Abstract
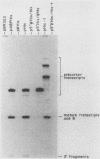
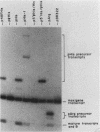
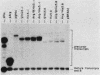
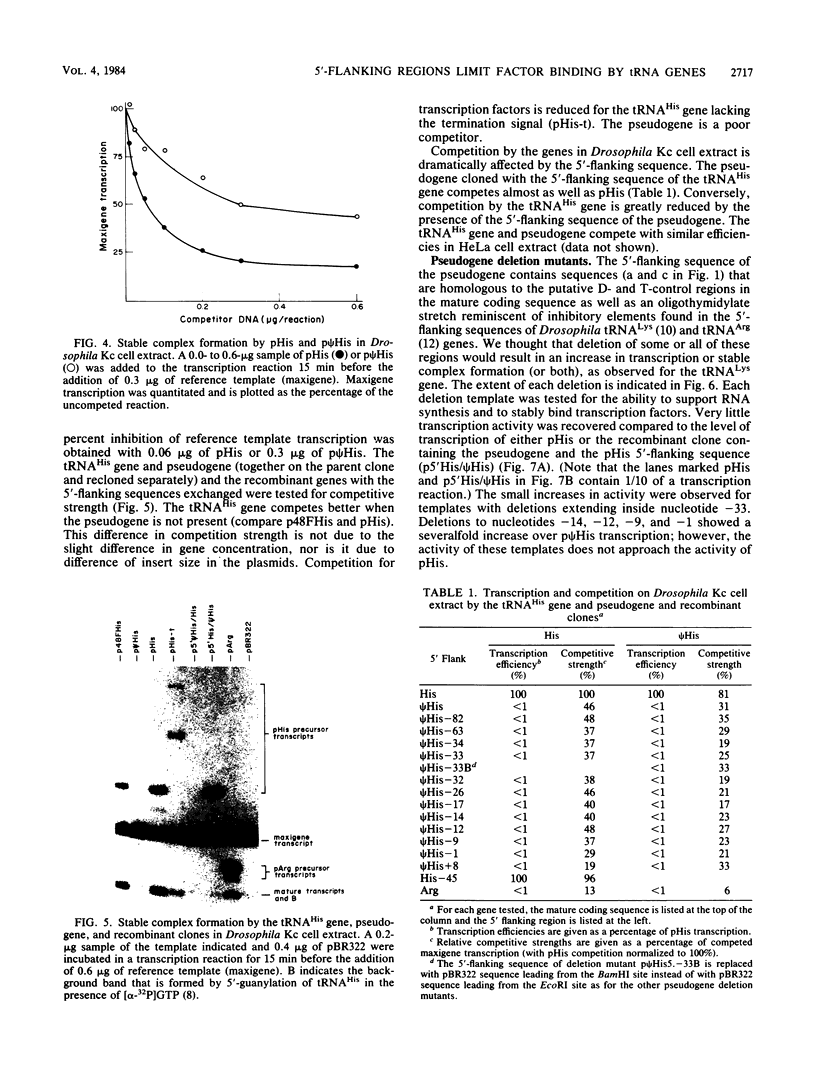
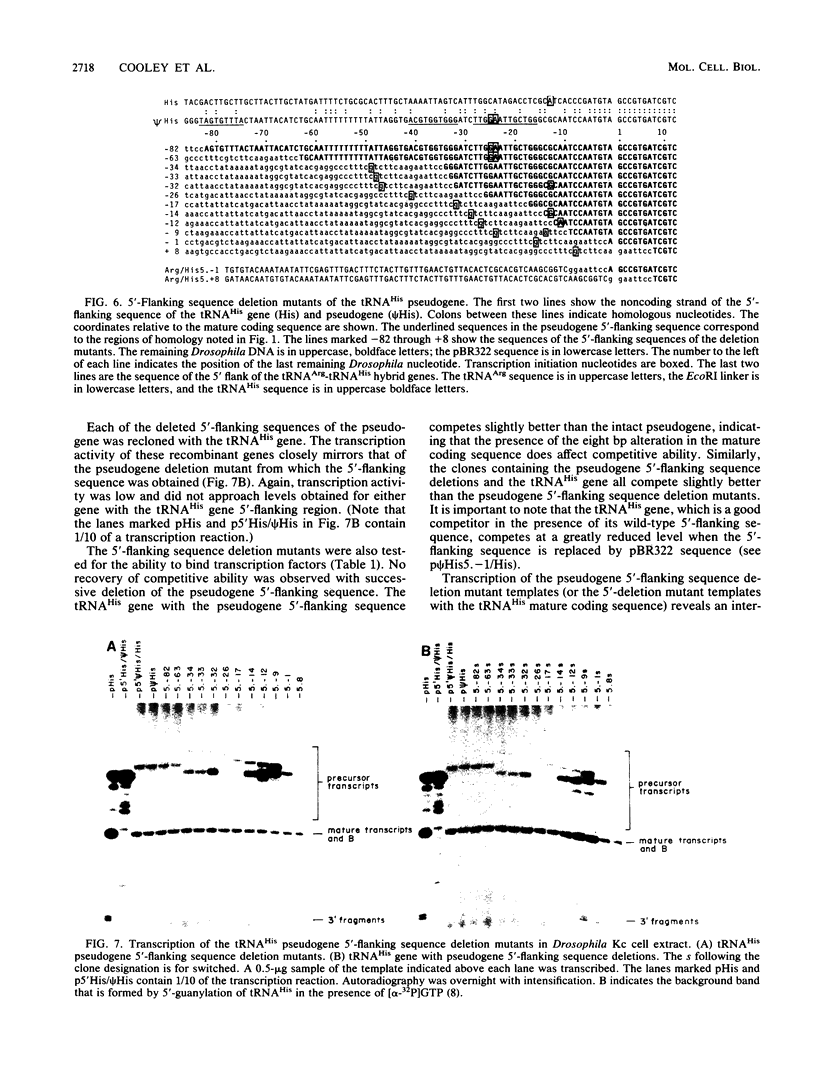
We determined the sequence of a Drosophila tRNA gene cluster containing a tRNAHis gene and a tRNAHis pseudogene in close proximity on the same DNA strand. The pseudogene contains eight consecutive base pairs different from the region of the bona fide gene which codes for the 3' portion of the anticodon stem of tRNAHis. The tRNAHis gene is transcribed efficiently in Drosophila Kc cell extract, whereas the pseudogene is not. The pseudogene is also a much poorer competitor than the real gene in a stable transcription complex formation assay, even though the sequence alteration in the pseudogene does not affect the sequence or spacing of the putative internal transcription control regions. Recombinant clones were constructed in which the 5'-flanking regions are exchanged. The transcription efficiencies and competitive abilities of the recombinant clones resemble those of the genes from which the 5' flank was derived; for example, the tRNAHis pseudogene with the 5'-flanking sequence of the tRNAHis gene is now efficiently transcribed. Deletion analysis of the pseudogene 5' flank failed to uncover an inhibitory element. Deletion analysis of the real gene showed very high dependence on the presence of the wild-type 5'-flanking sequence for factor binding to the internal control regions and stable complex formation. The 5'-flanking sequence of a Drosophila tRNAArg gene active in the Drosophila Kc cell extract does not restore transcriptional activity or stable complex formation. The tRNAHis gene and pseudogene behave atypically in HeLa cell extract. Both genes compete for HeLa transcription factors, but neither of them is efficiently transcribed. Removal of the 5'-flanking sequences of each gene and replacement with various sequences, including the tRNAArg gene 5' flank, does not allow increased transcription in HeLa cell extract.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Model P., Dixon G. H., Wosnick N. A. An E. coli gene coding for a protamine-like protein. Cell. 1981 Nov;26(3 Pt 1):299–304. doi: 10.1016/0092-8674(81)90198-7. [DOI] [PubMed] [Google Scholar]
