Abstract
Paclitaxel is a widely used chemotherapy drug for advanced laryngeal cancer patients. However, the fact that there are 20-40% of advanced laryngeal cancer patients do not response to paclitaxel makes it necessary to figure out potential biomarkers for paclitaxel sensitivity prediction. In this work, Hep2, a laryngeal cancer cell line, untreated or treated with lower dose of paclitaxel for 24 h, was applied to DNA microarray chips for gene and miR expression profile analysis. Expression of eight genes altered significantly following paclitaxel treatment, which was further validated by quantitative real-time PCR. Four up-regulated genes were ID2, BMP4, CCL4 and ACTG2, in which ID2 and BMP4 were implicated to be involved in several drugs sensitivity. While the down-regulated four genes, MAPK4, FASN, INSIG1 and SCD, were mainly linked to the endoplasmic reticulum and fatty acid biosynthesis, these two cell processes that are associated with drug sensitivity by increasing evidences. After paclitaxel treatment, expression of 49 miRs was significantly altered. Within these miRs, the most markedly expression-changed were miR-31-star, miR-1264, miR-3150b-5p and miR-210. While the miRs putatively modulated the mRNA expression of the most significantly expression-altered genes were miR-1264, miR-130a, miR-27b, miR-195, miR-1291, miR-214, miR-1277 and miR-1265, which were obtained by miR target prediction and miRNA target correlation. Collectively, our study might provide potential biomarkers for paclitaxel sensitivity prediction and drug resistance targets in laryngeal cancer patients.
Keywords: Laryngeal cancer, paclitaxel, gene expression profiles, miR expression profiles, cell lines
Introduction
Laryngeal cancer (LC) is a common head and neck cancer. In 2013, an estimated 12,260 new cases of LC will be diagnosed in the United States, accounting for 3,630 deaths (American Cancer Society). The vast majority of LCs are of squamous cell histology, and approximately 40% of patients will have advanced (stage III or IV) disease when first evaluated [1]. Surgery, radiation therapy or chemotherapy alone or combination therapy are widely used to treat LC. With the advances in treatment, not only survival but also larynx preservation becomes important goals for treatment of LC patients.
Chemotherapy has become more and more important since this treatment is able to preserve laryngeal function. Paclitaxel is one of the commonly used chemotherapeutic agents for LC. When used alone for head and neck cancer treatment, the objective response rate of LC patients to paclitaxel is 20-40% [2]. Induction chemotherapy or chemotherapy administered concomitantly with radiation provides a survival advantage as well as a significantly increased rate of organ preservation when compared with radiation alone [3,4]. In 2003, Pfreundner et al showed a paclitaxel-based induction chemotherapy and concurrent regimen produced an 84% 2-year organ preservation rate with acceptable toxicity in LC patients [4]. Others have reported similar results using taxane-based regimens [5-7]. In 2007, it was reported that a 74% 2-year organ preservation rate was obtained with paclitaxel-based concurrent chemoradiotherapy for resectable advanced larynx patients, two-year survival was 63%, and the objective response rate was 50% [8]. Above reports suggested that paclitaxel-based regimens, including induction chemotherapy and concurrent chemoradiotherapy, have good effect on patients’ survival and larynx preservation. However, there are still 20-40% of patients who do not respond to paclitaxel-based therapy. So, it is urgent to find potential biomarkers for paclitaxel sensitivity evaluation to improve the therapy effect of paclitaxel.
Many genes or microRNAs (miRs) have been reported to be involved in paclitaxel sensitivity or resistance of various cancers. For example, expression of β-tubulin isotypes [9], member or regulator of the actin cytoskeleton, such as γ-actin [10] and LIMK2 [11] and the extracellular matrix protein transforming growth factor-β induced (TGFBI) [12] was correlated with paclitaxel sensitivity in different cancers; paclitaxel sensitivity of various cancer cells was also associated to expression of miR-200c [13], miR-148a [14], miR-125b [15], miR-21 [16], miR-337-3p [17] and miR-34a [18]. However, there are few studies on biomarkers of laryngeal cancer cells for paclitaxel. Thus it is worth to observe the influence of genes and miRs above mentioned on paclitaxel sensitivity of laryngeal cancer cells.
