to p53 with a low affinity (2, 30, 53). E6 from high-risk types has also been shown to activate telomerase via upregulation of hTERT expression (13, 24, 36, 52). Furthermore, the high-risk E6 proteins have been shown to interact with a number of other cellular proteins, such as the putative calcium-binding protein E6-BP, the focal adhesion protein paxillin, p300/CBP, and PDZ domain proteins such as hDLG, MUPP-1, MAGI-1, and hScrib (1, 11, 12, 14, 23, 29, 39, 40, 48-51, 54). While the majority of these interactions have been shown to be specific for the high-risk forms of E6, several reports have described cellular binding partners for low-risk E6 proteins, such as zyxin, GPS2, Bak, and MCM7 (4, 5, 26, 27, 37, 47). In addition, it has been reported that high-risk and low-risk forms of E6 can bind to p73 (37), although this has not been seen in other studies (33). The physiological relevance of each of these interactions of the low-risk E6 proteins is not yet fully understood.
The HPV life cycle is closely linked to the differentiation status of its target epithelial tissue (28). Infection is thought to occur in cells in the basal layer of stratified epithelia where viral genomes are established as episomes and are replicated synchronously with chromosomal DNA. Following cell division, one of the infected daughter cells migrates away from the basal layer and begins to differentiate, resulting in the activation of late gene expression, amplification of the viral genomes, and virion assembly. In addition to their roles in immortalization, recent work has demonstrated that E6 and E7 play important roles in the HPV life cycle (9, 45). Using a genetic system involving transfection of cloned HPV genomes into keratinocytes, it has been demonstrated that expression of functional high-risk E6 and E7 proteins are required to maintain HPV-31 genomes as stably replicating episomes (45). Although HPV-31 genomes lacking E6 or E7 can be replicated in transient assays, they cannot be maintained in long-term stable replication assays. In addition, spontaneously immortalized keratinocytes transfected with HPV-16 genomes containing mutations in E7 are unable to amplify viral DNA upon differentiation, suggesting that E7 may also be involved in later stages of the viral life cycle (9).
The mechanism by which E6 and E7 contribute to the maintenance of episomes in the viral life cycle is not fully understood. HPV-31 genomes containing a point mutation in E6 that eliminates the ability to target p53 for degradation cannot be maintained episomally, suggesting a role for p53 in this process (45). However, when a second mutation in the E7 open reading frame (ORF) that converted its Rb binding affinity to that of the low-risk E7 was introduced into the genome, the ability to be stably maintained was restored (38). These findings suggest that a complex interplay between viral proteins can contribute to facilitating a cellular environment conducive to episomal maintenance. As with E6, genomes containing point mutations in E7 that abolish binding to Rb are unable to be stably maintained as episomes, suggesting that inactivation of Rb is also important for stable replication (45). Recent work has shown that the low-risk HPV-11 genome can also be stably maintained as an episome following transfection of normal human keratinocytes (46). Since high- and low-risk E6 and E7 proteins probably share some common functions in the productive life cycle, we have investigated the effects of inhibiting translation of these proteins in the context of the complete viral genome (Fig. 1).
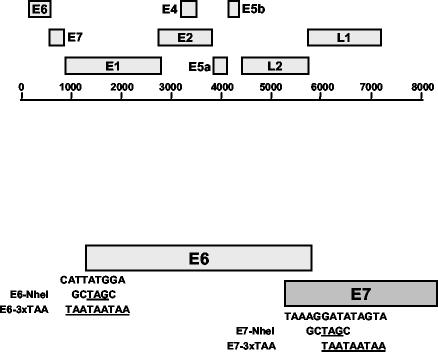
FIG. 1.