- Altwegg M., Kubli E. The nucleotide sequence of histidine tRNA gamma of Drosophila melanogaster. Nucleic Acids Res. 1980 Aug 11;8(15):3259–3262. doi: 10.1093/nar/8.15.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. J., Schaack J., Sharp S., Söll D. Partial purification of Drosophila Kc cell RNA polymerase III transcription components. Evidence for shared 5 S RNA and tRNA gene factors. J Biol Chem. 1983 Dec 25;258(24):15224–15231. [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Raugei G., Costanzo F., Dente L., Cortese R. Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase iii. Cell. 1983 Mar;32(3):725–733. doi: 10.1016/0092-8674(83)90058-2. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Traboni C., Cortese R. Relationship between the two components of the split promoter of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1921–1925. doi: 10.1073/pnas.79.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Appel B., Söll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5' terminus of histidine tRNA. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Denison R. A., Weiner A. M. Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol. 1982 Jul;2(7):815–828. doi: 10.1128/mcb.2.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Dingermann T., Sharp S., Appel B., DeFranco D., Mount S., Heiermann R., Pongs O., Söll D. Transcription of cloned tRNA and 5S RNA genes in a Drosophila cell free extract. Nucleic Acids Res. 1981 Aug 25;9(16):3907–3918. doi: 10.1093/nar/9.16.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingermann T., Sharp S., Schaack J., Söll D. Stable transcription complex formation of eukaryotic tRNA genes is dependent on a limited separation of the two intragenic control regions. J Biol Chem. 1983 Sep 10;258(17):10395–10402. [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Gage L. P. Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell. 1979 Nov;18(3):817–828. doi: 10.1016/0092-8674(79)90134-x. [DOI] [PubMed] [Google Scholar]
- Goddard J. P., Squire M., Bienz M., Smith J. D. A human tRNAGlu gene of high transcriptional activity. Nucleic Acids Res. 1983 May 11;11(9):2551–2562. doi: 10.1093/nar/11.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Hollis G. F., Hieter P. A., McBride O. W., Swan D., Leder P. Processed genes: a dispersed human immunoglobulin gene bearing evidence of RNA-type processing. Nature. 1982 Mar 25;296(5855):321–325. doi: 10.1038/296321a0. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Makowski D. R., Haas R. A., Dolan K. P., Grunberger D. Molecular cloning, sequence analysis and in vitro expression of a rat tRNA gene cluster. Nucleic Acids Res. 1983 Dec 20;11(24):8609–8624. doi: 10.1093/nar/11.24.8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Roeder R. G. Selective and accurate transcription of the Xenopus laevis 5S RNA genes in isolated chromatin by purified RNA polymerase III. Proc Natl Acad Sci U S A. 1977 Jan;74(1):44–48. doi: 10.1073/pnas.74.