Abstract
Enormous strides have recently been made in our understanding of the biology and pathobiology of mitochondria. Many diseases have been identified as caused by mitochondrial dysfunction, and many pharmaceuticals have been identified as previously unrecognized mitochondrial toxicants. A much smaller but growing literature indicates that mitochondria are also targeted by environmental pollutants. We briefly review the importance of mitochondrial function and maintenance for health based on the genetics of mitochondrial diseases and the toxicities resulting from pharmaceutical exposure. We then discuss how the principles of mitochondrial vulnerability illustrated by those fields might apply to environmental contaminants, with particular attention to factors that may modulate vulnerability including genetic differences, epigenetic interactions, tissue characteristics, and developmental stage. Finally, we review the literature related to environmental mitochondrial toxicants, with a particular focus on those toxicants that target mitochondrial DNA. We conclude that the fields of environmental toxicology and environmental health should focus more strongly on mitochondria.
Key Words: contaminants, mitochondria, mitochondrial DNA, mitochondrial toxicity, mitochondrial disease.
MITOCHONDRIA AND MITOCHONDRIAL DNA
Mitochondria are essential organelles best known for ATP generation and their involvement in apoptosis. They also play critical roles in other key processes including calcium, copper, and iron homeostasis; heme and iron-sulfur cluster assembly; synthesis of pyrimidines and steroids; thermogenesis and fever response; and calcium signaling. The great majority of the ~1000–1500 (Calvo and Mootha, 2010) proteins that carry out these functions are imported from the cytoplasm via mitochondria targeting sequences or other mechanisms (Bolender et al., 2008), and the proteins present vary significantly with tissue (Johnson et al., 2007). This variability probably reflects extensive variability in function, ranging from energy production in muscle mitochondria to steroid synthesis in adrenal mitochondria (Vafai and Mootha, 2012). A small number (13 in humans) is encoded in the mitochondrial genome, along with the tRNA and rRNAs required for their synthesis. These proteins are components of complexes I, III, IV, and V of the electron transport chain (ETC). Despite their small number, they are essential and are transcribed at high rates: mtRNA represents ~5% of total cellular RNA in many tissues but as high as 30% in heart cells (Mercer et al., 2011).
Mitochondrial morphology also varies with cell type, developmental stage, and environment (Jansen and de Boer, 1998; Rube and van der Bliek, 2004; Vafai and Mootha, 2012)(Fig. 1). Morphology ranges from highly fragmented to highly networked, is regulated via the processes of mitochondrial fusion and fission, and is responsive to a variety of stressors (Jendrach et al., 2008; Twig and Shirihai, 2011). Morphological dynamics are critical for maintenance of mitochondrial function (Green and Van Houten, 2011; Youle and van der Bliek, 2012). The amount of cellular mitochondria and their content are regulated via mitochondrial biogenesis, nuclear signaling–mediated nuclear and mitochondrial transcription, autophagy, and intraorganellar degradation processes (Diaz and Moraes, 2008; Mijaljica et al., 2007; Piantadosi and Suliman, 2012; Scarpulla, 2008). “Mitophagy” can selectively remove damaged mitochondria (Kim and Lemasters, 2011; Kim et al., 2007), and damaged cellular components can be degraded inside the mitochondria or exported to lysosomes and peroxisomes via mitochondria-derived vesicles (Fischer et al., 2012; Nunnari and Suomalainen, 2012). Mitochondria do not merely receive instructions from the nucleus, however, but rather can initiate signaling in a variety of ways. These include “retrograde signaling” (i.e., from the mitochondria to the nucleus; Chae et al., 2013; Liu and Butow, 2006) to coordinate nuclear transcription with mitochondrial needs via pathways including AMP-activated protein kinase, sirtuins, calcium signaling, peroxisome proliferator–activated receptor g coactivator 1a protein, and others (Piantadosi and Suliman, 2012); mitochondrial-nuclear reactive oxygen species (ROS)–mediated signaling (Storz, 2006); cell cycle arrest signaling that may involve proteins with similar functions in the nucleus (Green et al., 2011; Koczor et al., 2009; Kulawiec et al., 2009); and initiation of apoptosis (Tann et al., 2011; Raimundo et al., 2012), as opposed to the better-known amplification of an apoptotic process initiated elsewhere. There is evidence for a “mitocheckpoint” that senses mitochondrial dysfunction and triggers cell cycle arrest (Singh, 2006) although the mechanism for this response is not fully understood.
Fig. 1.
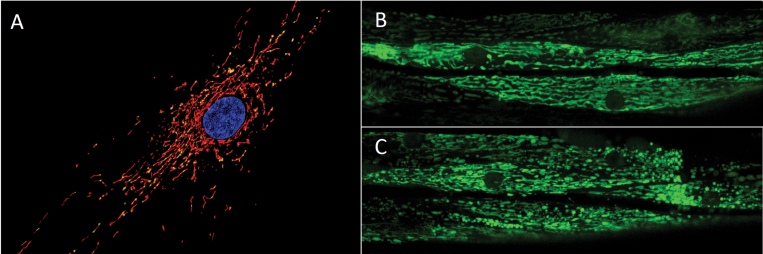
Mitochondrial morphology. Panel (A) shows the mitochondrial network (MitoTracker Red-stained) containing many mtDNA nucleoids (PicoGreen-stained) surrounding the nucleus (Hoescht-stained) of a human primary fibroblast (photo: Amanda Bess). Panels (B) and (C) show a transgenic strain of C. elegans that expresses a mitochondrial matrix-targeted green fluorescent protein in body wall muscle cells. The image in panel (B) is from a control nematode, and the image in panel (C) is from a nematode exposed to 1µM carbonyl cyanide m-chlorophenyl hydrazone, a potent ionophore and inhibitor of oxidative phosphorylation that leads to mitochondrial fragmentation; the mitochondrial matrix was visualized via expression of green fluorescent protein (photo: John Rooney).
Mitochondrial DNA (mtDNA) in many animal species is a circular intron-free genome consisting of 14,000–17,000 base pairs (16,569 in humans) although it is much larger and noncircular and contains introns in some species (Anderson et al., 1981; Burger et al., 2003). It is typically maternally inherited and present in multiple copies within each cell; the mtDNAs are grouped in DNA-protein complexes referred to as nucleoids (Bogenhagen, 2012; Kukat et al., 2011) that are anchored to the inner mitochondrial membrane. Most somatic cells in mammals have 103–104 mtDNAs (Shoubridge and Wai, 2007), but this number varies significantly by cell type and developmental stage, as discussed at more length below. mtDNA replication occurs independently of the cell cycle. mtDNA half-life has been reported as 2–4 days (Kai et al., 2006; Lipsky and Pedersen, 1981) but has not been characterized in a wide range of cell types. mtDNA replication and transcription are regulated in part by Tfam (Mitochondrial transcription factor A), a major protein component of the nucleoid (Campbell et al., 2012). From an evolutionary perspective, mtDNA has a high mutation rate compared with nuclear DNA (nDNA). Normally, all of the mtDNAs in all of the cells of an organism are of the same sequence; the presence of more than one sequence in a cell is referred to as “heteroplasmy.”
Recognition of the key role of mtDNA integrity and mitochondrial function in health has grown greatly in recent years (Chan, 2007; Chan et al., 2006; Christodoulou, 2000; Cohen, 2010; de Moura et al., 2010; DiMauro and Davidzon, 2005; Hirano et al., 2001; Jansen and Burton, 2004; Kwong et al., 2006; Manfredi and Beal, 2000; Nunnari and Suomalainen, 2012; Schapira, 2012; Schmidt, 2010; Shaughnessy et al., 2010; Taylor and Turnbull, 2005; Wallace, 2005), partly due to our increased understanding and awareness of mitochondrial diseases.
MITOCHONDRIAL DISEASES
mtDNA and nDNA Mutations Can Cause Mitochondrial Disease
Leber’s Hereditary Optical Neuropathy and mitochondrial myopathies were in 1988 the first diseases demonstrated to be caused by mtDNA mutations (Holt et al., 1988; Wallace et al., 1988). There are now more than 200, including MELAS (myopathy, encephalopathy, lactic acidosis, stroke-like episodes), MERRF (myoclonus epilepsy with ragged red fibers), and many others (Holt, 2010). Importantly, cellular dysfunction and clinical disease do not occur until a threshold proportion of mtDNAs carrying mutations (heteroplasmy) is reached (DiMauro and Schon, 2003; Rossignol et al., 2003). Together, these diseases affect at least 1/10,000 people clinically (Chinnery et al., 2012), establishing mtDNA mutations as a major cause of disease.
Because most mitochondrial proteins are nuclear encoded, it is not surprising that mutations in many nuclear genes also cause mitochondrial diseases. For example, mutations in DNA polymerase γ, the only mtDNA polymerase and thus responsible for all mtDNA replication and repair, cause a wide range of mitochondrial diseases including progressive external ophthalmoplegia, Alper’s syndrome, and Leigh’s syndrome (Copeland, 2012). The disease Friedreich’s ataxia is caused by mutations in the gene encoding frataxin, a protein involved in iron-sulfur cluster assembly (Marmolino, 2011).
Although individually rare, diseases caused by mtDNA and nDNA mutations are estimated to collectively have an incidence of ~1/4000 individuals (Chinnery et al., 2004; DiMauro and Davidzon, 2005; Howell et al., 2005; Wallace, 2005). There are currently no pharmaceutical cures for any mitochondrial disease (Nunnari and Suomalainen, 2012).
Mitochondrial Involvement in Common Diseases: Causation Versus Correlation
The diseases described above are unambiguously caused by mitochondrial dysfunction. There are also many very common diseases for which there is strong correlative evidence, suggesting that mitochondrial dysfunction is at least partially causative (Wallace, 2005). These are frequently diseases of old age or energy homeostasis and include neurodegenerative diseases such as Parkinson’s Disease and Alzheimer’s Disease (Coskun et al., 2012), many cancers (Brandon et al., 2006; Kulawiec et al., 2010; Penta et al., 2001), diabetes (Rolo and Palmeira, 2006), metabolic syndrome (Mercer et al., 2010), cardiovascular disease (Ballinger, 2005), and others (Schon et al., 2012). Considerable effort is currently devoted to investigate whether these relationships are causal. There is experimental evidence demonstrating that defective mtDNA replication and repair can accelerate organismal aging (Bratic and Larsson, 2013; Trifunovic et al., 2004).
Gene-Environment Interactions
In addition to the genetic diseases described so far, several gene-environment interactions have been identified (Cohen, 2010; Finsterer and Segall, 2010). For example, valproic acid is fatal in the context of some mitochondrial diseases (Krähenbühl et al., 2000; Silva et al., 2008), aminoglycosides (e.g., gentamycin) can cause deafness in the context of specific mtDNA mutations (Guan, 2011), and cigarette smoking and alcohol intake are major risk factors for vision loss in people who are homoplasmic for a mutation causing Leber’s hereditary optic neuropathy (Kirkman et al., 2009).
Lessons from Mitochondrial Diseases
The existence of so many mitochondrial diseases illustrates the critical importance of maintenance of mitochondrial and mtDNA integrity for health. It also raises an important question with implications for environment-mediated mitochondrial toxicity: Why did it take us so long to realize that many diseases are in fact mitochondrial diseases? Part of the answer is surely that these diseases are very complicated. Although they often target tissues that have high energy use or contain nonreplicating (and therefore irreplaceable) cells, this is somewhat oversimplified (Vafai and Mootha, 2012), and there is significant variability in their presentation. Some mtDNA mutations result in single-tissue diseases, others affect a wide spectrum of tissues, and others affect different tissues in different patients, with varying age of onset (Vafai and Mootha, 2012; Nunnari and Suomalainen, 2012). Furthermore, as illustrated by the valproic acid and aminoglycoside examples, the same environmental exposure may be innocuous in some people but deadly in others, depending on genetic background. These complications suggest that identifying the etiology of an environmentally induced mitochondrial dysfunction is likely to be challenging with both laboratory and epidemiological approaches, as borne out by the observation that, currently, the causes of only ~50% of severe cases of mitochondrial dysfunction are identified (Vafai and Mootha, 2012).
DRUG-INDUCED MITOCHONDRIAL AND MTDNA TOXICITY
Drugs Can Cause Many Off-Target Mitochondrial Effects
Recognition that mitochondrial toxicity is a key “off-target” effect of many drugs is also a relatively recent development. Drugs have now been identified that inhibit and uncouple the ETC; inhibit mitochondrial transport pathways, fatty acid oxidation or uptake, the citric acid cycle, mtDNA replication, and mitochondrial protein synthesis (a common mode of toxicity of antibiotics that target bacterial ribosomes, to which mitochondrial ribosomes are similar due to mitochondria’s evolutionary origins); generate (or exacerbate generation of) ROS; and more (Begriche et al., 2011; Cohen, 2010; Dykens and Will, 2007; Kovacic et al., 2005; Mehta et al., 2008; Neustadt and Pieczenik, 2008; Pacheu-Grau et al., 2010; Rowe et al., 2001).