In present study, to systematically understand the roles of genes or miRs in paclitaxel sensitivity, Hep2 cells, untreated or treated with lower dose of paclitaxel for 24 h, were applied to DNA microarray chips for gene and miR expression profile analysis. The differentially expressed genes and miRs were identified and the relationships between significantly expression-altered miRs and genes were observed.
Materials and methods
Cell culture
Hep2 cell line was purchased from ATCC (CCL-23) and maintained in DMEM medium supplemented with 10% FBS (Hyclone), penicillin (100 IU/ml) and Streptomycin (100 μg/ml) (Life Technologies). Cells in the exponential growth phase were used for all the experiments.
MTS assay for Hep2 cell viability
Hep2 cells (4×103) were cultured in 100 μl of DMEM medium each well in a 96-well plate. After 24 h, the cells were treated with paclitaxel (0, 2, 6.3, 20, 63, 200, 630, 2000 nmol/L, respectively) for 72 h. Every treatment was triplicate in the same experiment. Then 20 μl of MTS (CellTiter 96 AQueous One Solution Reagent; Promega) was added to each well for 2 h at 37°C. After incubation, the absorbance was read at a wavelength of 490 nm according to the manufacturer’s instructions. The IC50 calculation was performed with GraphPad Prism 5.0 software.
The concentration of paclitaxel at which Hep2 cell viability was suppressed by 10% or so in 24 h was determined as follow: Hep2 cells were treated with paclitaxel (0, 0.2, 0.63, 2.0, 6.3 and 20 nmol/L, respectively) for 24 h. Every treatment was triplicate in the same experiment. The cell viability was examined as above mentioned.
The time-course of paclitaxel treatment was carried out as follow: Hep2 cells were left untreated or treated with paclitaxel (2 nmol/L) for 24, 48 and 72 h, respectively. Every treatment was triplicate in the same experiment. The cell viability was calculated relatively to the untreated cells at every time point.
Microarray analysis: gene and miR expression profile
Hep2 cells (8×104) were grown in 2 ml of DMEM medium (10% FBS) each well in a 6-well plate. After 24 h, the cells were treated with paclitaxel (2 nmol/L) for 24 h or left untreated, respectively. Every treatment was duplicated in the same experiment. All the samples were homogenized with 1 ml Trizol (Invitrogen, Life Technologies) and total RNAs were extracted according to the manufacturer’s instructions.
500 ng total RNA was used to synthesize double-strand cDNA and in vitro transcribed to cRNA, purified 10 μg cRNA was used to synthesize 2nd-cycle cDNA and then hydrolyzed by RNase H and purified. Above steps were performed with Ambion WT Expression Kit. 5.5 μg 2nd-cycle cDNA was fragmented and the single-stranded cDNA was labeled with GeneChip2 WT Terminal Labeling Kit and Controls Kit (Affymetrix, PN 702880). About 700 ng fragmented and labeled single-stranded cDNA were hybridized to an Affymetrix GeneChip Human Gene 1.0 ST array, which was washed and stained with GeneChip2 Hybridization, Wash and Stain kit (Affymetrix).
Total RNA from Hep2 cells, untreated or treated with lower dose of paclitaxel for 24 h, was processed and hybridized to Affymetrix GeneChip® miRNA 2.0 Array, which recognizes 1,105 separate human miRs in accordance with the Sanger Institute miRBase version 15. Each sample was duplicated for miR expression profile.
Microarray data analysis was done using Significance Analysis of Microarrays (SAM) method, as described before [19]. Gene set enrichment analysis (GSEA) was performed to the differential expression genes with DAVID 6.7 online software.
Quantitative real-time PCR (qPCR)
Total RNA above isolated was synthesized to cDNA using PrimeScript RT reagent kit with gDNA Eraser (Takara, RR074A) for RT-PCR with mixture of oligo-dT and Random Primer (9 mer). The primers used for qPCR validation were list in Table 1. Real-time qPCR was performed on CFX-96 (Bio-lab), with endogenous control hActb. Gene expression was calculated relative to expression of hActb endogenous control and adjusted relative to expression in untreated control cells.