Schematic of HPV-11 genome and HPV11E6 and HPV11E7 mutations. (Top) Linear representation of the HPV-11 genome is shown. (Bottom) HPV11E6-NheI and HPV11E6-3xTAA mutants contain the TAG and TAA stop codons, respectively, in place of the ATG start codon in the E6 ORF. HPV11E7-NheI and HPV11E7-3xTAA mutants contain the TAG and TAA stop codons 25 and 27 nucleotides, respectively, downstream of the E7 initiation codon. In HPV11E6-NheI, the E6 start codon was replaced with an NheI site containing a TAG stop codon (TTATGG→GCTAGC), while HPV11E6-3xTAA contains three TAA stop codons in place of the E6 start codon (CATTATGGA→TAATAATAA). HPV11E7-NheI contains an NheI site 25 nucleotides downstream of the E7 initiation codon (AGGATA→GCTAGC). HPV11E7-3xTAA contains three TAA stop codons 27 nucleotides downstream of the E7 initiation codon (GATATAGTA→TAATAATAA). The parental HPV11 plasmid contains the HPV-11 genome inserted into the BamHI site of pBR322 (46). All mutant genomes were generated using the QuikChange XL mutagenesis kit according to the manufacturer's instructions (Stratagene, La Jolla, Calif.). Following mutagenesis, an AgeI-SwaI fragment containing the mutated region was digested and religated into the original HPV11 plasmid. The complete AgeI-SwaI fragment was sequenced to confirm the presence of the desired mutation and lack of any other sequence changes.
Loss of episomal maintenance in cells containing HPV-11 E6 and E7 mutant genomes.
We initially constructed two mutant HPV-11 genomes that inhibited E6 or E7 function. In HPV11E6-NheI, the E6 start codon was substituted with a TAG stop codon that then generates an NheI site. In HPV-11, the start codon for E7 is located within the E6 ORF, so in order to inhibit E7 function, we inserted a translation termination codon 25 nucleotides downstream of the end of the E6 coding sequence to generate HPV11E7-NheI. Wild-type and mutant HPV-11 genomes were cotransfected with pSV2Neo into normal human foreskin keratinocytes (HFKs) and selected for neomycin resistance as previously described (8, 46). Within each experiment, a single donor isolate was used. Following selection for 8 days, resistant colonies were pooled and expanded for analyses. Cells were grown to confluence and passaged once per week. Each passage of confluent cells represents approximately 3 or 4 population doublings. In previous studies, using similar transfection methods, Frattini et al. demonstrated that all clonal isolates of HPV-31-transfected cells examined contained HPV DNA at copy numbers similar to those observed for pooled cultures (10). Therefore, we believe it is likely that the majority of cells within our transfected pools contain HPV-11 DNA. However, we cannot totally exclude the possibility that some neomycin-resistant cells contain no HPV-11 DNA.
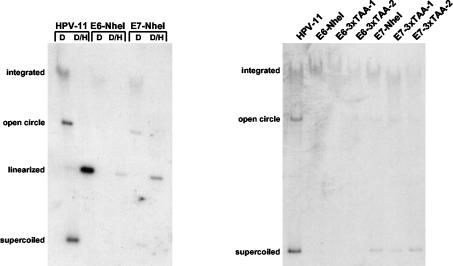
A representative Southern blot analysis of cells transfected with HPV11, HPV11E6-NheI, or HPV11E7-NheI plasmid is shown in Fig. 2. Cells transfected with the wild-type HPV-11 genome were found to harbor both episomal and integrated forms of the genome at the second passage after transfection (Fig. 2, left). However, no extrachromosomal copies of the genome were detected in cells at the same passage transfected with the HPV11E6-NheI genome, and these cells maintained only small numbers of integrated copies. This suggests that HPV-11 E6 expression is important for episomal maintenance. In cells transfected with the HPV11E7-NheI genome, there was a significant decrease observed in episomal copy number, although low levels of episomes were still detected. Similar results were seen in three separate transfections using different donor keratinocytes. These findings suggest that low-risk E6 and E7 play significant roles in episomal maintenance, as measured at the second passage following transfection.
FIG. 2.
Southern blot analysis of HPV11E6 and HPV11E7 mutant genomes. Genomic DNA was extracted at the second passage following transfection and examined by Southern blot analysis. HFKs were transfected after isolation from foreskins, and the cells were split once or twice during the selection period. (Left) Southern blot analysis of HPV11E6 and HPV11E7 mutant genomes. Samples contained 5 μg of genomic DNA and were digested with DpnI alone (D) to remove any residual input DNA or DpnI and HindIII (D/H) to linearize HPV DNA. (Right) Southern blot analysis of HPV11E6-3xTAA and HPV11E7-3xTAA mutant genomes. Two separate plates were transfected with the same mutant genome and are shown in lanes HPV11E6-3xTAA-1 and HPV11E6-3xTAA-2 and in lanes HPV11E7-3xTAA-1 and HPV11E7-3xTAA-2. Samples contained 8 μg of genomic DNA and were digested with DpnI to remove any residual input DNA. Southern blot analysis was performed as described previously (8, 46). Within each experiment, DNA samples were taken from equivalent passages and quantitated by spectrophotometry. Equal amounts of DNA were electrophoresed on agarose gels, and following electrophoresis, gels were stained with ethidium bromide to confirm that there were no significant differences in total DNA.