One well-studied example is adriamycin (doxorubicin), a chemotherapeutic whose clinical use is limited by the fact that it also causes irreversible and cumulative cardiomyopathy (Wallace, 2007). It appears to act largely by poisoning mitochondria, both via redox cycling to generate ROS and by inhibiting ATP production via uncoupling of oxidative phosphorylation. Although its antineoplastic function is largely a result of DNA intercalation and topoisomerase II inhibition in the nucleus, a substantial amount reaches mitochondria, where it has a high binding affinity for cardiolipin, a lipid found only in the mitochondrial inner membrane (Mustonen and Kinnunen, 1993).
Another example is the nucleoside reverse transcriptase inhibitors (NRTIs) that are used to combat human immunodeficiency virus (HIV) infection. NRTIs act by inhibiting the reverse transcriptase activity required for viral replication. They have been highly successful in treating adults and in preventing transmission of HIV from pregnant mothers to their children, but unfortunately many NRTIs also inhibit the mtDNA polymerase γ. This has resulted mtDNA depletion- and mutation-mediated mitochondrial toxicity, and even death, in patients and in animal models (Benhammou et al., 2007; Blanche et al., 1999; Chan, 2007; Claessens et al., 2003; Divi et al., 2010; Kohler and Lewis, 2007). Similar effects have been observed with nucleoside analogs intended for other viruses as well (McKenzie et al., 1995). Thus, chemicals that damage mtDNA or alter its copy number can have very serious health consequences.
Such toxicities have led to failed clinical trials, “Black Box” warnings and withdrawal of drugs from the market, and have driven the pharmaceutical industry to develop better methods for detection of drug-induced mitochondrial dysfunction (Begriche et al., 2011; Dykens et al., 2007; Marroquin et al., 2007; Nadanaciva et al., 2009; Pereira et al., 2009).
Lessons from Drug-Induced Mitochondrial Toxicity
Mechanisms of drug-induced toxicity offer lessons in terms of how other chemicals might damage mitochondria; these are discussed at more length below. As in the case of mitochondrial diseases, it is instructive to consider why it took us so long to realize that many drugs target mitochondria. One reason is that frequently only a small percentage of people have adverse reactions (Dykens and Will, 2007). For example, only ~1% of children of HIV-treated mothers showed mitochondrial toxicity (Barret et al., 2003). The reason for this variability is not entirely clear. In some cases, gene-environment interactions are likely the explanation. In others, environment-environment interactions, epigenetic factors (see below), or the stochasticity associated with mtDNA replication and allocation may be involved. In any case, it is sobering from an environmental toxicology perspective that it took years to recognize the mitochondrial toxicity of many drugs because drugs are in general much better studied for toxicity than most environmental contaminants. This suggests that many environmental pollutants may be undiscovered mitochondrial toxicants.
REASONS FOR MITOCHONDRIAL VULNERABILITY, AND FACTORS THAT MAY MODULATE VULNERABILITY
Reasons for mitochondrial vulnerability are presented graphically in Figure 2 and discussed in more detail below.
Fig. 2.
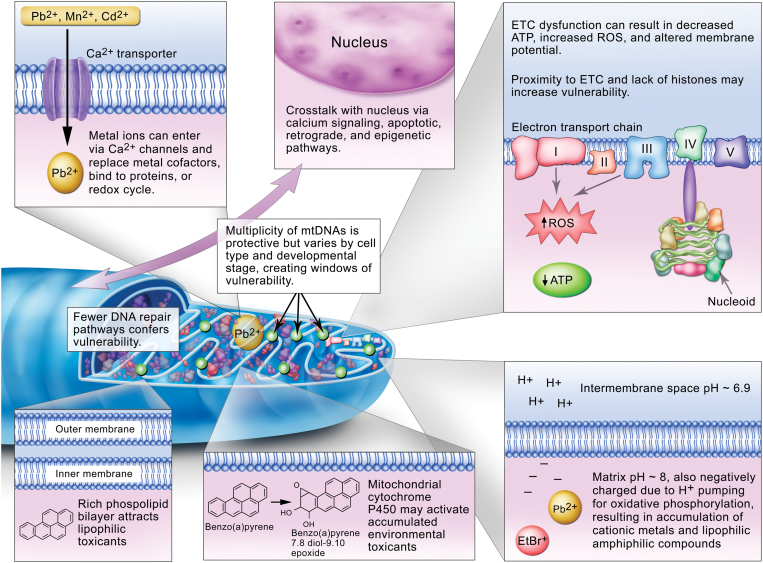
Factors that affect mitochondrial vulnerability to environmental toxicants, as discussed in the text.
Accumulation of Pollutants
The high lipid content of mitochondrial membranes facilitates accumulation of lipophilic compounds such as polycyclic aromatic hydrocarbons (PAHs) (Backer and Weinstein, 1982) and some alkylating agents (Wunderlich et al., 1972). Cationic metals, such as lead, cadmium, mercury, and manganese, have also been shown to accumulate in mitochondria preferentially (Atchison and Hare, 1994; Bucio et al., 1999; Castellino and Aloj, 1969; Gavin et al., 1992; Sokolova et al., 2005a). These metals may accumulate in mitochondria due to both entry via calcium transporters (i.e., molecular mimicry) and chemical behavior resulting from their interactions with mitochondrial pH and charge. The matrix has a negative charge and slightly alkaline pH (~8 vs. ~6.9 for the intermembrane space) due to the proton pumping associated with oxidative phosphorylation. This also results in accumulation of certain organic chemicals, particularly amphiphilic xenobiotics including well-known mitochondrial toxicants such as ethidium bromide, paraquat, 1-methyl-4-phenylpyridinium (MPP+), and others (Cohen, 2010; Mehta et al., 2008). It is also a property that is exploited to target toxic (Modica-Napolitano and Aprille, 2001), diagnostic (Ross et al., 2005), or therapeutic (Murphy, 2008; Murphy and Smith, 2007) chemicals to mitochondria.
Ability to Metabolically Activate Pollutants
Another factor contributing to the mitochondrial susceptibility is the presence of cytochrome P450s in mitochondria, which can activate chemicals that are relatively nonreactive prior to metabolism, such as PAHs and mycotoxins (Dong et al., 2009; Genter et al., 2006; Omura, 2006). In addition, the existence of close contacts between mitochondria and the endoplasmic reticulum (de Brito and Scorrano, 2010) raises the possibility that metabolites generated in the endoplasmic reticulum could reach the mitochondria.
mtDNA Exhibits Unique Vulnerabilities But May Be Buffered by High Copy Number
mtDNA is more vulnerable than nDNA to some environmental genotoxins although the opposite is also true in some cases (see below). A number of explanations for this observation have been presented, including mtDNA’s physical location, its reduced protein packaging compared with nDNA, and its reduced repair capacity. These, along with the potential for protection due to high copy number, are described in more detail below.
Physically, mtDNA nucleoids are anchored to the matrix side of the inner membrane, in close proximity to the ETC and many proteins that contain transition metal ions. The ETC normally generates a significant amount of ROS by electron loss; this has been estimated at ~0.1% of O2 converted to superoxide anion (Beckman and Ames, 1998; Fridovich, 2004) although the amount may decrease with mild uncoupling (Miwa and Brand, 2003). Much of this appears to be produced by Complex I, with release to the matrix, and Complex III, with release to the matrix and intermembrane space (Brand et al., 2004). Manganese superoxide dismutase (SOD) in the matrix, and copper-zinc SOD in the intermembrane space, convert superoxide anion to the longer lived and uncharged hydrogen peroxide; additional antioxidant enzymes are present and regulated as well (Valle et al., 2005). Mitochondrial H2O2 may interact with nearby metals to produce hydroxyl radicals (Giulivi et al., 1995) in close proximity to mtDNA. The many chemicals, disease states, etc., that cause an increase in ROS generation may increase the amount of superoxide produced: This includes redox cycling agents that can intercept electrons such as adriamycin; inhibitors of the ETC such as antimycin A and sodium azide that lead to a super-reduced state of upstream complexes, increasing the tendency to release electrons; dietary and exercise-related ETC dysfunction; hyperoxia and hypoxia-reoxygenation; and others (Bailey et al., 2006; Barrett et al., 2004; Begriche et al., 2006; Davies and Doroshow, 1986; Hasegawa et al., 1990; Indo et al., 2007; Ishiguro et al., 2001; Li et al., 2003a; Powell and Jackson, 2003; Senft et al., 2002a , b; Turrens et al., 1982a , b). Furthermore, the Fenton reaction may be facilitated by redox cycling of transition metal ions that are either chelated (Dean and Nicholson, 1994) or released via ROS-induced degradation of metal-containing proteins (Mladenka et al., 2006).
mtDNA lacks histones, which has frequently been cited as a potential reason for increased vulnerability. However, recent work (Bogenhagen, 2012) has shown that in fact a large number of proteins associate with mtDNA, some of which are explicitly protective (Valle et al., 2005), so mtDNA vulnerability to reactive chemicals due to “nakedness” may be less than previously speculated. Recent work has shown that nucleoid structure is not static but can also be remodeled in response to exposure to DNA intercalators including ethidium bromide and doxorubicin (Ashley and Poulton, 2009a , b) and oxidative stress (Rebelo et al., 2009). Rebelo et al. (2009) presented evidence suggesting that different areas of the mitochondrial genome were differentially packaged, so different regions may be more or less “naked.” The nucleoid remodeling observed by Ashley and Poulton (2009a) was protective against mtDNA depletion that resulted from intercalator exposure. Furthermore, the remodeling was p53 mediated, increasing the evidence for a role of p53 in mitochondrial homeostasis (Szczepanek et al., 2012). mtDNA occurs in a surprising array of structures in cells from a wide range of tissues and species (Kolesar et al., 2012). This may reflect to some extent significant variation in the packaging function of proteins such as Tfam (Campbell et al., 2012) and the transcriptional status of the individual genomes (Kolesar et al., 2012). Finally, Chen et al. (2005) showed that aconitase is a nucleoid component that assists in mtDNA maintenance and protection against ethidium bromide exposure, raising the possibility that nucleoid repackaging may respond to bioenergetics and redox perturbations via aconitase mobilization (Shadel, 2005). Thus, mtDNAs likely have differential susceptibility to damage, both in a cell type–specific fashion and even within single cells.
A very large difference between nDNA and mtDNA is the relative lack of repair pathways present in mitochondria, at least in humans (there is species variability). Although our understanding of mtDNA repair pathways is evolving and includes more proteins and pathways than previously recognized (Croteau et al., 2012; Kamenisch et al., 2010; Liu and Demple, 2010; Scheibye-Knudsen et al., 2012; Sykora et al., 2011; Valentin-Vega et al., 2012), many types of damage are either irreparable or only very slowly repaired. Base excision repair is present in mitochondria, but nucleotide excision repair (NER) is not, and recombinational and mismatch repairs are either absent or quite limited (Druzhyna et al., 2008; Gredilla et al., 2010; Kazak et al., 2012; Kraytsberg et al., 2004). Furthermore, mtDNA is a major target for oxidative stress (Van Houten et al., 2006), despite the relatively robust capacity for repair of oxidative damage. The sole replicative mtDNA polymerase, DNA polymerase γ, appears to have very little translesion synthesis capacity (Graziewicz et al., 2004, 2007; Kasiviswanathan et al., 2012), so that this damage tolerance mechanism is also lacking or very limited. The absence of NER is of particular relevance from an environmental toxicology perspective because it means that many important environmental genotoxicants including some PAHs, mycotoxins, and ultraviolet light will cause damage that is irreparable in mtDNA. Furthermore, because a subset of DNA damage caused by ROS is repaired by NER (Brooks, 2008; Cline et al., 2010), any of the many factors that increase ROS in mitochondria (see above) could also create some highly persistent damage.
What might the effects of persistent mtDNA damage be? In addition to inhibiting replication, there is strong evidence that such damage inhibits RNA transcription (Cline, 2012) and mitochondrial function (Bess et al., 2012; Brar et al., 2012; Furda et al., 2012b; Leung et al., 2013; Rachek et al., 2009). Although most of the measures of mitochondrial function so far have focused on energetics (ATP levels, oxygen consumption, etc.), it may be useful to consider other endpoints. For example, might mitochondrial dysfunction in the context of steroid-producing cells result in endocrine disruption? mtDNA damage likely contributes to generation of mtDNA mutations although this process is currently poorly understood (Copeland, 2012; Krishnan et al., 2008; Kulawiec et al., 2010; Loeb et al., 2005). Improved sequencing methods support the relatively high (compared with the rest of the mtDNA) mutability of the D loop region (Schmitt et al., 2012) and will improve our understanding of both mutagenesis and normal occurrence of heteroplasmy, which is now detectable at much lower levels than previous methods permitted. The “vicious cycle” hypothesis suggests that mitochondrial damage can lead to greater mitochondrial dysfunction, in turn leading to greater release of ROS and damage, etc. (Yakes and Van Houten, 1997).