Table 1.
Primers for qPCR validation
| Gene | Forward | Reverse |
|---|---|---|
| Actb | GCATCCCCCAAAGTTCACAA | GGACTTCCTGTAACAACGCATCT |
| SCD | GGAGCCACCGCTCTTACAAA | ACGAGCCCATTCATAGACATCA |
| INSIG1 | AGTGCTGCCTGGAGGAAACTC | GAGGGTTCAGTGGAGGGTGTAA |
| MAPK4 | TGAGAAGGGTGACTGCATCG | ACCAAACCATTGACACCGAAG |
| TGM2 | GGGCCACTTCATTTTGCTCTT | ACTCCTGCCGCTCCTCTTC |
| FASN | CGCTCGGCATGGCTATCT | CTCGTTGAAGAACGCATCCA |
| SREBF1 | GCCCCTGTAACGACCACTG | CAGCGAGTCTGCCTTGATG |
| FAM83A | AGGGACGGGAGGCATGA | GGACCCACTGGCTCTTGACA |
| OCLM | CTCCTGTGGTGTAAATCATTGCTT | AATCCTGAAACCTGCAATGCA |
| HOXB5 | TCCTTCCATGCTCCCAACTC | CACAGACACAAACATTCAGAAACACT |
| TTK | GACTTTCCACCTGCTTGTCAGTT | AGTGGCAAGTATTTGATGCTGTTG |
| AGPAT1 | TGAGGGTCTGGGTGTTTCCT | CACGTTTGAAGGGCAGCAT |
| PDK4 | TCAGGACACTTTACGGGATCAA | TGGAGGAAACAAGGGTTCACA |
| MED10 | AAGGCAAGATCGACACCATGA | CCCCGGATGCTTCGATACTT |
| PSG4 | TTGCTGGCTACATTTGGTACAA | CATCCTCCTGCGTGACATTC |
| IGFBP3 | TCCAATAGTCCCCAAGCAGTACA | TTCCACCCCCTCCATTCAA |
| SEMA3D | GCTCATAAGGAAAGTGCAGACCAT | GTCATAGGTTTTGCTTGGACATGT |
| CCL4 | TGGGTCCAGGAGTACGTGTATG | CTTCCCTGAAGACTTCCTGTCTCT |
| ID2 | CAGTCCCGTGAGGTCCGTTA | GGGTTTTGCTCCGGGAGAT |
| ACTG2 | TTGATGTCTCGCACAATTTCTCT | ATTGTGCGAGACATCAAGGAG |
| BMP4 | CACACGACTACTGGACACGAGACT | AGGGCTCACATCAAAAGTTTCC |
miR target prediction and miRNA target correlation
miR target prediction was performed with miRWalk online software. The comparative analysis was done by 5 prediction programs: miRanda, miRDB, miRWalk, RNA22 and TargetScan. 8 genes induced or repressed by paclitaxel were selected to perform miR target prediction. miRs predicted by greater than or equal to 3 programs were selected as putative upstream target of some gene. The putative upstream miRs were done intersection with miRs that expression level altered significantly (FDR<10%) following paclitaxel treatment. The overlapped miRs were selected to construct miR-gene networks with the aid of Cytoscape 2.8 software.
Statistical analysis
R2 values were calculated using Pearson’s correlation coefficient. The significant difference was calculated using Student’s t-test.
Results
Hep2 cell line is sensitive to paclitaxel
To determine chemosensitivity of Hep2 cells to paclitaxel, Hep2 cells were treated with paclitaxel at different concentrations for 72 h, cell viability was examined by MTS assay and IC50 dose to paclitaxel was calculated. IC50 dose of Hep2 to paclitaxel at 72 h is 0.018 μmol/L (R2=0.94) (Figure 1A). According to data reported in DTP Data Search, the mean IC50 of NCI-60 cell panel to paclitaxel is 0.009-0.035 μmol/L. So, Hep2 cell line is sensitive to paclitaxel.
Figure 1.