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M. D., Hendrick J. P., Jr, Lerner M. R., Steitz J. A., Reichlin M. A mammalian tRNAHis-containing antigen is recognized by the polymyositis-specific antibody anti-Jo-1. Nucleic Acids Res. 1983 Feb 11;11(3):853–870. doi: 10.1093/nar/11.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J., Egan J., Hudson L., Landy A. The tyrT locus: termination and processing of a complex transcript. Cell. 1981 Nov;26(3 Pt 1):305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Burke D. J., Cooley L., Söll D. The extent of a eukaryotic tRNA gene. 5'- and 3'-flanking sequence dependence for transcription and stable complex formation. J Biol Chem. 1984 Feb 10;259(3):1461–1467. [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Sege R., Söll D., Ruddle F. H., Queen C. A conversational system for the computer analysis of nucleic acid sequences. Nucleic Acids Res. 1981 Jan 24;9(2):437–444. doi: 10.1093/nar/9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Silberklang M., Hosbach H. A., Schmidt T., Kubli E., Gergen J. P., Wensink P. C., Söll D. The initiator tRNA genes of Drosophila melanogaster: evidence for a tRNA pseudogene. Nucleic Acids Res. 1981 Nov 25;9(22):5867–5882. doi: 10.1093/nar/9.22.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Schaack J., Sharp J. A., Burke D. J., DeRobertis E. M., Söll D. Each element of the Drosophila tRNAArg gene split promoter directs transcription in Xenopus oocytes. Nucleic Acids Res. 1983 Dec 20;11(24):8677–8690. doi: 10.1093/nar/11.24.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Shibuya K., Noguchi S., Nishimura S., Sekiya T. Characterization of a rat tRNA gene cluster containing the genes for tRNAAsp, tRNAGly and tRNAGlu, and pseudogenes. Nucleic Acids Res. 1982 Jul 24;10(14):4441–4448. doi: 10.1093/nar/10.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S., Schmidt O., Söll D., Hovemann B. The nucleotide sequence of a cloned Drosophila arginine tRNA gene and its in vitro transcription in Xenopus germinal vesicle extracts. J Biol Chem. 1979 Oct 25;254(20):10290–10294. [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Denison R. A. Either gene amplification or gene conversion may maintain the homogeneity of the multigene family encoding human U1 small nuclear RNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1141–1149. doi: 10.1101/sqb.1983.047.01.129. [DOI] [PubMed] [Google Scholar]
- Wilde C. D., Crowther C. E., Cripe T. P., Gwo-Shu Lee M., Cowan N. J. Evidence that a human beta-tubulin pseudogene is derived from its corresponding mRNA. Nature. 1982 May 6;297(5861):83–84. doi: 10.1038/297083a0. [DOI] [PubMed] [Google Scholar]