On the other hand, the mitochondrial genome is present in multiple copies, potentially conferring buffering via redundancy. As described earlier, mitochondrial dysfunction resulting from mtDNA mutations is not observed until a certain threshold of heteroplasmy is reached. This likely results from the combination of multiple copies and the fact that mitochondrial fusion permits swapping of contents, sometimes referred to as complementation or functional buffering. Presumably, such buffering would also occur for mtDNA damage. We recently found that similar levels of mtDNA damage led to much greater inhibition of mitochondrial function in live Caenorhabditis elegans than cells in culture (Bess et al., 2012, 2013b; Leung et al., 2013). Although there are multiple possible explanations for this observation, it could result from the much higher mtDNA copy number in human cells in culture compared with C. elegans. The high copy number also means that irreparable—or even repairable—DNA damage can be handled by degrading damaged mtDNAs by one of a number of possible mechanisms including degradation of the genomes themselves or of entire mitochondria along with mtDNA (i.e., autophagy or mitophagy)(Bess et al., 2012, 2013a, b; Rouzier et al., 2012; Shokolenko et al., 2009). mtDNAs that have been removed can then be replaced by mtDNA replication. The high copy number and potential functional buffering also suggests that mtDNA damage may be an excellent early biomarker of mitochondrial damage: High levels of mtDNA damage will occur and persist and therefore be detectable, yet will precede late-stage collapse of mitochondrial function associated with clinical detection.
Further complicating the picture is evidence that inhibition of mitochondrial function may, at intermediate levels and under some circumstances, have at least some beneficial effects. For example, mild inhibition of mitochondrial function extends lifespan and increases resistance to a variety of stressors in C. elegans (Dillin et al., 2002; Rea et al., 2007) and mice (Liu et al., 2005). In many cases, this effect is only observed if the inhibition occurs during development and not if it occurs later in life. However, it is important to note that greater inhibition leads to pathological effects, and in general, developmental exposures that alter programming of mitochondrial function later in life are not always be beneficial (Knudsen and Green, 2004).
Cell Type Modulates Sensitivity
Mitochondria vary dramatically from tissue to tissue (Calvo and Mootha, 2010; Vafai and Mootha, 2012), so it is not surprising that evidences from mitochondrial disease and mitochondrial drug toxicity indicate that different cell types are differentially susceptible to mitochondrial toxicants. There are many factors that may sensitize specific cell types to mitochondrial toxicity. For example, high reliance on mitochondrial function may mean that dysfunction more easily causes cell death (e.g., as often seen in mitochondrial diseases that target high energy-use cells) because the increased function may result in higher ROS production in some circumstances. An interesting example here is the aforementioned case of adriamycin, which causes principally mitochondrial toxicity in heart, where much electron flow occurs in mitochondria, and much more endoplasmic reticulum damage in liver, where electron flow is much higher in the ER (Berthiaume and Wallace, 2007; Brown and Borutaite, 2012). Certain cell types (e.g., some neurons) also have low antioxidant defenses and/or high inherent oxidative stress (Crouch et al., 2007). Proliferating cells, both cancerous and noncancerous, generate a great deal of ATP via aerobic glycolysis rather than oxidative phosphorylation, apparently in order to preferentially utilize carbon for biosynthesis required for cell division (Vander Heiden et al., 2009). Any functional deficits associated with mitochondrial dysfunction in such cells may result more from loss of biosynthetic than energy-generating capacity. Finally, there is evidence that mtDNA repair capacity varies with cell type, with brain tissue in particular showing low repair (Karahalil et al., 2002; Szczesny et al., 2010). Nonetheless, it is clear that the full spectrum of factors that result in cell-type sensitivity is not well understood, and short-term effects may differ from long-term effects, in part due to the occurrence of secondary responses (Vafai and Mootha, 2012).
Developmental Stage Modulates Sensitivity
Some developmental stages are likely more sensitive to mitochondrial toxicants due to age-dependent differences in mitochondrial content, mitochondrial metabolism, and mitochondrial defense/repair mechanisms.
Mitochondrial count/volume and mtDNA copy number vary dramatically with age in humans and other species that have been examined. Although most cells thus far examined have 103–104 copies of mtDNA, somatic and primordial germ cells in the early embryo may contain as few as 10s or 100s of copies, constituting a “bottleneck” in mtDNA copy number and/or transferral (Carling et al., 2011; Jansen, 2000; Jansen and de Boer, 1998; Pikó and Taylor, 1987; Shoubridge and Wai, 2007). Lower copy number is likely to reduce the protective effect of redundant copies described above. However, the bottleneck may also offer an opportunity for removal of severely damaged mitochondria or aberrant mtDNAs (Fan et al., 2008; Stewart et al., 2008), although it is not entirely clear when this process of purifying selection acts or how it may vary by mutation type (Freyer et al., 2012), in part because the mitochondrial genome is not believed to be very active early in development when the bottleneck occurs. Furthermore, although newly fertilized eggs have a very high mtDNA copy number (~105 (Shoubridge and Wai, 2007); but highly variable (May-Panloup et al., 2007)), the evidence suggests that in many species those mtDNAs are simply allocated into daughter cells during early development, without additional replication, resulting in much lower copy number after initial rounds of cell division (Artuso et al., 2012; Bratic et al., 2009; Chase and Dawid, 1972; Leung et al., 2013; Rubenstein et al., 1977; Tsang and Lemire, 2002; Wai et al., 2008). Because mtDNA allocation into daughter cells at mitosis (“mitotic segregation”) is apparently random, and the bottleneck phenomenon results in low copy number at least in some cell types (most have not been examined), it is possible for some cells to end up with a high percentage of damaged or mutant mtDNAs after multiple cycles of cell division. This high level of heteroplasmy or damaged mtDNAs would then be inherited by all of the cells derived from the progenitor cell. As previously pointed out in other contexts (Krishnan et al., 2008), mitochondrial alterations such as mutations in cell types that do undergo a bottleneck would not manifest until the second (in the case of somatic cells) or even third (in the case of primordial germ cells) generation, relative to an exposed mother. The effects might be somatic alterations or fertility difficulties in the children of the exposed mother or mitochondrial defects in the grandchildren. There is also a lower copy number in old age (Hebert et al., 2010; Meyer et al., 2007), suggesting vulnerability in aged individuals.
Age-related changes in mitochondrial metabolism are also well documented. Early life stages often display more anaerobic metabolism and spherical morphologies, and older individuals typically show signs of mitochondrial dysfunction although these effects are frequently tissue specific (Braeckman et al., 2002; Brand et al., 2004; Brys et al., 2007, 2010; Dillin et al., 2002; Hebert et al., 2010; Jansen and de Boer, 1998; Knudsen and Green, 2004; Meyer et al., 2007; Tsang and Lemire, 2003; Tsang et al., 2001; Yasuda et al., 2006). How reduced aerobic mitochondrial function will influence the effect of mitochondrial toxicants is not certain. On the one hand, reduced dependence on oxidative phosphorylation can result in less sensitivity to mitochondrial toxicity, as observed in at least some cells in culture (Marroquin et al., 2007). On the other hand, reduced capacity to generate energy might inhibit repair processes and stress responses. Furthermore, the age dependence of nonenergy-related mitochondrial functions (calcium homeostasis, heme and iron-sulfur cluster assembly, synthesis of pyrimidines and steroids, etc.) is less well understood. Finally, there is evidence for an age-related decrease in base excision repair capacity in mtDNA (Anson et al., 2006; Druzhyna et al., 2008; Gredilla et al., 2010; Maynard et al., 2009; Szczesny and Mitra, 2005; Szczesny et al., 2003).
We speculate that these early-life vulnerabilities make mitochondria important candidates for playing a role in diseases and pathologies resulting from early environmental exposures, an important area of current environmental health research (Kai et al., 2006; Knudsen and Green, 2004; Landrigan et al., 2005). Similarly, late-life vulnerabilities suggest that environmental toxicants may play an important role in the mitochondrial dysfunction commonly observed in aging and diseases of aging (Ames et al., 1995; Chomyn and Attardi, 2003; Fridovich, 2004; Melov et al., 1995; Nicholls, 2002).
Conclusion: Vulnerability and Robustness
Many of the physical, chemical, and biological characteristics of mitochondria make them vulnerable to toxicant exposure (Fig. 2). At the same time, mitochondria are generally robust (Vafai and Mootha, 2012) and are protected in a number of ways, including great redundancy of content, ability to replace defective components, and complementation. However, genetic deficiencies in the mitochondrial processes responsible for these defenses may sensitize some people to exposures (Meyer and Bess, 2012).
MITOCHONDRIAL SUSCEPTIBILITY TO ENVIRONMENTAL TOXICANTS
The pathologies associated with mitochondrial disease and drug-induced toxicity highlight the importance of mitochondrial function for health. The demonstrated mitochondrial effects of many drugs along with the theoretical considerations described above lead to the concern that mitochondria may be an important target for many environmental toxicants, presumably via many or all of the same mechanisms described above for drugs. The possibility that environmental contaminants may target mitochondria has been noted previously (Shaughnessy et al., 2010). Nonetheless, which contaminants are important is far from clear (Schmidt, 2010). We review some of the literature on pollutants that target mitochondria, focusing in particular on mtDNA as a target. We also discuss promising research approaches and propose important directions for future research.
Environmental Mitochondrial Toxicants
Although environmental contaminants have in general been less well studied than drugs from the mitochondrial toxicity perspective, there are nonetheless some well-understood examples. Paraquat, for example, appears to act largely by redox cycling in a fashion analogous to adriamycin (Martinez and Greenamyre, 2012). Rotenone is a Complex I inhibitor that results in increased ROS production, mtDNA damage, and apoptosis (Ishiguro et al., 2001; Li et al., 2003b; Shokolenko et al., 2009). Carbon monoxide and cyanide act as Complex IV inhibitors (Ninomiya-Tsuji, 2008). An example of an environment-environment interaction is provided by the unfortunate co-occurrence of folate deficiency and exposure to either cyanide or methanol, which appears to have resulted in mitochondrial toxicity–mediated optic neuropathy in ~50,000 people (Sadun, 1998). There are also many other contaminants that are less well characterized as mitochondrial toxicants but are reported to cause at least some of their effects via mitochondrial toxicity. These include particulate matter (Janssen et al., 2012; Hou et al., 2010; Xia et al., 2004), lipopolysaccharide (Suliman et al., 2003), cadmium and copper (Garceau et al., 2010; Sokolova et al., 2005a, b), PAH quinones (Babu et al., 2005), dioxin (Biswas et al., 2008; Shertzer et al., 2006), acrolein (Jia et al., 2007), acrylamide (Lee et al., 2012), perfluorinated compounds (Starkov and Wallace, 2002; Walters et al., 2009), usnic acid (Guo et al., 2008; Joseph et al., 2009), methoxychlor (Gupta et al., 2006), 3-nitropropionate (Sabri, 1998), cigarette smoke (Ballinger et al., 1996; Cakir et al., 2007; Westbrook et al., 2010), manganese and lead (Bowman et al., 2011; Sabri, 1998; Zheng et al., 1998), arsenic (Dopp et al., 2008; Echaniz-Laguna et al., 2012; Naranmandura et al., 2011), mercury (Belyaeva et al., 2012; Farina et al., 2012; O’Hara et al., 2002), and pentachlorophenol (Valmas et al., 2008). It is important to note that many if not all of the exposures are likely to affect targets other than mitochondria, and in some cases, mitochondrial effects may be secondary to effects elsewhere.
However, although many of these chemicals cause effects other than mitochondrial toxicity, the importance of the mitochondrial effects is in some cases supported by the fact that there is overlap in the pathologies observed after exposures and mitochondrial diseases. For example, the optic neuropathy described by Sadun (1998) was very similar to Leber’s Hereditary Optical Neuropathy, and rotenone, paraquat, and manganese are all associated with Parkinson’s Disease or related symptoms (Cannon and Greenamyre, 2011; Tanner et al., 2011).
Environmental Mitochondrial Genotoxicants
Supplementary table 1 lists studies that have directly compared nDNA with mtDNA damage after chemical exposure. A few studies are included that examined only mtDNA damage or deletions. Studies that have examined chemical effects on mtDNA copy number only, while important, were too numerous to tabulate. Although a full review of the observed effects is beyond the scope of this review, it is important to note that chemical exposures have resulted in both decreased and increased mtDNA copy number, and this may depend not just on the chemical but also on the dose and time point assessed. For example, oxidative stress appears to be able to stimulate mitochondrial biogenesis (including mtDNA replication) at low doses but depletion at higher doses. This may be a function of competing effects of ROS, resulting in both removal of damaged genomes (Shokolenko et al., 2009) and induction of biogenesis (Suliman et al., 2005). In addition, short-term depletion can result in compensatory responses (Suliman et al., 2003; Leung et al., 2013) over the longer term.
Some genotoxicants cause more damage in mtDNA (e.g., PAHs and oxidative stress), whereas others (e.g., methylmethanesulfonate) cause more damage in nDNA (Supplementary table 1). In some cases (e.g., cisplatin, aflatoxin B1), both have been reported (Supplementary table 1). For some toxicants, the degree of differential vulnerability has been quite variable in different reports. In a dramatic example, PAHs and aflatoxin B1 have been reported to cause more mtDNA than nDNA damage, but the ratio has been as low as less than 2:1 and as high as 500:1 (Supplementary table 1). Similarly, mtDNA:nDNA damage ratios after cisplatin exposure have varied from much less than 1 to several hundred. The reasons for these large differences are unclear but may reflect different experimental designs, methods for measuring DNA damage, or cell type differences such as nucleoid packaging (Campbell et al., 2012). Of note, the number of genotoxicants that have been tested for relative mitochondrial versus nuclear genotoxicity is relatively limited.