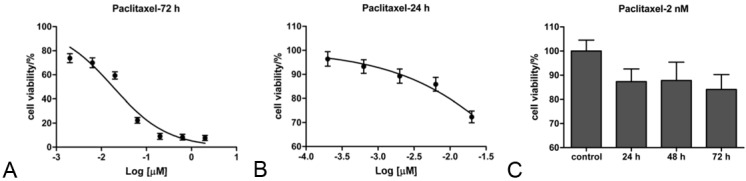
Hep2 cells are sensitive to paclitaxel. A. MTS assay for Hep2 cells treated with paclitaxel (0, 2, 6.3, 20, 63, 200, 630, 2000 nmol/L, respectively) for 72 h. B. MTS assay for Hep2 cells treated with paclitaxel (0, 0.2, 0.63, 2.0, 6.3 and 20 nmol/L, respectively) for 24 h. C. Time-course effect of lower dose treatment of paclitaxel. Hep2 cells were left untreated or treated with paclitaxel (2 nmol/L) for 24, 48 and 72 h, respectively. Every treatment was triplicate in the same experiment. Error bars represent the standard deviation (SD).
To find a suitable dose to inhibit Hep2 cells growth by 10% or so, we used a narrower range of paclitaxel concentrations to treat Hep2 cells for 24 h. The IC50 of Hep2 to paclitaxel at 24 h is 0.143 μmol/L (R2=0.97). As Hep2 cells viability was repressed by 10.7% at the concentration of 2 nmol/L (Figure 1B), this concentration is suitable for study on paclitaxel sensitivity. The concentration was far lower than the corresponding IC50 dose.
And then, Hep2 cells were treated with 2 nmol/L of paclitaxel for 24, 48 and 72 h or left untreated. The time-course effect of paclitaxel treatment was present in Figure 1C. The results showed that when treated with the lower concentration of paclitaxel for 24 h, Hep2 cell viability was suppressed by 15% or so.
Gene expression analysis
Hep2 cells, treated with a lower dose of paclitaxel for 24 h or left untreated, were applied to DNA microarray chips. The results of bioinformatics analysis showed that when cells were treated with this moderate condition, gene expression did not alter dramatically. Expression of 36 genes was increased by higher than 30% (Table 2), and that of 19 genes was decreased by higher than 30% (Table 3) after paclitaxel treatment. The most markedly expression-altered genes were ID2 (up to 1.85 fold), CCL4 (up to 1.77 fold), HOXB5 (down to 0.53 fold), RBMY2EP (down to 0.61 fold) and FASN (down to 0.61 fold). Since amount of the significantly expression-altered genes was too little, only few pathways/cell processes were obtained after GSEA. Up-regulated genes were enriched in regulation of protein amino acid phosphorylation (BMP4, TTK and IGFBP3, p=0.04, FDR=44%), while down-regulated genes were enriched in carboxylic acid biosynthetic process (SCD, FASN, LGSN and AGPAT1, p=3.01E-04, FDR=0.4%) and the endoplasmic reticulum (ER) (SREBF1, SCD, INSIG1, UBQLN4 and AGPAT1, p=0.003, FDR=2.7%).
Table 2.