In order to confirm that these findings were not the result of cis effects due to the sequences used to generate the mutations, we examined the effects of a second set of mutant genomes in our transfection assays. In HPV11E6-3xTAA, three TAA stop codons were inserted in place of the E6 start codon, while HPV11E7-3xTAA contains three TAA stop codons 27 nucleotides downstream of the E7 initiation codon (Fig. 1). HFKs were transfected with wild-type HPV-11 genomes or the HPV11E6-NheI, HPV11E6-3xTAA, HPV11E7-NheI, and HPV11E7-3xTAA mutant genomes. As shown in Fig. 2, cells transfected with wild-type HPV-11 maintained high levels of episomes along with some integrated forms. In contrast, at the same passage number, no episomal copies of the genome were detected in cells transfected with either the HPV11E6-NheI or HPV11E6-3xTAA mutant genome, and these cells had only low levels of integrated DNA. These findings confirm that E6 function is critical for episomal maintenance of the low-risk HPV-11 genome. In cells transfected with either of the HPV11E7 mutant genomes, we consistently detected a significantly lower level of episomal DNA than that seen with cells transfected with wild-type HPV-11. These findings indicate that in the absence of E7 expression, cells were impaired in the ability to maintain episomes, although low levels could still be detected. Similar effects were observed in three independent experiments at similar passage numbers.
In this study, our use of two types of mutations to eliminate E6 and E7 function ensured that the phenotype we observed was truly due to the lack of E6 or E7 protein and not due to the alteration of a cis element important for other viral functions. Although we have previously shown that disruption of splice donor and acceptor sites within the E6 ORF in the high-risk HPV-31 genome leads to a loss of E1 and/or E2 expression, no such splice sites or splicing patterns in E6 have been found in the low-risk HPV-11 genome (17, 45). Thus, while we cannot completely exclude the possibility that expression of other viral genes may have been affected by these mutations, we believe that the similarity of our findings with two different types of mutations makes this possibility less likely.
HPV11E6 and HPV11E7 mutant genomes cannot be maintained episomally over extended time in culture.
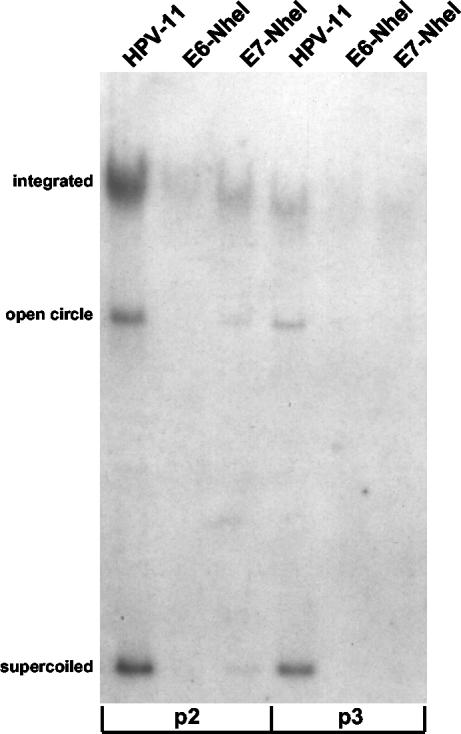
The above experiments demonstrated that HPV11E6 mutant genomes could no longer be maintained extrachromosomally within two passages following transfection. Although episomal maintenance of HPV11E7 mutant genomes was also impaired, low levels of episomal DNA could still be detected at the same passage. We thus examined whether the reduced levels of viral episomes seen in cells transfected with HPV11E7 mutant genomes could be maintained upon further passage in culture. As shown in Fig. 3, wild-type HPV-11-transfected cells harbored both episomal and integrated copies of the genome at two and three passages after transfection. Consistent with the results seen at earlier passages, no episomal DNA was detected at either passage for the HPV11E6-NheI genome. In cells transfected with the HPV11E7-NheI genome, a very low level of episomal DNA could be detected at the second passage, but by the following passage, there were no longer any episomal copies detected, suggesting that the lack of E7 function leads to a more severe defect in episomal maintenance over time. We did not extensively analyze the state of episomal DNA beyond passage three, but in all passages where the wild-type HPV-11 genome was compared with the HVP11E6 or HVP11E7 mutant genome, we detected levels of episomal DNA that were significantly higher for the wild-type genome. In the experiment shown in Fig. 3, we were unable to detect episomal DNA at passage six for the wild-type HPV-11 genome. In a previous study, Thomas et al. detected episomal forms of HPV-11 at later passages (46). We believe that the different results observed in this study are due to the specific host keratinocytes used for each experiment. Similar findings were observed with the HPV11E6-3xTAA and HPV11E7-3xTAA mutants (data not shown), and comparable results were found in six other independent experiments comparing the wild-type HPV-11 genome with the HPV11E6-NheI mutant genome. These findings indicate that E6 and E7 play an important role in the episomal maintenance of the HPV-11 genome, such that genomes lacking either E6 or E7 function cannot be maintained beyond three passages following transfection.