The differential mtDNA versus nDNA susceptibilities likely reflect a combination of the different factors reviewed earlier: toxicokinetics (i.e., where the contaminant and/or its metabolites compartmentalize within the cell), differential packaging of the two genomes, lack of repair of some types of damage in mtDNA, etc.
Approaches for Investigating mtDNA Damage
As can be seen from the examples above, it can be challenging to determine that a specific chemical causes toxicity only via mitochondrial effects. Most chemicals, even those that clearly directly cause mitochondrial toxicity, will also cause some other effects. In some cases, mitochondrial toxicity may be a secondary effect. How then can we isolate the effects of mitochondrial or mtDNA toxicity?
Several approaches have been used. One is to remove mtDNA from the picture altogether via the use of rho zero (mtDNA-depleted) cells (Brar et al., 2012; Davermann et al., 2002; Huang et al., 2012). This approach is clean but has the disadvantage of creating cells that are highly abnormal. A second approach is to create mtDNA damage by targeting a restriction endonuclease to the mitochondria, so that it will only cut mtDNA. This is also a clean approach, with the disadvantage that it only permits the study of doublestrand breaks, rather than any of the types of DNA damage that are more commonly caused by toxicant exposure. A third approach is genetic; for example, Tann et al. (2011) recently used siRNA to specifically deplete the mtDNA repair enzyme ExoG, Green et al. (2011) studied the effects of p53 deficiency on cellular response to rotenone, and Simsek et al. (2011) compared the rescue capacity of nuclear- versus mitochondrial-targeted DNA Ligase III in DNA Ligase III–deficient mice. Several groups have demonstrated mtDNA-mediated effects by targeting DNA repair enzymes to mitochondria, improving DNA repair, and reducing or rescuing the toxic effects observed (Cai et al., 2005; Koczor et al., 2009; Rachek et al., 2009; Shokolenko et al., 2010). The use of a suite of toxicants that all impact a similar mitochondrial target (e.g., ETC complex I) has also been employed (Kulawiec et al., 2009). Finally, it is possible to use a genotoxicant that causes much more damage to mtDNA than nDNA (Anson et al., 1998, 2006; Herraiz et al., 2003; Rachek et al., 2009; van der Eb et al., 2003) or that is repaired in nDNA but not in mtDNA (Bess et al., 2012; Leung et al., 2013). These approaches are more environmentally realistic but less clean because it is never possible to entirely avoid some damage to macromolecules other than mtDNA.
A methodological prerequisite for such studies is an unbiased method for comparing DNA damage in the two genomes. One approach is to measure specific adducts after extracting and isolating two genomes. However, this may lead to artifacts (Anson et al., 2000). An alternative approach is to use a sequence-based approach that permits analysis of damage in the two genomes without differential extraction. The two best described methods are the Southern blot (Anson et al., 2006) and QPCR (Furda et al., 2012a; Hunter et al., 2010; Meyer, 2010) assays.
Another critical methodological consideration, especially for in vitro (cell culture) work, is ensuring that the cells are actually using their mitochondria. Marroquin et al. (2007) showed that cells grown on typical high glucose media, which derive nearly all of their ATP from anaerobic metabolism, show much less susceptibility to a variety of mitochondrial toxicants than do the same cells grown on galactose, which forces them to use oxidative phosphorylation. This also means that the typical concerns about using cancer cells are especially pertinent because many cancer cells are not only grown on glucose but exhibit very abnormal mitochondrial metabolism in any case (Nakajima and Van Houten, 2012).
Future Directions
There are important areas of basic mitochondrial biology that are growing rapidly and represent important opportunities for interdisciplinary research for environmental health scientists. For example, the growing understanding of nuclear-mitochondrial signaling and coordination described earlier has important implications for mitochondrial responses to stressors. Three additional emerging research areas are addressed briefly below.
Our understanding of repair and removal of mtDNA damage has increased greatly. Nonetheless, much remains to be learned about repair pathways per se (Gredilla et al., 2010; Liu and Demple, 2010), how mtDNA damage leads to mtDNA mutations (Copeland, 2012; Krishnan et al., 2008; Loeb et al., 2005), and the role of mitochondrial dynamics in mtDNA maintenance. Recent results demonstrate that in addition to functioning in removing mtDNA damage (Bess et al., 2012, 2013a, b; Rouzier et al., 2012) and mtDNA mutations (Fan et al., 2008; Stewart et al., 2008), mitochondrial dynamics processes may be critical even for normal function of DNA repair pathways that are present in the mitochondria (Rouzier et al., 2012) and for maintenance of nDNA integrity (Qian et al., 2012). As discussed above, a key future area of research will be to investigate how these functions vary by cell type and developmental stage.
Second, there is growing understanding of the importance of epigenetic-mitochondrial interactions (Chinnery et al., 2012; Wallace and Fan, 2010). Most directly, there is now evidence for methylation and hydroxymethylation at some CpGs in mtDNA, probably carried out via a mitochondrial-targeted form of DNA methyltransferase 1 (DNMT1), where it may serve as both a de novo and maintenance methyltransferase (Shock et al., 2011). Furthermore, the levels of mitochondrial DNMT1 were regulated by stress-responsive transcription factors involved in mitochondrial biogenesis (Shock et al., 2011), and one study reports that exposure to metal-rich particulate matter can alter mtDNA methylation (Byun et al., in press). It will be important to clarify both how common pollutant-mediated alterations in mtDNA methylation are and what the functional consequences of mtDNA methylation are. More indirect evidence for a mitochondria-epigenetics relationship was provided by Carugno et al. (2012), who found in an epidemiological study that benzene exposure resulted in increased mtDNA copy number, and that this correlated with altered methylation patterns in four nuclear loci (LINE-1, Alu, p15, and MAGEA1). There are also examples of nuclear-encoded mitochondrial proteins whose epigenetic regulation affects mitochondrial homeostasis and mitochondrial changes affecting nDNA methylation. For example, in vitro methylation of the Tfam promoter site led to reduced transcription of mtDNA (Choi et al., 2004); Tfam function is absolutely required for mtDNA maintenance and mitochondrial function, as shown using various mouse Tfam knockout strains (Dogan and Trifunovic, 2011). Methylation of the mtDNA polymerase γ regulates mtDNA copy number (Kelly et al., 2012). Loss of mtDNA altered nDNA methylation at multiple specific loci in cancer cell lines (Smiraglia et al., 2008; Xie et al., 2007). The mechanisms for mitochondrial-nuclear influences are unclear but may include retrograde signaling (Bungard et al., 2010), oxidative stress–induced alterations in DNA methylation (Chia et al., 2011), and alterations in mitochondrial function that reduce availability of mitochondria-produced molecules required and potentially rate limiting for epigenetic modification (e.g., methyl groups for DNA methylation or cofactors such as flavin adenine dinucleotide, NAD+, acetyl-CoA, and α-ketoglutarate required for histone modification; Cyr and Domann, 2011; Donohue and Bultman, 2012; Minocherhomji et al., 2012).
Finally, in addition to laboratory work, we hope that epidemiologists will consider mitochondrial endpoints. Although there are diseases where we know mitochondrial dysfunction is causal, more research is needed for many more where there is only correlative evidence or where such association may be theoretical only. Epidemiological work may be particularly difficult in the context of mitochondrial dysfunction for a variety of reasons, including (1) the unusual patterns of heritability, (2) heteroplasmy and the requirement that a threshold be reached prior to clinical detection of effects, (3) the potential for latency of effects, especially when a damaging event occurs early in development, and (4) potential epigenetic influences.
In conclusion, we argue that mitochondria are likely an underappreciated target of environmental contaminants and hope that future work from the environmental toxicology and environmental health communities will focus more on mitochondria.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences (1R01-ES017540-01A2, P42 ES010356-10A2).
Supplementary Material
REFERENCES
- Ames B. N., Shigenaga M. K., Hagen T. M. (1995). Mitochondrial decay in aging. Biochim. Biophys. Acta 1271, 165–170 [DOI] [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., et al. (1981). Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- Anson R. M., Croteau D. L., Stierum R. H., Filburn C., Parsell R., Bohr V. A. (1998). Homogenous repair of singlet oxygen-induced DNA damage in differentially transcribed regions and strands of human mitochondrial DNA. Nucleic Acids Res. 26, 662–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson R. M., Hudson E., Bohr V. A. (2000). Mitochondrial endogenous oxidative damage has been overestimated. FASEB J. 14, 355–360 [DOI] [PubMed] [Google Scholar]
- Anson R. M., Mason P. A., Bohr V. A. (2006). Gene-specific and mitochondrial repair of oxidative DNA damage. Methods Mol. Biol. 314, 155–181 [DOI] [PubMed] [Google Scholar]
- Artuso L., Romano A., Verri T., Domenichini A., Argenton F., Santorelli F. M., Petruzzella V. (2012). Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio). Biochim. Biophys. Acta. 1817, 1002–1011 [DOI] [PubMed] [Google Scholar]
- Ashley N., Poulton J. (2009a). Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene 28, 3880–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley N., Poulton J. (2009b). Mitochondrial DNA is a direct target of anti-cancer anthracycline drugs. Biochem. Biophys. Res. Commun. 378, 450–455 [DOI] [PubMed] [Google Scholar]
- Atchison W. D., Hare M. F. (1994). Mechanisms of methylmercury-induced neurotoxicity. FASEB J. 8, 622–629 [DOI] [PubMed] [Google Scholar]
- Babu T. S., Tripuranthakam S., Greenberg B. M. (2005). Biochemical responses of the aquatic higher plant Lemna gibba to a mixture of copper and 1,2-dihydroxyanthraquinone: Synergistic toxicity via reactive oxygen species. Environ. Toxicol. Chem. 24, 3030–3036 [DOI] [PubMed] [Google Scholar]
- Backer J. M., Weinstein I. B. (1982). Interaction of benzo(a)pyrene and its dihydrodiol-epoxide derivative with nuclear and mitochondrial DNA in C3H10T ½ cell cultures. Cancer Res. 42, 2764–2769 [PubMed] [Google Scholar]
- Bailey S. M., Robinson G., Pinner A., Chamlee L., Ulasova E., Pompilius M., Page G. P., Chhieng D., Jhala N., Landar A., et al. (2006). S-adenosylmethionine prevents chronic alcohol-induced mitochondrial dysfunction in the rat liver. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G857–G867 [DOI] [PubMed] [Google Scholar]
- Ballinger S. W. (2005). Mitochondrial dysfunction in cardiovascular disease. Free Radic. Biol. Med. 38, 1278–1295 [DOI] [PubMed] [Google Scholar]
- Ballinger S. W., Bouder T. G., Davis G. S., Judice S. A., Nicklas J. A., Albertini R. J. (1996). Mitochondrial genome damage associated with cigarette smoking. Cancer Res. 56, 5692–5697 [PubMed] [Google Scholar]
- Barret B., Tardieu M., Rustin P., Lacroix C., Chabrol B., Desguerre I., Dollfus C., Mayaux M. J., Blanche S. (2003). Persistent mitochondrial dysfunction in HIV-1-exposed but uninfected infants: Clinical screening in a large prospective cohort. AIDS 17, 1769–1785 [DOI] [PubMed] [Google Scholar]
- Barrett M. J., Alones V., Wang K. X., Phan L., Swerdlow R. H. (2004). Mitochondria-derived oxidative stress induces a heat shock protein response. J. Neurosci. Res. 78, 420–429 [DOI] [PubMed] [Google Scholar]
- Beckman K. B., Ames B. N. (1998). Mitochondrial aging: Open questions. Ann. N. Y. Acad. Sci. 854, 118–127 [DOI] [PubMed] [Google Scholar]
- Begriche K., Igoudjil A., Pessayre D., Fromenty B. (2006). Mitochondrial dysfunction in NASH: Causes, consequences and possible means to prevent it. Mitochondrion 6, 1–28 [DOI] [PubMed] [Google Scholar]
- Begriche K., Massart J., Robin M. A., Borgne-Sanchez A., Fromenty B. (2011). Drug-induced toxicity on mitochondria and lipid metabolism: Mechanistic diversity and deleterious consequences for the liver. J. Hepatol. 54, 773–794 [DOI] [PubMed] [Google Scholar]