The most significantly up-regulated genes
| Gene Symbol | fold change | p value | Gene Description |
|---|---|---|---|
| ID2 | 1.851 | 0.01 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| CCL4 | 1.773 | 0.03 | chemokine (C-C motif) ligand 4 |
| SEMA3D | 1.471 | 0.01 | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| PSG4 | 1.464 | 0.04 | pregnancy specific beta-1-glycoprotein 4 |
| DUSP5P | 1.423 | 0.02 | dual specificity phosphatase 5 pseudogene |
| ACTG2 | 1.415 | 0.001 | actin, gamma 2, smooth muscle, enteric |
| MED10 | 1.414 | 0.001 | mediator complex subunit 10 |
| IGFBP3 | 1.408 | 0.01 | insulin-like growth factor binding protein 3 |
| SCN9A | 1.389 | 0.04 | sodium channel, voltage-gated, type IX, alpha subunit |
| LYPD6B | 1.386 | 0.02 | LY6/PLAUR domain containing 6B |
| DEPDC7 | 1.384 | 0.001 | DEP domain containing 7 |
| BMP4 | 1.378 | 0.04 | bone morphogenetic protein 4 |
| NDUFV2 | 1.373 | 0.002 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| OLR1 | 1.363 | 0.01 | oxidized low density lipoprotein (lectin-like) receptor 1 |
| NDUFV2 | 1.360 | 0.01 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| DLEU2 | 1.348 | 0.02 | deleted in lymphocytic leukemia 2 (non-protein coding) |
| RPL23AP53 | 1.344 | 0.01 | ribosomal protein L23a pseudogene 53 |
| PDK4 | 1.341 | 0.04 | pyruvate dehydrogenase kinase, isozyme 4 |
| TTPA | 1.340 | 0.05 | tocopherol (alpha) transfer protein |
| TIMM8A | 1.334 | 0.02 | translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| RLIM | 1.331 | 0.02 | ring finger protein, LIM domain interacting |
| GIN1 | 1.329 | 0.04 | gypsy retrotransposon integrase 1 |
| TTK | 1.329 | 0.001 | TTK protein kinase |
| TUBB8 | 1.327 | 0.03 | tubulin, beta 8 class VIII |
| SASS6 | 1.327 | 0.0003 | spindle assembly 6 homolog (C. elegans) |
| SLC7A11 | 1.326 | 0.001 | solute carrier family 7 (anionic amino acid transporter light chain, xc-system), member 11 |
| GTF2F2 | 1.321 | 0.01 | general transcription factor IIF, polypeptide 2, 30kDa |
| BCAT1 | 1.320 | 0.01 | branched chain amino-acid transaminase 1, cytosolic |
| FKTN | 1.314 | 0.02 | fukutin |
| KIAA1143 | 1.314 | 0.02 | KIAA1143 |
| DENND2C | 1.310 | 0.03 | DENN/MADD domain containing 2C |
| ZNF791 | 1.310 | 0.02 | zinc finger protein 791 |
| ZNF33B | 1.307 | 0.05 | zinc finger protein 33B |
| ARHGAP11A | 1.307 | 0.01 | Rho GTPase activating protein 11A |
| FBXO16 | 1.303 | 0.04 | F-box protein 16 |
| C3orf26 | 1.301 | 0.01 | chromosome 3 open reading frame 26 |
Table 3.
The most significantly down-regulated genes
| Gene Symbol | fold change | p value | Gene Description |
|---|---|---|---|
| HOXB5 | 0.531 | 0.02 | homeobox B5 |
| RBMY2EP | 0.606 | 0.02 | RNA binding motif protein, Y-linked, family 2, member E pseudogene |
| FASN | 0.613 | 0.0002 | fatty acid synthase |
| TGM2 | 0.630 | 0.003 | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) |
| OCLM | 0.632 | 0.01 | oculomedin |
| FAM83A | 0.646 | 0.003 | family with sequence similarity 83, member A |
| MAPK4 | 0.650 | 0.01 | mitogen-activated protein kinase 4 |
| UBQLN4 | 0.655 | 0.01 | ubiquilin 4 |
| FAM127A | 0.673 | 0.01 | family with sequence similarity 127, member A |
| SREBF1 | 0.674 | 0.001 | sterol regulatory element binding transcription factor 1 |
| INSIG1 | 0.675 | 0.005 | insulin induced gene 1 |
| GK3P | 0.676 | 0.04 | glycerol kinase 3 pseudogene |
| LGSN | 0.685 | 0.01 | lengsin, lens protein with glutamine synthetase domain |
| AGPAT1 | 0.685 | 0.01 | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| WBP2 | 0.686 | 0.01 | WW domain binding protein 2 |
| SCD | 0.688 | 0.002 | stearoyl-CoA desaturase (delta-9-desaturase) |
| GPR141 | 0.690 | 0.03 | G protein-coupled receptor 141 |
| SSBP4 | 0.694 | 0.01 | single stranded DNA binding protein 4 |
| ADAMTSL4 | 0.700 | 0.01 | ADAMTS-like 4 |
qPCR validation of gene expression
Then 20 genes were selected for further validating the fold change determined by microarray. Within these genes, expression of 11 genes was up-regulated and that of 9 genes was down-regulated following paclitaxel treatment. As showed in Figure 2A, the expression trends of 17 genes were consistent between microarray data and qPCR results following paclitaxel treatment, although expression fold change varied to some extents. For AGPAT1, qPCR data showed its expression was increased, which was in contrast to microarray data. While for TTK and HOXB5, their expression determined by qPCR had no significant difference (p>0.05) before and after paclitaxel treatment, which was not in line with microarray data. In terms of expression trends, 17 of 20 (85%) genes induced or repressed following paclitaxel treatment were positively validated by qPCR. And then, the correlation between both data sets was observed using R2. As shown in Figure 2B, an R2 value of 0.84 (p<0.05) was calculated for the fold change determined by these two methods. Collectively, these data suggested that microarray data were mostly reliable. The genes whose expression altered the most markedly were ID2, CCL4, BMP4, ACTG2, FASN, INSIG1, MAPK4 and SCD.