FIG. 3.
Analysis of cells transfected with HPV11E6 and HPV11E7 mutant genomes over extended time in culture. Genomic DNA was extracted at passage two (p2) or passage three (p3) after transfection and examined by Southern blot analysis. All samples contained 8 μg of genomic DNA and were digested with DpnI to remove any residual input DNA.
Stable replication of HPV-11 genomes containing point mutations in E6.
We next sought to investigate the mechanisms by which the HPV-11 E6 protein contributes to episomal maintenance. We focused on E6, since elimination of its activity led to rapid loss of episomes. In order to identify domains within the HPV-11 E6 protein that are important for episomal maintenance, a series of substitution mutations within the E6 ORF in the context of the HPV-11 genome was made. We reasoned that since both the high-risk HPV-31 E6 protein and the low-risk HPV-11 E6 protein are important for stable episomal maintenance, they might bind to a common partner or act by a shared pathway through conserved amino acids. Comparison of the amino acid sequences of the high-risk HPV-31, HPV-16, and HPV-18 E6 types with the low-risk HPV-11 and HPV-6 E6 types indicated a lack of extensive homology between the two sets of proteins (Fig. 4). Indeed, the majority of amino acids common between all five types are scattered within the protein, rather than clustered into domains. The two major motifs conserved between all five forms of the protein are the two zinc finger regions (Cys-X-X-Cys-29 amino acids-Cys-X-X-Cys). However, previous mutational analysis with the high-risk E6 proteins suggested that these residues are important for structural integrity rather than protein-protein interaction domains (3, 31). We identified several amino acids that are conserved in the different forms of E6 (amino acids W133, CC67/137, L111) that have already been studied in the context of the high-risk E6 proteins. Mutation of amino acid 133 (W133R) has been shown to inhibit the ability of HPV-16 E6 to both bind p53 and target it for degradation (3), while the CC67/137GG mutation blocks p53- or p73-dependent transactivation (37). The L111Q mutation eliminates most E6 activities, including binding and degradation of p53, as well as binding to E6-AP and E6-BP (31). In addition, in the context of the complete HPV-16 genome, the L111Q mutation inhibits episomal maintenance (37, 38). Finally, we identified two additional amino acids (R56 and R78) that are conserved in low-risk and high-risk forms of E6 but have not been examined previously.
FIG. 4.
Sequence homology between high-risk and low-risk forms of E6. Amino acid sequences of the high-risk HPV-16, HPV-18, and HPV-31 E6 proteins were aligned with amino acid sequences of the low-risk HPV-6b and HPV-11 E6 proteins using MegAlign. Consensus residues are depicted above the aligned sequences, with black indicating strong conservation and white indicating weak conservation. Mutated residues are boxed, and changes are indicated. All mutations were constructed using the QuikChange XL mutagenesis kit. Gaps introduced to maximize sequence alignment are indicated by dashes. E6.pro, E6 protein.