- Belyaeva E. A., Sokolova T. V., Emelyanova L. V., Zakharova I. O. (2012). Mitochondrial electron transport chain in heavy metal-induced neurotoxicity: Effects of cadmium, mercury, and copper. ScientificWorldJournal 2012, 136063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhammou V., Tardieu M., Warszawski J., Rustin P., Blanche S. (2007). Clinical mitochondrial dysfunction in uninfected children born to HIV-infected mothers following perinatal exposure to nucleoside analogues. Environ. Mol. Mutagen. 48, 173–178 [DOI] [PubMed] [Google Scholar]
- Berthiaume J. M., Wallace K. B. (2007). Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol. Toxicol. 23, 15–25 [DOI] [PubMed] [Google Scholar]
- Bess A. S., Crocker T. L., Ryde I. T., Meyer J. N. (2012). Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 40, 7916–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bess A. S., Leung M. C. K., Ryde I. T., Rooney J. P., Hinton D. E., Meyer J. N.(2013a). Effects of mutations in mitochondrial dynamics-related genes on the mitochondrial response to ultraviolet C radiation in developing Caenorhabditis elegans. Worm 2, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bess A. S., Ryde I. T., Hinton D. E., Meyer J. N. (2013b). UVC-induced mitochondrial degradation via autophagy correlates with mtDNA damage removal in primary human fibroblasts. J. Biochem. Mol. Toxicol. 27, 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Srinivasan S., Anandatheerthavarada H. K., Avadhani N. G. (2008). Dioxin-mediated tumor progression through activation of mitochondria-to-nucleus stress signaling. Proc. Natl. Acad. Sci. U. S. A. 105, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche S., Tardieu M., Rustin P., Slama A., Barret B., Firtion G., Ciraru-Vigneron N., Lacroix C., Rouzioux C., Mandelbrot L., et al. (1999). Persistent mitochondrial dysfunction and perinatal exposure to antiretroviral nucleoside analogues. Lancet 354, 1084–1089 [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F. (2012). Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta. 1819, 914–920 [DOI] [PubMed] [Google Scholar]
- Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008). Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A. B., Kwakye G. F., Hernández E. H., Aschner M. (2011). Role of manganese in neurodegenerative diseases. J. Trace Elem. Med. Biol. 25, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeckman B. P., Houthoofd K., De Vreese A., Vanfleteren J. R. (2002). Assaying metabolic activity in ageing Caenorhabditis elegans. Mech. Ageing Dev. 123, 105–119 [DOI] [PubMed] [Google Scholar]
- Brand M. D., Buckingham J. A., Esteves T. C., Green K., Lambert A. J., Miwa S., Murphy M. P., Pakay J. L., Talbot D. A., Echtay K. S. (2004). Mitochondrial superoxide and aging: Uncoupling-protein activity and superoxide production. Biochem. Soc. Symp. 71, 203–213 [DOI] [PubMed] [Google Scholar]
- Brandon M., Baldi P., Wallace D. C. (2006). Mitochondrial mutations in cancer. Oncogene 25, 4647–4662 [DOI] [PubMed] [Google Scholar]
- Brar S. S., Meyer J. N., Bortner C. D., Van Houten B., Martin W. J., 2nd. (2012). Mitochondrial DNA-depleted A549 cells are resistant to bleomycin. Am. J. Physiol. Lung Cell. Mol. Physiol. 303, L413–L424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic I., Hench J., Henriksson J., Antebi A., Bürglin T. R., Trifunovic A. (2009). Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic Acids Res. 37, 1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A., Larsson N. G. (2013). The role of mitochondria in aging. J. Clin. Invest. 123, 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P. J. (2008). The 8,5’-cyclopurine-2’-deoxynucleosides: Candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst). 7, 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. C., Borutaite V. (2012). There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion 12, 1–4 [DOI] [PubMed] [Google Scholar]
- Brys K., Castelein N., Matthijssens F., Vanfleteren J. R., Braeckman B. P. (2010). Disruption of insulin signalling preserves bioenergetic competence of mitochondria in ageing Caenorhabditis elegans. BMC Biol. 8, 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brys K., Vanfleteren J. R., Braeckman B. P. (2007). Testing the rate-of-living/oxidative damage theory of aging in the nematode model Caenorhabditis elegans. Exp. Gerontol. 42, 845–851 [DOI] [PubMed] [Google Scholar]
- Bucio L., García C., Souza V., Hernández E., González C., Betancourt M., Gutiérrez-Ruiz M. C. (1999). Uptake, cellular distribution and DNA damage produced by mercuric chloride in a human fetal hepatic cell line. Mutat. Res. 423, 65–72 [DOI] [PubMed] [Google Scholar]
- Bungard D., Fuerth B. J., Zeng P. Y., Faubert B., Maas N. L., Viollet B., Carling D., Thompson C. B., Jones R. G., Berger S. L. (2010). Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329, 1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Gray M. W., Lang B. F. (2003). Mitochondrial genomes: Anything goes. Trends Genet. 19, 709–716 [DOI] [PubMed] [Google Scholar]
- Byun H. M., Panni T., Motta V., Hou L., Nordio F., Apostoli P., Bertazzi P. A., Baccarelli A. A. (in press). Effects of airborne pollutants on mitochondrial DNA methylation. Part. Fibre Toxicol. 10:18, 10.1186/1743-8977-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Xu Y., Cooper R. J., Ferkowicz M. J., Hartwell J. R., Pollok K. E., Kelley M. R. (2005). Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res. 65, 3319–3327 [DOI] [PubMed] [Google Scholar]
- Cakir Y., Yang Z., Knight C. A., Pompilius M., Westbrook D., Bailey S. M., Pinkerton K. E., Ballinger S. W. (2007). Effect of alcohol and tobacco smoke on mtDNA damage and atherogenesis. Free Radic. Biol. Med. 43, 1279–1288 [DOI] [PubMed] [Google Scholar]
- Calvo S. E., Mootha V. K. (2010). The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 11, 25–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. T., Kolesar J. E., Kaufman B. A. (2012). Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta. 1819, 921–929 [DOI] [PubMed] [Google Scholar]
- Cannon J. R., Greenamyre J. T. (2011). The role of environmental exposures in neurodegeneration and neurodegenerative diseases. Toxicol. Sci. 124, 225–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling P. J., Cree L. M., Chinnery P. F. (2011). The implications of mitochondrial DNA copy number regulation during embryogenesis. Mitochondrion 11, 686–692 [DOI] [PubMed] [Google Scholar]
- Carugno M., Pesatori A. C., Dioni L., Hoxha M., Bollati V., Albetti B., Byun H. M., Bonzini M., Fustinoni S., Cocco P., et al. (2012). Increased mitochondrial DNA copy number in occupations associated with low-dose benzene exposure. Environ. Health Perspect. 120, 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino N., Aloj S. (1969). Intracellular distribution of lead in the liver and kidney of the rat. Br. J. Ind. Med. 26, 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S., Ahn B. Y., Byun K., Cho Y. M., Yu M. H., Lee B., Hwang D., Park K. S. (2013). A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 6, rs4 [DOI] [PubMed] [Google Scholar]
- Chan D., Frank S., Rojo M. (2006). Mitochondrial dynamics in cell life and death. Cell Death Differ. 13, 680–684 [DOI] [PubMed] [Google Scholar]
- Chan D. C. (2007). Mitochondrial dynamics in disease. N. Engl. J. Med. 356, 1707–1709 [DOI] [PubMed] [Google Scholar]
- Chase J. W., Dawid I. B. (1972). Biogenesis of mitochondria during Xenopus laevis development. Dev. Biol. 27, 504–518 [DOI] [PubMed] [Google Scholar]
- Chen X. J., Wang X., Kaufman B. A., Butow R. A. (2005). Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307, 714–717 [DOI] [PubMed] [Google Scholar]
- Chia N., Wang L., Lu X., Senut M. C., Brenner C., Ruden D. M. (2011). Hypothesis: Environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 6, 853–856 [DOI] [PubMed] [Google Scholar]
- Chinnery P. F., DiMauro S., Shanske S., Schon E. A., Zeviani M., Mariotti C., Carrara F., Lombes A., Laforet P., Ogier H., et al. (2004). Risk of developing a mitochondrial DNA deletion disorder. Lancet 364, 592–596 [DOI] [PubMed] [Google Scholar]
- Chinnery P. F., Elliott H. R., Hudson G., Samuels D. C., Relton C. L. (2012). Epigenetics, epidemiology and mitochondrial DNA diseases. Int. J. Epidemiol. 41, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. S., Kim S., Kyu Lee H., Lee K. U., Pak Y. K. (2004). In vitro methylation of nuclear respiratory factor-1 binding site suppresses the promoter activity of mitochondrial transcription factor A. Biochem. Biophys. Res. Commun. 314, 118–122 [DOI] [PubMed] [Google Scholar]
- Chomyn A., Attardi G. (2003). MtDNA mutations in aging and apoptosis. Biochem. Biophys. Res. Commun. 304, 519–529 [DOI] [PubMed] [Google Scholar]
- Christodoulou J. (2000). Genetic defects causing mitochondrial respiratory chain disorders and disease. Hum. Reprod. 15(Suppl. 2), 28–43 [DOI] [PubMed] [Google Scholar]
- Claessens Y. E., Chiche J. D., Mira J. P., Cariou A. (2003). Bench-to-bedside review: Severe lactic acidosis in HIV patients treated with nucleoside analogue reverse transcriptase inhibitors. Crit. Care. 7, 226–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. D. (2012). Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim. Biophys. Acta. 1819, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. D., Lodeiro M. F., Marnett L. J., Cameron C. E., Arnold J. J. (2010). Arrest of human mitochondrial RNA polymerase transcription by the biological aldehyde adduct of DNA, M1dG. Nucleic Acids Res. 38, 7546–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. H. (2010). Pharmacologic effects on mitochondrial function. Dev. Disabil. Res. Rev. 16, 189–199 [DOI] [PubMed] [Google Scholar]
- Copeland W. C. (2012). Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 47, 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun P., Wyrembak J., Schriner S. E., Chen H. W., Marciniack C., Laferla F., Wallace D. C. (2012). A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim. Biophys. Acta. 1820, 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D. L., Rossi M. L., Canugovi C., Tian J., Sykora P., Ramamoorthy M., Wang Z. M., Singh D. K., Akbari M., Kasiviswanathan R., et al. (2012). RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell. 11, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch P. J., Cimdins K., Duce J. A., Bush A. I., Trounce I. A. (2007). Mitochondria in aging and Alzheimer’s disease. Rejuvenation Res. 10, 349–357 [DOI] [PubMed] [Google Scholar]
- Cyr A. R., Domann F. E. (2011). The redox basis of epigenetic modifications: From mechanisms to functional consequences. Antioxid. Redox Signal. 15, 551–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davermann D., Martinez M., McKoy J., Patel N., Averbeck D., Moore C. W. (2002). Impaired mitochondrial function protects against free radical-mediated cell death. Free Radic. Biol. Med. 33, 1209–1220 [DOI] [PubMed] [Google Scholar]
- Davies K. J., Doroshow J. H. (1986). Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J. Biol. Chem. 261, 3060–3067 [PubMed] [Google Scholar]
- de Brito O. M., Scorrano L. (2010). An intimate liaison: Spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 29, 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura M. B., dos Santos L. S., Van Houten B. (2010). Mitochondrial dysfunction in neurodegenerative diseases and cancer. Environ. Mol. Mutagen. 51, 391–405 [DOI] [PubMed] [Google Scholar]
- Dean R. T., Nicholson P. (1994). The action of nine chelators on iron-dependent radical damage. Free Radic. Res. 20, 83–101 [DOI] [PubMed] [Google Scholar]
- Diaz F., Moraes C. T. (2008). Mitochondrial biogenesis and turnover. Cell Calcium. 44, 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Hsu A. L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A. G., Kamath R. S., Ahringer J., Kenyon C. (2002). Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 [DOI] [PubMed] [Google Scholar]
- Dimauro S., Davidzon G. (2005). Mitochondrial DNA and disease. Ann. Med. 37, 222–232 [DOI] [PubMed] [Google Scholar]
- DiMauro S., Schon E. A. (2003). Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668 [DOI] [PubMed] [Google Scholar]
- Divi R. L., Einem T. L., Fletcher S. L., Shockley M. E., Kuo M. M., St Claire M. C., Cook A., Nagashima K., Harbaugh S. W., Harbaugh J. W., et al. (2010). Progressive mitochondrial compromise in brains and livers of primates exposed in utero to nucleoside reverse transcriptase inhibitors (NRTIs). Toxicol. Sci. 118, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan S. A., Trifunovic A. (2011). Modelling mitochondrial dysfunction in mice. Physiol. Res. 60(Suppl. 1), S61–S70 [DOI] [PubMed] [Google Scholar]
- Donohue D. R., Bultman S. J. (2012). Metaboloepigenetics: Interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell. Physiol. 227, 3169–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Dalton T. P., Miller M. L., Chen Y., Uno S., Shi Z., Shertzer H. G., Bansal S., Avadhani N. G., Nebert D. W. (2009). Knock-in mouse lines expressing either mitochondrial or microsomal CYP1A1: Differing responses to dietary benzo[a]pyrene as proof of principle. Mol. Pharmacol. 75, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopp E., von Recklinghausen U., Hartmann L. M., Stueckradt I., Pollok I., Rabieh S., Hao L., Nussler A., Katier C., Hirner A. V., et al. (2008). Subcellular distribution of inorganic and methylated arsenic compounds in human urothelial cells and human hepatocytes. Drug Metab. Dispos. 36, 971–979 [DOI] [PubMed] [Google Scholar]
- Druzhyna N. M., Wilson G. L., LeDoux S. P. (2008). Mitochondrial DNA repair in aging and disease. Mech. Ageing Dev. 129, 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykens J. A., Marroquin L. D., Will Y. (2007). Strategies to reduce late-stage drug attrition due to mitochondrial toxicity. Expert Rev. Mol. Diagn. 7, 161–175 [DOI] [PubMed] [Google Scholar]
- Dykens J. A., Will Y. (2007). The significance of mitochondrial toxicity testing in drug development. Drug Discov. Today. 12, 777–785 [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A., Benoilid A., Vinzio S., Fornecker L. M., Lannes B., Goullé J. P., Broly F., Mousson de Camaret B. (2012). Mitochondrial myopathy caused by arsenic trioxide therapy. Blood 119, 4272–4274 [DOI] [PubMed] [Google Scholar]
- Fan W., Waymire K. G., Narula N., Li P., Rocher C., Coskun P. E., Vannan M. A., Narula J., Macgregor G. R., Wallace D. C. (2008). A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319, 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina M., Avila D. S., da Rocha J. B., Aschner M. (2012). Metals, oxidative stress, and neurodegeneration: A focus on iron, manganese, and mercury. Neurochem. Int. 62, 575–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J., Segall L. (2010). Drugs interfering with mitochondrial disorders. Drug Chem. Toxicol. 33, 138–151 [DOI] [PubMed] [Google Scholar]
- Fischer F., Hamann A., Osiewacz H. D. (2012). Mitochondrial quality control: An integrated network of pathways. Trends Biochem. Sci. 37, 284–292 [DOI] [PubMed] [Google Scholar]
- Freyer C., Cree L. M., Mourier A., Stewart J. B., Koolmeister C., Milenkovic D., Wai T., Floros V. I., Hagström E., Chatzidaki E. E., et al. (2012). Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat. Genet. 44, 1282–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. (2004). Mitochondria: Are they the seat of senescence? Aging Cell. 3, 13–16 [DOI] [PubMed] [Google Scholar]
- Furda A. M., Bess A. S., Meyer J. N., Van Houten B. (2012a). Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol. Biol. 920, 111–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furda A. M., Marrangoni A. M., Lokshin A., Van Houten B. (2012b). Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair (Amst). 11, 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau N., Pichaud N., Couture P. (2010). Inhibition of goldfish mitochondrial metabolism by in vitro exposure to Cd, Cu and Ni. Aquat. Toxicol. 98, 107–112 [DOI] [PubMed] [Google Scholar]
- Gavin C. E., Gunter K. K., Gunter T. E. (1992). Mn2+ sequestration by mitochondria and inhibition of oxidative phosphorylation. Toxicol. Appl. Pharmacol. 115, 1–5 [DOI] [PubMed] [Google Scholar]
- Genter M. B., Clay C. D., Dalton T. P., Dong H., Nebert D. W., Shertzer H. G. (2006). Comparison of mouse hepatic mitochondrial versus microsomal cytochromes P450 following TCDD treatment. Biochem. Biophys. Res. Commun. 342, 1375–1381 [DOI] [PubMed] [Google Scholar]
- Giulivi C., Boveris A., Cadenas E. (1995). Hydroxyl radical generation during mitochondrial electron transfer and the formation of 8-hydroxydesoxyguanosine in mitochondrial DNA. Arch. Biochem. Biophys. 316, 909–916 [DOI] [PubMed] [Google Scholar]
- Graziewicz M. A., Bienstock R. J., Copeland W. C. (2007). The DNA polymerase gamma Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2’-deoxyguanosine. Hum. Mol. Genet. 16, 2729–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz M. A., Sayer J. M., Jerina D. M., Copeland W. C. (2004). Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 32, 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R., Bohr V. A., Stevnsner T. (2010). Mitochondrial DNA repair and association with aging–an update. Exp. Gerontol. 45, 478–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D. R., Van Houten B. (2011). SnapShot: Mitochondrial quality control. Cell 147, 950, 950.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. L., Singh A. V., Ruest L. B., Pisano M. M., Prough R. A., Knudsen T. B. (2011). Differential programming of p53-deficient embryonic cells during rotenone block. Toxicology 290, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M. X. (2011). Mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. Mitochondrion 11, 237–245 [DOI] [PubMed] [Google Scholar]
- Guo L., Shi Q., Fang J. L., Mei N., Ali A. A., Lewis S. M., Leakey J. E., Frankos V. H. (2008). Review of usnic acid and Usnea barbata toxicity. J. Environ. Sci. Health. C. Environ. Carcinog. Ecotoxicol. Rev. 26, 317–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. K., Schuh R. A., Fiskum G., Flaws J. A. (2006). Methoxychlor causes mitochondrial dysfunction and oxidative damage in the mouse ovary. Toxicol. Appl. Pharmacol. 216, 436–445 [DOI] [PubMed] [Google Scholar]
- Hasegawa E., Takeshige K., Oishi T., Murai Y., Minakami S. (1990). 1-Methyl-4-phenylpyridinium (MPP+) induces NADH-dependent superoxide formation and enhances NADH-dependent lipid peroxidation in bovine heart submitochondrial particles. Biochem. Biophys. Res. Commun. 170, 1049–1055 [DOI] [PubMed] [Google Scholar]
- Hebert S. L., Lanza I. R., Nair K. S. (2010). Mitochondrial DNA alterations and reduced mitochondrial function in aging. Mech. Ageing Dev. 131, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herraiz M., Beraza N., Solano A., Sangro B., Montoya J., Qian C., Prieto J., Bustos M. (2003). Liver failure caused by herpes simplex virus thymidine kinase plus ganciclovir therapy is associated with mitochondrial dysfunction and mitochondrial DNA depletion. Hum. Gene Ther. 14, 463–472 [DOI] [PubMed] [Google Scholar]
- Hirano M., Davidson M., DiMauro S. (2001). Mitochondria and the heart. Curr. Opin. Cardiol. 16, 201–210 [DOI] [PubMed] [Google Scholar]
- Holt I. J. (2010). Zen and the art of mitochondrial DNA maintenance. Trends Genet. 26, 103–109 [DOI] [PubMed] [Google Scholar]
- Holt I. J., Harding A. E., Morgan-Hughes J. A. (1988). Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 [DOI] [PubMed] [Google Scholar]
- Hou L., Zhu Z. Z., Zhang X., Nordio F., Bonzini M., Schwartz J., Hoxha M., Dioni L., Marinelli B., Pegoraro V., et al. (2010). Airborne particulate matter and mitochondrial damage: A cross-sectional study. Environ. Health. 9, 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N., Elson J. L., Chinnery P. F., Turnbull D. M. (2005). mtDNA mutations and common neurodegenerative disorders. Trends Genet. 21, 583–586 [DOI] [PubMed] [Google Scholar]
- Huang S. X., Partridge M. A., Ghandhi S. A., Davidson M. M., Amundson S. A., Hei T. K. (2012). Mitochondria-derived reactive intermediate species mediate asbestos-induced genotoxicity and oxidative stress-responsive signaling pathways. Environ. Health Perspect. 120, 840–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. E., Jung D., Di Giulio R. T., Meyer J. N. (2010). The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods 51, 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indo H. P., Davidson M., Yen H. C., Suenaga S., Tomita K., Nishii T., Higuchi M., Koga Y., Ozawa T., Majima H. J. (2007). Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion 7, 106–118 [DOI] [PubMed] [Google Scholar]
- Ishiguro H., Yasuda K., Ishii N., Ihara K., Ohkubo T., Hiyoshi M., Ono K., Senoo-Matsuda N., Shinohara O., Yosshii F., et al. (2001). Enhancement of oxidative damage to cultured cells and Caenorhabditis elegans by mitochondrial electron transport inhibitors. IUBMB Life. 51, 263–268 [DOI] [PubMed] [Google Scholar]
- Jansen R. P. (2000). Germline passage of mitochondria: Quantitative considerations and possible embryological sequelae. Hum. Reprod. 15(Suppl. 2), 112–128 [DOI] [PubMed] [Google Scholar]
- Jansen R. P., Burton G. J. (2004). Mitochondrial dysfunction in reproduction. Mitochondrion 4, 577–600 [DOI] [PubMed] [Google Scholar]
- Jansen R. P., de Boer K. (1998). The bottleneck: Mitochondrial imperatives in oogenesis and ovarian follicular fate. Mol. Cell. Endocrinol. 145, 81–88 [DOI] [PubMed] [Google Scholar]
- Janssen B. G., Munters E., Pieters N., Smeets K., Cox B., Cuypers A., Fierens F., Penders J., Vangronsveld J., Gyselaers W., et al. (2012). Placental mitochondrial DNA content and particulate air pollution during in utero life. Environ. Health Perspect. 120, 1346–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrach M., Mai S., Pohl S., Vöth M., Bereiter-Hahn J. (2008). Short- and long-term alterations of mitochondrial morphology, dynamics and mtDNA after transient oxidative stress. Mitochondrion 8, 293–304 [DOI] [PubMed] [Google Scholar]
- Jia L., Liu Z., Sun L., Miller S. S., Ames B. N., Cotman C. W., Liu J. (2007). Acrolein, a toxicant in cigarette smoke, causes oxidative damage and mitochondrial dysfunction in RPE cells: protection by (R)-alpha-lipoic acid. Invest. Ophthalmol. Vis. Sci. 48, 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. T., Harris R. A., French S., Blair P. V., You J., Bemis K. G., Wang M., Balaban R. S. (2007). Tissue heterogeneity of the mammalian mitochondrial proteome. Am. J. Physiol. Cell Physiol. 292, C689–C697 [DOI] [PubMed] [Google Scholar]
- Joseph A., Lee T., Moland C. L., Branham W. S., Fuscoe J. C., Leakey J. E., Allaben W. T., Lewis S. M., Ali A. A., Desai V. G. (2009). Effect of (+)-usnic acid on mitochondrial functions as measured by mitochondria-specific oligonucleotide microarray in liver of B6C3F1 mice. Mitochondrion 9, 149–158 [DOI] [PubMed] [Google Scholar]
- Kai Y., Takamatsu C., Tokuda K., Okamoto M., Irita K., Takahashi S. (2006). Rapid and random turnover of mitochondrial DNA in rat hepatocytes of primary culture. Mitochondrion 6, 299–304 [DOI] [PubMed] [Google Scholar]
- Kamenisch Y., Fousteri M., Knoch J., von Thaler A. K., Fehrenbacher B., Kato H., Becker T., Dollé M. E., Kuiper R., Majora M., et al. (2010). Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 207, 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahalil B., Hogue B. A., de Souza-Pinto N. C., Bohr V. A. (2002). Base excision repair capacity in mitochondria and nuclei: Tissue-specific variations. FASEB J. 16, 1895–1902 [DOI] [PubMed] [Google Scholar]
- Kasiviswanathan R., Gustafson M. A., Copeland W. C., Meyer J. N. (2012). Human mitochondrial DNA polymerase γ exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 287, 9222–9229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L., Reyes A., Holt I. J. (2012). Minimizing the damage: Repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 13, 659–671 [DOI] [PubMed] [Google Scholar]
- Kelly R. D., Mahmud A., McKenzie M., Trounce I. A., St John J. C. (2012). Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A. Nucleic Acids Res. 40, 10124–10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Lemasters J. J. (2011). Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid. Redox Signal. 14, 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007). Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman M. A., Yu-Wai-Man P., Korsten A., Leonhardt M., Dimitriadis K., De Coo I. F., Klopstock T., Chinnery P. F. (2009). Gene-environment interactions in Leber hereditary optic neuropathy. Brain 132(Pt 9), 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen T. B., Green M. L. (2004). Response characteristics of the mitochondrial DNA genome in developmental health and disease. Birth Defects Res. C. Embryo Today. 72, 313–329 [DOI] [PubMed] [Google Scholar]
- Koczor C. A., Shokolenko I. N., Boyd A. K., Balk S. P., Wilson G. L., Ledoux S. P. (2009). Mitochondrial DNA damage initiates a cell cycle arrest by a Chk2-associated mechanism in mammalian cells. J. Biol. Chem. 284, 36191–36201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler J. J., Lewis W. (2007). A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ. Mol. Mutagen. 48, 166–172 [DOI] [PubMed] [Google Scholar]
- Kolesar J. E., Wang C. Y., Taguchi Y. V., Chou S. H., Kaufman B. A. (2012). Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res. 41, e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic P., Pozos R. S., Somanathan R., Shangari N., O’Brien P. J. (2005). Mechanism of mitochondrial uncouplers, inhibitors, and toxins: Focus on electron transfer, free radicals, and structure-activity relationships. Curr. Med. Chem. 12, 2601–2623 [DOI] [PubMed] [Google Scholar]