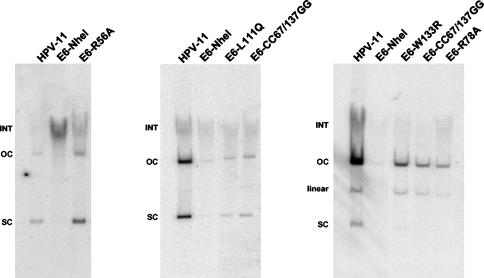
Mutant genomes that incorporated these mutations individually into the E6 ORF in the context of the complete HPV-11 genome were constructed. Wild-type and mutant HPV-11 genomes were then cotransfected with pSV2Neo into HFKs and selected for neomycin resistance as in previous experiments. Resistant colonies were pooled and expanded for Southern blot analyses. As shown in Fig. 5, cells transfected with the HPV11E6-NheI mutant genome contained little to no episomal DNA at the second passage after transfection. In contrast, cells transfected with the HPV11E6-R56A mutant genome contained episomal DNA at levels similar to those of cells transfected with the wild-type HPV-11 genome, suggesting that this mutation has little effect on episomal maintenance at this passage number. In contrast, Southern blot analysis performed at the second passage following transfection of genomes containing the L111Q, CC66/137GG, W133R, and R78A mutations revealed impaired episomal maintenance (Fig. 5). Cells transfected with these mutant genomes contained little to no episomal DNA compared to cells transfected with the wild-type HPV-11 genome. These findings suggest that mutation of these residues significantly impairs, but does not totally inhibit, the ability of E6 to facilitate HPV-11 stable replication at this passage number. Similar effects were seen in three independent experiments for each mutant genome with the exception of the HPV11E6-R56A mutant, which was examined once. In the rightmost Southern blot shown in Fig. 5, we detected high levels of open circle and linear HPV-11 DNA as opposed to supercoiled forms, which we believe was due to nicking that occurred during the DNA extraction process. At later passages, cells transfected with the HPV11E6-L111Q, -CC66/137GG, -W133R, and -R78A mutant genomes lost the ability to maintain episomes, while cells transfected with wild-type HPV-11 still retained significant levels of extrachromosomal viral DNA (data not shown). We have therefore identified four mutations in E6 that impair its function in episomal maintenance.
FIG. 5.
Analysis of HPV-11 genomes containing substitution mutations in the E6 ORF. Genomic DNA was extracted at the second passage following transfection and examined by Southern blot analysis. All samples contained 15 μg of genomic DNA and were digested with DpnI to remove any residual input DNA. The positions of integrated (INT), open circle (OC), linear, and supercoiled (SC) forms of HPV DNA are indicated to the left of the blots. (Left) HFKs were transfected with the wild-type HPV-11, HPV11E6-NheI, or HPV11E6-R56A genome. (Middle) HFKs were transfected with the wild-type HPV-11, HPV11E6-NheI, HPV11E6-L111Q, or HPV11E6-CC67/137GG genome. (Right) HFKs were transfected with the wild-type HPV-11, HPV11E6-NheI, HPV11E6-W133R, HPV11E6-CC67/137GG, or HPV11E6-R78A genome. Increased levels of open circle and linear DNA in these samples are likely due to nicking that occurred during DNA extraction.
In this study, we have identified a function for the low-risk HPV-11 E6 protein in modulating the stable maintenance of viral episomes. This is one of the first functions that has been demonstrated for low-risk E6 in the context of the viral life cycle. As E6 proteins have shown only weak nonspecific DNA binding activity, we do not believe that E6 acts directly to modulate episome replication or segregation. Rather, we suspect that E6 acts to alter the cellular environment to allow for the long-term maintenance of extrachromosomal elements. The presence of extrachromosomal DNAs in normal cells is not tolerated and is likely sensed as a DNA damage stimulus. Papillomaviruses must therefore block these normal cellular checkpoints to allow for persistence of viral episomes. Given that high-risk E6 proteins have been shown to act in a manner similar to low-risk E6 in modulating the maintenance of viral episomes, they may target similar cellular pathways. It is likely that these functions are mediated through the binding of E6 proteins to cellular factors, and the studies described provide important tools to identify these factors.
Our analysis of the HPV-11 E6 protein identified four amino acid substitution mutations that resulted in significantly impaired episomal maintenance. These mutants will be of particular importance in the future in facilitating the identification of cellular binding partners that mediate these functions of E6. The four substitution mutations induced a significant reduction in episomal copy number, but not a total loss of episomal maintenance at early passage. These individual mutations therefore did not induce as severe a phenotype as did the complete translation stop mutants. This could indicate that multiple activities of E6 are required for its role in episomal maintenance or that some of the mutations we have examined induce structural changes in E6 that indirectly affect its function leading to a loss of episomes. Finally, we cannot exclude the possibility that some of these mutations alter protein stability, but our inability to immunoprecipitate the HPV-11 E6 protein from cells precludes testing this hypothesis. Our observation that some substitution mutations in E6 result in a phenotype similar to that seen with stop codon mutations further suggests that these effects are not due to nonsense-mediated decay. Further work characterizing the ability of these different E6 mutants to bind to cellular proteins is necessary to better elucidate the mechanism by which the low-risk E6 protein facilitates episomal maintenance.