- Kulawiec M., Ayyasamy V., Singh K. K. (2009). p53 regulates mtDNA copy number and mitocheckpoint pathway. J. Carcinog. 8, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krähenbühl S., Brandner S., Kleinle S., Liechti S., Straumann D. (2000). Mitochondrial diseases represent a risk factor for valproate-induced fulminant liver failure. Liver 20, 346–348 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Schwartz M., Brown T. A., Ebralidse K., Kunz W. S., Clayton D. A., Vissing J., Khrapko K. (2004). Recombination of human mitochondrial DNA. Science 304, 981 [DOI] [PubMed] [Google Scholar]
- Krishnan K. J., Reeve A. K., Samuels D. C., Chinnery P. F., Blackwood J. K., Taylor R. W., Wanrooij S., Spelbrink J. N., Lightowlers R. N., Turnbull D. M. (2008). What causes mitochondrial DNA deletions in human cells? Nat. Genet. 40, 275–279 [DOI] [PubMed] [Google Scholar]
- Kukat C., Wurm C. A., Spåhr H., Falkenberg M., Larsson N. G., Jakobs S. (2011). Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U. S. A. 108, 13534–13539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulawiec M., Salk J. J., Ericson N. G., Wanagat J., Bielas J. H. (2010). Generation, function, and prognostic utility of somatic mitochondrial DNA mutations in cancer. Environ. Mol. Mutagen. 51, 427–439 [DOI] [PubMed] [Google Scholar]
- Kwong J. Q., Beal M. F., Manfredi G. (2006). The role of mitochondria in inherited neurodegenerative diseases. J. Neurochem. 97, 1659–1675 [DOI] [PubMed] [Google Scholar]
- Landrigan P. J., Sonawane B., Butler R. N., Trasande L., Callan R., Droller D. (2005). Early environmental origins of neurodegenerative disease in later life. Environ. Health Perspect. 113, 1230–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Manjanatha M. G., Aidoo A., Moland C. L., Branham W. S., Fuscoe J. C., Ali A. A., Desai V. G. (2012). Expression analysis of hepatic mitochondria-related genes in mice exposed to acrylamide and glycidamide. J. Toxicol. Environ. Health. A. 75, 324–339 [DOI] [PubMed] [Google Scholar]
- Leung M. C., Rooney J. P., Ryde I. T., Bernal A. J., Bess A. S., Crocker T. L., Ji A. Q., Meyer J. N. (2013). Effects of early life exposure to ultraviolet C radiation on mitochondrial DNA content, transcription, ATP production, and oxygen consumption in developing Caenorhabditis elegans. BMC Pharmacol. Toxicol. 14, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J. A., Robinson J. P. (2003a). DPI induces mitochondrial superoxide-mediated apoptosis. Free Radic. Biol. Med. 34, 465–477 [DOI] [PubMed] [Google Scholar]
- Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J. A., Robinson J. P. (2003b). Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278, 8516–8525 [DOI] [PubMed] [Google Scholar]
- Lipsky N. G., Pedersen P. L. (1981). Mitochondrial turnover in animal cells. Half-lives of mitochondria and mitochondrial subfractions of rat liver based on [14C]bicarbonate incorporation. J. Biol. Chem. 256, 8652–8657 [PubMed] [Google Scholar]
- Liu P., Demple B. (2010). DNA repair in mammalian mitochondria: Much more than we thought? Environ. Mol. Mutagen. 51, 417–426 [DOI] [PubMed] [Google Scholar]
- Liu X., Jiang N., Hughes B., Bigras E., Shoubridge E., Hekimi S. (2005). Evolutionary conservation of the clk-1-dependent mechanism of longevity: Loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 19, 2424–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185 [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Wallace D. C., Martin G. M. (2005). The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc. Natl. Acad. Sci. U. S. A. 102, 18769–18770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi G., Beal M. F. (2000). The role of mitochondria in the pathogenesis of neurodegenerative diseases. Brain Pathol. 10, 462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmolino D. (2011). Friedreich’s ataxia: Past, present and future. Brain Res. Rev. 67, 311–330 [DOI] [PubMed] [Google Scholar]
- Marroquin L. D., Hynes J., Dykens J. A., Jamieson J. D., Will Y. (2007). Circumventing the Crabtree effect: Replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci. 97, 539–547 [DOI] [PubMed] [Google Scholar]
- Martinez T. N., Greenamyre J. T. (2012). Toxin models of mitochondrial dysfunction in Parkinson’s disease. Antioxid. Redox Signal. 16, 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Panloup P., Chretien M. F., Malthiery Y., Reynier P. (2007). Mitochondrial DNA in the oocyte and the developing embryo. Curr. Top. Dev. Biol. 77, 51–83 [DOI] [PubMed] [Google Scholar]
- Maynard S., Schurman S. H., Harboe C., de Souza-Pinto N. C., Bohr V. A. (2009). Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 30, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie R., Fried M. W., Sallie R., Conjeevaram H., Di Bisceglie A. M., Park Y., Savarese B., Kleiner D., Tsokos M., Luciano C. (1995). Hepatic failure and lactic acidosis due to fialuridine (FIAU), an investigational nucleoside analogue for chronic hepatitis B. N. Engl. J. Med. 333, 1099–1105 [DOI] [PubMed] [Google Scholar]
- Mehta R., Chan K., Lee O., Tafazoli S., O’Brien P. J. (2008). Drug-associated mitochondrial toxicity. In Drug-Induced Mitochondrial Dysfunction. (Dykens J. A., Will Y, Eds.), pp. 71–126 John Wiley & Sons, Inc., Hoboken, NJ: [Google Scholar]
- Melov S., Shoffner J. M., Kaufman A., Wallace D. C. (1995). Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res. 23, 4122–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J. R., Cheng K. K., Figg N., Gorenne I., Mahmoudi M., Griffin J., Vidal-Puig A., Logan A., Murphy M. P., Bennett M. (2010). DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ. Res. 107, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Neph S., Dinger M. E., Crawford J., Smith M. A., Shearwood A. M., Haugen E., Bracken C. P., Rackham O., Stamatoyannopoulos J. A., et al. (2011). The human mitochondrial transcriptome. Cell 146, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. N. (2010). QPCR: A tool for analysis of mitochondrial and nuclear DNA damage in ecotoxicology. Ecotoxicology 19, 804–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. N., Bess A. S. (2012). Involvement of autophagy and mitochondrial dynamics in determining the fate and effects of irreparable mitochondrial DNA damage. Autophagy 8, 1822–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. N., Boyd W. A., Azzam G. A., Haugen A. C., Freedman J. H., Van Houten B. (2007). Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 8, R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R. J. (2007). Different fates of mitochondria: Alternative ways for degradation? Autophagy 3, 4–9 [DOI] [PubMed] [Google Scholar]
- Minocherhomji S., Tollefsbol T. O., Singh K. K. (2012). Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics 7, 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa S., Brand M. D. (2003). Mitochondrial matrix reactive oxygen species production is very sensitive to mild uncoupling. Biochem. Soc. Trans. 31(Pt 6), 1300–1301 [DOI] [PubMed] [Google Scholar]
- Mladenka P., Simůnek T., Hübl M., Hrdina R. (2006). The role of reactive oxygen and nitrogen species in cellular iron metabolism. Free Radic. Res. 40, 263–272 [DOI] [PubMed] [Google Scholar]
- Modica-Napolitano J. S., Aprille J. R. (2001). Delocalized lipophilic cations selectively target the mitochondria of carcinoma cells. Adv. Drug Deliv. Rev. 49, 63–70 [DOI] [PubMed] [Google Scholar]
- Murphy M. P. (2008). Targeting lipophilic cations to mitochondria. Biochim. Biophys. Acta. 1777, 1028–1031 [DOI] [PubMed] [Google Scholar]
- Murphy M. P., Smith R. A. (2007). Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 47, 629–656 [DOI] [PubMed] [Google Scholar]
- Mustonen P., Kinnunen P. K. (1993). On the reversal by deoxyribonucleic acid of the binding of adriamycin to cardiolipin-containing liposomes. J. Biol. Chem. 268, 1074–1080 [PubMed] [Google Scholar]
- Nadanaciva S., Willis J. H., Barker M. L., Gharaibeh D., Capaldi R. A., Marusich M. F., Will Y. (2009). Lateral-flow immunoassay for detecting drug-induced inhibition of mitochondrial DNA replication and mtDNA-encoded protein synthesis. J. Immunol. Methods. 343, 1–12 [DOI] [PubMed] [Google Scholar]
- Nakajima E. C., Van Houten B. (2012). Metabolic symbiosis in cancer: Refocusing the Warburg lens. Mol. Carcinog. 52, 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranmandura H., Xu S., Sawata T., Hao W. H., Liu H., Bu N., Ogra Y., Lou Y. J., Suzuki N. (2011). Mitochondria are the main target organelle for trivalent monomethylarsonous acid (MMA(III))-induced cytotoxicity. Chem. Res. Toxicol. 24, 1094–1103 [DOI] [PubMed] [Google Scholar]
- Neustadt J., Pieczenik S. R. (2008). Medication-induced mitochondrial damage and disease. Mol. Nutr. Food Res. 52, 780–788 [DOI] [PubMed] [Google Scholar]
- Nicholls D. G. (2002). Mitochondrial function and dysfunction in the cell: Its relevance to aging and aging-related disease. Int. J. Biochem. Cell Biol. 34, 1372–1381 [DOI] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J. (2008). Mitochondrial dysfunction. In Molecular and Biochemical Toxicology. 4th ed (Smart R. C., Hodgson R, Eds.), pp. 319–332 John Wiley & Sons, Hoboken, NJ: [Google Scholar]
- Nunnari J., Suomalainen A. (2012). Mitochondria: In sickness and in health. Cell 148, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara M. F., Charlap J. H., Craig R. C., Knudsen T. B. (2002). Mitochondrial transduction of ocular teratogenesis during methylmercury exposure. Teratology 65, 131–144 [DOI] [PubMed] [Google Scholar]
- Omura T. (2006). Mitochondrial P450s. Chem. Biol. Interact. 163, 86–93 [DOI] [PubMed] [Google Scholar]
- Pacheu-Grau D., Gómez-Durán A., López-Pérez M. J., Montoya J., Ruiz-Pesini E. (2010). Mitochondrial pharmacogenomics: Barcode for antibiotic therapy. Drug Discov. Today. 15, 33–39 [DOI] [PubMed] [Google Scholar]
- Penta J. S., Johnson F. M., Wachsman J. T., Copeland W. C. (2001). Mitochondrial DNA in human malignancy. Mutat. Res. 488, 119–133 [DOI] [PubMed] [Google Scholar]
- Pereira C. V., Moreira A. C., Pereira S. P., Machado N. G., Carvalho F. S., Sardão V. A., Oliveira P. J. (2009). Investigating drug-induced mitochondrial toxicity: A biosensor to increase drug safety? Curr. Drug Saf. 4, 34–54 [DOI] [PubMed] [Google Scholar]
- Piantadosi C. A., Suliman H. B. (2012). Redox regulation of mitochondrial biogenesis. Free Radic. Biol. Med. 53, 2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikó L., Taylor K. D. (1987). Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev. Biol. 123, 364–374 [DOI] [PubMed] [Google Scholar]
- Powell C. S., Jackson R. M. (2003). Mitochondrial complex I, aconitase, and succinate dehydrogenase during hypoxia-reoxygenation: Modulation of enzyme activities by MnSOD. Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L189–L198 [DOI] [PubMed] [Google Scholar]
- Qian W., Choi S., Gibson G. A., Watkins S. C., Bakkenist C. J., Van Houten B. (2012). Mitochondrial hyperfusion induced by loss of fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J. Cell Sci. 125, 5745–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachek L. I., Yuzefovych L. V., Ledoux S. P., Julie N. L., Wilson G. L. (2009). Troglitazone, but not rosiglitazone, damages mitochondrial DNA and induces mitochondrial dysfunction and cell death in human hepatocytes. Toxicol. Appl. Pharmacol. 240, 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo N., Song L., Shutt T. E., McKay S. E., Cotney J., Guan M. X., Gilliland T. C., Hohuan D., Santos-Sacchi J., Shadel G. S. (2012). Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell 148, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. L., Ventura N., Johnson T. E. (2007). Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5, e259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo A. P., Williams S. L., Moraes C. T. (2009). In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 37, 6701–6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolo A. P., Palmeira C. M. (2006). Diabetes and mitochondrial function: Role of hyperglycemia and oxidative stress. Toxicol. Appl. Pharmacol. 212, 167–178 [DOI] [PubMed] [Google Scholar]
- Ross M. F., Kelso G. F., Blaikie F. H., James A. M., Cochemé H. M., Filipovska A., Da Ros T., Hurd T. R., Smith R. A., Murphy M. P. (2005). Lipophilic triphenylphosphonium cations as tools in mitochondrial bioenergetics and free radical biology. Biochemistry. (Mosc). 70, 222–230 [DOI] [PubMed] [Google Scholar]
- Rossignol R., Faustin B., Rocher C., Malgat M., Mazat J. P., Letellier T. (2003). Mitochondrial threshold effects. Biochem. J. 370(Pt 3), 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzier C., Bannwarth S., Chaussenot A., Chevrollier A., Verschueren A., Bonello-Palot N., Fragaki K., Cano A., Pouget J., Pellissier J. F., et al. (2012). The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain 135(Pt 1), 23–34 [DOI] [PubMed] [Google Scholar]
- Rowe T. C., Weissig V., Lawrence J. W. (2001). Mitochondrial DNA metabolism targeting drugs. Adv. Drug Deliv. Rev. 49, 175–187 [DOI] [PubMed] [Google Scholar]
- Rube D. A., van der Bliek A. M. (2004). Mitochondrial morphology is dynamic and varied. Mol. Cell. Biochem. 256-257, 331–339 [DOI] [PubMed] [Google Scholar]
- Rubenstein J. L., Brutlag D., Clayton D. A. (1977). The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell 12, 471–482 [DOI] [PubMed] [Google Scholar]
- Sabri M. I. (1998). Toxin induced mitochondrial dysfunction and neurodegeneration. In Mitochondrial DNA Mutations in Aging, Disease, and Cancer. (Singh K. K, Ed.), pp. 297–317, Springer-Verlag and Landes Bioscience; Austin, TX; [Google Scholar]
- Sadun A. (1998). Acquired mitochondrial impairment as a cause of optic nerve disease. Trans. Am. Ophthalmol. Soc. 96, 881–923 [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C. (2008). Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- Schapira A. H. (2012). Mitochondrial diseases. Lancet 379, 1825–1834 [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M., Ramamoorthy M., Sykora P., Maynard S., Lin P. C., Minor R. K., Wilson D. M., 3rd., Cooper M., Spencer R., de Cabo R., et al. (2012). Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med. 209, 855–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. W. (2010). A closer look at climate change skepticism. Environ. Health Perspect. 118, a536–a540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. W., Kennedy S. R., Salk J. J., Fox E. J., Hiatt J. B., Loeb L. A. (2012). Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 109, 14508–14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E. A., DiMauro S., Hirano M. (2012). Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat. Rev. Genet. 13, 878–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft A. P., Dalton T. P., Nebert D. W., Genter M. B., Hutchinson R. J., Shertzer H. G. (2002a). Dioxin increases reactive oxygen production in mouse liver mitochondria. Toxicol. Appl. Pharmacol. 178, 15–21 [DOI] [PubMed] [Google Scholar]
- Senft A. P., Dalton T. P., Nebert D. W., Genter M. B., Puga A., Hutchinson R. J., Kerzee J. K., Uno S., Shertzer H. G. (2002b). Mitochondrial reactive oxygen production is dependent on the aromatic hydrocarbon receptor. Free Radic. Biol. Med. 33, 1268–1278 [DOI] [PubMed] [Google Scholar]
- Shadel G. S. (2005). Mitochondrial DNA, aconitase ‘wraps’ it up. Trends Biochem. Sci. 30, 294–296 [DOI] [PubMed] [Google Scholar]
- Shaughnessy D. T., Worth L., Jr, Lawler C. P., McAllister K. A., Longley M. J., Copeland W. C. (2010). Meeting report: Identification of biomarkers for early detection of mitochondrial dysfunction. Mitochondrion 10, 579–581 [DOI] [PubMed] [Google Scholar]
- Shertzer H. G., Genter M. B., Shen D., Nebert D. W., Chen Y., Dalton T. P. (2006). TCDD decreases ATP levels and increases reactive oxygen production through changes in mitochondrial F(0)F(1)-ATP synthase and ubiquinone. Toxicol. Appl. Pharmacol. 217, 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock L. S., Thakkar P. V., Peterson E. J., Moran R. G., Taylor S. M. (2011). DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U. S. A. 108, 3630–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko I., Venediktova N., Bochkareva A., Wilson G. L., Alexeyev M. F. (2009). Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 37, 2539–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko I. N., Alexeyev M. F., LeDoux S. P., Wilson G. L. (2010). The approaches for manipulating mitochondrial proteome. Environ. Mol. Mutagen. 51, 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge E. A., Wai T. (2007). Mitochondrial DNA and the mammalian oocyte. Curr. Top. Dev. Biol. 77, 87–111 [DOI] [PubMed] [Google Scholar]
- Silva M. F., Aires C. C., Luis P. B., Ruiter J. P., IJlst L., Duran M., Wanders R. J., Tavares de Almeida I. (2008). Valproic acid metabolism and its effects on mitochondrial fatty acid oxidation: A review. J. Inherit. Metab. Dis. 31, 205–216 [DOI] [PubMed] [Google Scholar]
- Simsek D., Furda A., Gao Y., Artus J., Brunet E., Hadjantonakis A. K., Van Houten B., Shuman S., McKinnon P. J., Jasin M. (2011). Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471, 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. K. (2006). Mitochondria damage checkpoint, aging, and cancer. Ann. N. Y. Acad. Sci. 1067, 182–190 [DOI] [PubMed] [Google Scholar]
- Smiraglia D. J., Kulawiec M., Bistulfi G. L., Gupta S. G., Singh K. K. (2008). A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Ther. 7, 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova I. M., Ringwood A. H., Johnson C. (2005a). Tissue-specific accumulation of cadmium in subcellular compartments of eastern oysters Crassostrea virginica Gmelin (Bivalvia: Ostreidae). Aquat. Toxicol. 74, 218–228 [DOI] [PubMed] [Google Scholar]
- Sokolova I. M., Sokolov E. P., Ponnappa K. M. (2005b). Cadmium exposure affects mitochondrial bioenergetics and gene expression of key mitochondrial proteins in the eastern oyster Crassostrea virginica Gmelin (Bivalvia: Ostreidae). Aquat. Toxicol. 73, 242–255 [DOI] [PubMed] [Google Scholar]
- Starkov A. A., Wallace K. B. (2002). Structural determinants of fluorochemical-induced mitochondrial dysfunction. Toxicol. Sci. 66, 244–252 [DOI] [PubMed] [Google Scholar]
- Stewart J. B., Freyer C., Elson J. L., Wredenberg A., Cansu Z., Trifunovic A., Larsson N. G. (2008). Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 6, e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P. (2006). Reactive oxygen species-mediated mitochondria-to-nucleus signaling: A key to aging and radical-caused diseases. Sci. STKE. 332, re3 [DOI] [PubMed] [Google Scholar]
- Suliman H. B., Carraway M. S., Piantadosi C. A. (2003). Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am. J. Respir. Crit. Care Med. 167, 570–579 [DOI] [PubMed] [Google Scholar]
- Suliman H. B., Welty-Wolf K. E., Carraway M. S., Schwartz D. A., Hollingsworth J. W., Piantadosi C. A. (2005). Toll-like receptor 4 mediates mitochondrial DNA damage and biogenic responses after heat-inactivated E. coli. FASEB J. 19, 1531–1533 [DOI] [PubMed] [Google Scholar]
- Sykora P., Croteau D. L., Bohr V. A., Wilson D. M., 3rd. (2011). Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc. Natl. Acad. Sci. U. S. A. 108, 7437–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek K., Lesnefsky E. J., Larner A. C. (2012). Multi-tasking: Nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 22, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B., Hazra T. K., Papaconstantinou J., Mitra S., Boldogh I. (2003). Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc. Natl. Acad. Sci. U. S. A. 100, 10670–10675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B., Mitra S. (2005). Effect of aging on intracellular distribution of abasic (AP) endonuclease 1 in the mouse liver. Mech. Ageing Dev. 126, 1071–1078 [DOI] [PubMed] [Google Scholar]
- Szczesny B., Tann A. W., Mitra S. (2010). Age- and tissue-specific changes in mitochondrial and nuclear DNA base excision repair activity in mice: Susceptibility of skeletal muscles to oxidative injury. Mech. Ageing Dev. 131, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tann A. W., Boldogh I., Meiss G., Qian W., Van Houten B., Mitra S., Szczesny B. (2011). Apoptosis induced by persistent single-strand breaks in mitochondrial genome: Critical role of EXOG (5’-EXO/endonuclease) in their repair. J. Biol. Chem. 286, 31975–31983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner C. M., Kamel F., Ross G. W., Hoppin J. A., Goldman S. M., Korell M., Marras C., Bhudhikanok G. S., Kasten M., Chade A. R., et al. (2011). Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Turnbull D. M. (2005). Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 [DOI] [PubMed] [Google Scholar]
- Tsang W. Y., Lemire B. D. (2002). Mitochondrial genome content is regulated during nematode development. Biochem. Biophys. Res. Commun. 291, 8–16 [DOI] [PubMed] [Google Scholar]
- Tsang W. Y., Lemire B. D. (2003). The role of mitochondria in the life of the nematode, Caenorhabditis elegans. Biochim. Biophys. Acta. 1638, 91–105 [DOI] [PubMed] [Google Scholar]
- Tsang W. Y., Sayles L. C., Grad L. I., Pilgrim D. B., Lemire B. D. (2001). Mitochondrial respiratory chain deficiency in Caenorhabditis elegans results in developmental arrest and increased life span. J. Biol. Chem. 276, 32240–32246 [DOI] [PubMed] [Google Scholar]
- Turrens J. F., Freeman B. A., Crapo J. D. (1982a). Hyperoxia increases H2O2 release by lung mitochondria and microsomes. Arch. Biochem. Biophys. 217, 411–421 [DOI] [PubMed] [Google Scholar]
- Turrens J. F., Freeman B. A., Levitt J. G., Crapo J. D. (1982b). The effect of hyperoxia on superoxide production by lung submitochondrial particles. Arch. Biochem. Biophys. 217, 401–410 [DOI] [PubMed] [Google Scholar]
- Twig G., Shirihai O. S. (2011). The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 14, 1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai S. B., Mootha V. K. (2012). Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383 [DOI] [PubMed] [Google Scholar]
- Valentin-Vega Y. A., Maclean K. H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F. C., Cleveland J. L., Green D. R., Kastan M. B. (2012). Mitochondrial dysfunction in ataxia-telangiectasia. Blood 119, 1490–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle I., Alvarez-Barrientos A., Arza E., Lamas S., Monsalve M. (2005). PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc. Res. 66, 562–573 [DOI] [PubMed] [Google Scholar]
- Valmas N., Zuryn S., Ebert P. R. (2008). Mitochondrial uncouplers act synergistically with the fumigant phosphine to disrupt mitochondrial membrane potential and cause cell death. Toxicology 252, 33–39 [DOI] [PubMed] [Google Scholar]
- van der Eb M. M., Geutskens S. B., van Kuilenburg A. B., van Lenthe H., van Dierendonck J. H., Kuppen P. J., van Ormondt H., van de Velde C. J., Wanders R. J., van Gennip A. H., et al. (2003). Ganciclovir nucleotides accumulate in mitochondria of rat liver cells expressing the herpes simplex virus thymidine kinase gene. J. Gene Med. 5, 1018–1027 [DOI] [PubMed] [Google Scholar]
- Van Houten B., Woshner V., Santos J. H. (2006). Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst). 5, 145–152 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T., Teoli D., Shoubridge E. A. (2008). The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 40, 1484–1488 [DOI] [PubMed] [Google Scholar]
- Wallace D. C. (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Fan W. (2010). Energetics, epigenetics, mitochondrial genetics. Mitochondrion 10, 12–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Singh G., Lott M. T., Hodge J. A., Schurr T. G., Lezza A. M., Elsas L. J., 2nd., Nikoskelainen E. K. (1988). Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 242, 1427–1430 [DOI] [PubMed] [Google Scholar]
- Wallace K. B. (2007). Adriamycin-induced interference with cardiac mitochondrial calcium homeostasis. Cardiovasc. Toxicol. 7, 101–107 [DOI] [PubMed] [Google Scholar]
- Walters M. W., Bjork J. A., Wallace K. B. (2009). Perfluorooctanoic acid stimulated mitochondrial biogenesis and gene transcription in rats. Toxicology 264, 10–15 [DOI] [PubMed] [Google Scholar]
- Westbrook D. G., Anderson P. G., Pinkerton K. E., Ballinger S. W. (2010). Perinatal tobacco smoke exposure increases vascular oxidative stress and mitochondrial damage in non-human primates. Cardiovasc. Toxicol. 10, 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich V., Tetzlaff I., Graffi A. (1972). Studies on nitrosodimethylamine: Preferential methylation of mitochondrial DNA in rats and hamsters. Chem. Biol. Interact. 4, 81–89 [DOI] [PubMed] [Google Scholar]
- Xia T., Korge P., Weiss J. N., Li N., Venkatesen M. I., Sioutas C., Nel A. (2004). Quinones and aromatic chemical compounds in particulate matter induce mitochondrial dysfunction: Implications for ultrafine particle toxicity. Environ. Health Perspect. 112, 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C. H., Naito A., Mizumachi T., Evans T. T., Douglas M. G., Cooney C. A., Fan C. Y., Higuchi M. (2007). Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem. Biophys. Res. Commun. 364, 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes F. M., Van Houten B. (1997). Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 94, 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Ishii T., Suda H., Akatsuka A., Hartman P. S., Goto S., Miyazawa M., Ishii N. (2006). Age-related changes of mitochondrial structure and function in Caenorhabditis elegans. Mech. Ageing Dev. 127, 763–770 [DOI] [PubMed] [Google Scholar]
- Youle R. J., van der Bliek A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Ren S., Graziano J. H. (1998). Manganese inhibits mitochondrial aconitase: A mechanism of manganese neurotoxicity. Brain Res. 799, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.