We also observed an effect on episomal maintenance when E7 function was inhibited. The effects we observed with HPV11E7 were, however, less severe than those seen with HPV11E6 mutant genomes, resulting initially in only a partial reduction in the levels of viral episomes. However, within three passages following transfection, episomal DNA was no longer detected. As in the case of E6, the specific manner in which E7 contributes to the HPV-11 viral life cycle is unclear. Previous studies have shown that low-risk forms of E7 bind to Rb, although this occurs with a greatly reduced affinity compared to that of high-risk forms of E7 (15). Therefore, it is possible that binding of Rb family members is responsible for the role of E7 in facilitating episomal maintenance; this is supported by studies in HPV-31 (45). Two early promoters are present in the HPV-11 genome, and one directs transcription of messages that contain E7 as the first ORF (6, 44). In contrast, in the high-risk HPV-31 genome, there is only a single early promoter, and E7 is found on transcripts as the second ORF with E6 or E6* upstream (21, 22). It is therefore possible that due to the configuration of early transcripts in low-risk viruses, the low-risk E7 proteins are synthesized at higher levels than their high-risk counterparts, leading to similar effects on Rb action. The testing of this hypothesis, however, will require the development of efficient antibodies to HPV-11 E7 to accurately determine levels in cells.
The study of episomal maintenance in the low-risk HPV-11 types is complicated by the lack of immortalizing capability of these viral types. This makes it important to compare the effects of various HPV-11 mutants on episomal maintenance as a function of passage number, as we have done in this study. For instance, inhibition of HPV-11 E7 function did not result in an immediate loss of episomal maintenance, but rather this was manifested only over extended passage in culture. However, at all passages examined, wild-type HPV-11 genomes were maintained extrachromosomally at significantly higher copy numbers than episomal forms of E7 mutant genomes. We are confident that the comparisons made in this study represent physiologically important activities of the low-risk E6 and E7 gene products. Upon extended passage of cells in culture, we also observed that the levels of wild-type HPV-11 episomes decreased, although they consistently remained higher than the levels of HPV11E6 or HPV11E7 mutant genomes at the same passage. Thus, we believe the differences we observed reflect true differences in episomal maintenance. We also observed some variation in the levels of wild-type HPV-11 episomal DNA between transfection experiments that we suspect was due to differences in the donor keratinocytes used. However, in experiments using the same donor isolate, we saw consistent differences between wild-type and mutant genomes. Thus, we believe that accurate comparisons can be made between wild-type and mutant genomes within a single experiment. In summary, we have identified novel functions for the low-risk E6 and E7 proteins in the HPV life cycle. These activities are similar to those previously observed for high-risk HPV proteins and may reflect common pathways utilized by both types of viruses during their productive life cycles.
Acknowledgments
We thank members of the Laimins laboratory for helpful comments and technical advice.
This study was supported by a grant from the NCI and a grant from the NIAID-supported Sexually Transmitted Disease Cooperative Research Center to L.A.L. (CA74202). S.T.O. was supported by a NIH/NRSA Carcinogenesis Training Grant (5 T32 CA09560-15) to Northwestern University.
REFERENCES
- 1.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 2.Crook, T., J. A. Tidy, and K. H. Vousden. 1991. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67:547-556. [DOI] [PubMed] [Google Scholar]
- 3.Dalal, S., Q. Gao, E. J. Androphy, and V. Band. 1996. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J. Virol. 70:683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degenhardt, Y. Y., and S. Silverstein. 2001. Interaction of zyxin, a focal adhesion protein, with the E6 protein from human papillomavirus type 6 results in its nuclear translocation. J. Virol. 75:11791-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degenhardt, Y. Y., and S. J. Silverstein. 2001. Gps2, a protein partner for human papillomavirus E6 proteins. J. Virol. 75:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiLorenzo, T. P., and B. M. Steinberg. 1995. Differential regulation of human papillomavirus type 6 and 11 early promoters in cultured cells derived from laryngeal papillomas. J. Virol. 69:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 8.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiol, D., S. Galizzi, and L. Banks. 2002. Mutational analysis of the discs large tumour suppressor identifies domains responsible for human papillomavirus type 18 E6-mediated degradation. J. Gen. Virol. 83:283-289. [DOI] [PubMed] [Google Scholar]
- 12.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 13.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 75:7198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heck, D. V., C. L. Yee, P. M. Howley, and K. Munger. 1992. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 89:4442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 947-978. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 17.Hubert, W. G., and L. A. Laimins. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse, J. M., and S. L. Beaudenon. 1996. Mechanism of HPV E6 proteins in cellular transformation. Semin. Cancer Biol. 7:317-326. [DOI] [PubMed] [Google Scholar]
- 19.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hummel, M., H. B. Lim, and L. A. Laimins. 1995. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J. Virol. 69:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 25.Kubbutat, M. H., and K. H. Vousden. 1996. Role of E6 and E7 oncoproteins in HPV-induced anogenital malignancies. Semin. Virol. 7:295-304. [Google Scholar]
- 26.Kuhne, C., and L. Banks. 1998. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 273:34302-34309. [DOI] [PubMed] [Google Scholar]
- 27.Kukimoto, I., S. Aihara, K. Yoshiike, and T. Kanda. 1998. Human papillomavirus oncoprotein E6 binds to the C-terminal region of human minichromosome maintenance 7 protein. Biochem. Biophys. Res. Commun. 249:258-262. [DOI] [PubMed] [Google Scholar]
- 28.Laimins, L. A. 1993. The biology of human papillomaviruses: from warts to cancer. Infect. Agents Dis. 2:74-86. [PubMed] [Google Scholar]
- 29.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X., and P. Coffino. 1996. High-risk human papillomavirus E6 protein has two distinct binding sites within p53, of which only one determines degradation. J. Virol. 70:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantovani, F., and L. Banks. 1999. The interaction between p53 and papillomaviruses. Semin. Cancer Biol. 9:387-395. [DOI] [PubMed] [Google Scholar]
- 33.Marin, M. C., C. A. Jost, M. S. Irwin, J. A. DeCaprio, D. Caput, and W. G. Kaelin, Jr. 1998. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol. Cell. Biol. 18:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, G., H. Lu, C. Calef, and T. Leitner. 1996. Heterogeneity of papillomaviruses. Semin. Cancer Biol. 7:349-358. [DOI] [PubMed] [Google Scholar]
- 36.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 75:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J. S., E. J. Kim, J. Y. Lee, H. S. Sin, S. E. Namkoong, and S. J. Um. 2001. Functional inactivation of p73, a homolog of p53 tumor suppressor protein, by human papillomavirus E6 proteins. Int. J. Cancer 91:822-827. [DOI] [PubMed] [Google Scholar]
- 38.Park, R. B., and E. J. Androphy. 2002. Genetic analysis of high-risk E6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J. Virol. 76:11359-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pim, D., M. Thomas, R. Javier, D. Gardiol, and L. Banks. 2000. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene 19:719-725. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner, M., J. M. Huibregtse, and P. M. Howley. 1994. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc. Natl. Acad. Sci. USA 91:8797-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 43.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 44.Smotkin, D., H. Prokoph, and F. O. Wettstein. 1989. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J. Virol. 63:1441-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, J. T., S. T. Oh, S. S. Terhune, and L. A. Laimins. 2001. Cellular changes induced by low-risk human papillomavirus type 11 in keratinocytes that stably maintain viral episomes. J. Virol. 75:7564-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, M., and L. Banks. 1999. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 80:1513-1517. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, M., B. Glaunsinger, D. Pim, R. Javier, and L. Banks. 2001. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20:5431-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, X., R. Salgia, J. L. Li, J. D. Griffin, and P. M. Howley. 1997. The bovine papillomavirus E6 protein binds to the LD motif repeats of paxillin and blocks its interaction with vinculin and the focal adhesion kinase. J. Biol. Chem. 272:33373-33376. [DOI] [PubMed] [Google Scholar]
- 51.Vande Pol, S. B., M. C. Brown, and C. E. Turner. 1998. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 16:43-52. [DOI] [PubMed] [Google Scholar]
- 52.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 54.Zimmermann, H., R. Degenkolbe, H. U. Bernard, and M. J. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]
- 56.zur Hausen, H., and E. M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]