Figure 2.
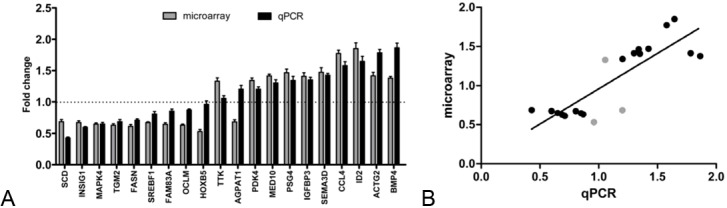
qPCR validation of microarray results. A. Fold changes of 20 gene expression levels following paclitaxel treatment, as determined by microarray and qPCR analysis were shown in histogram. Gene expression was calculated relative to expression of hActb endogenous control and adjusted relative to expression in untreated control cells. Every treatment was triplicate in the same experiment. Error bars represent the standard deviation (SD). B. Correlation of fold change in gene expression levels following paclitaxel treatment as determined by microarray and qPCR. The 3 grey dots represent the 3 genes whose expression treands did not match between microarray data and qPCR results.
miR expression analysis
Bioinformatics analysis showed that miR expression profile markedly altered. When false discovery rate (FDR) was set as 1%, the cutoff value of relative normalized difference for up-regulated genes was 1.73 (Figure 3C), for down-regulated genes was -2.00 (Figure 3A). According to this criterion (FDR<1%), expression of 5 miRs was significantly increased and that of 4 miRs was markedly decreased (Figure 3B). When FDR was set as 10%, the cutoff values were 1.20 and -1.35, respectively, for up-regulated and down-regulated miRs. According to this criterion (FDR<10%), 27 miRs showed increased expression and 22 miRs showed decreased expression (Table 4). Within these miRs, the most significantly expression-altered were miR-31-star, miR-1264, miR-3150b-5p and miR-210.
Figure 3.
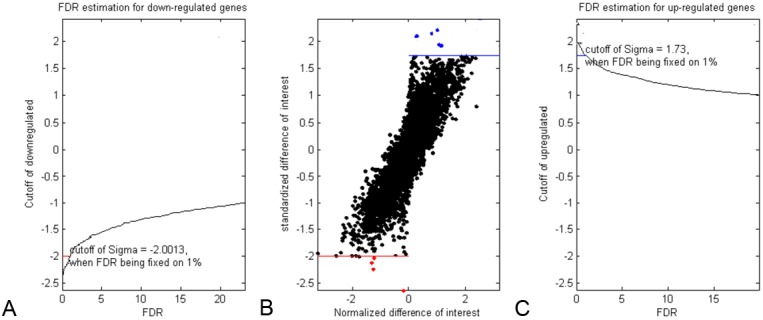
Selection of significantly altered miRs. The normalized relative difference of miRs expression following paclitaxel treatment from 0 to 24 h was calculated by SAM method. A. False discovery rate (FDR) estimation for down-regulated miRs. When FDR was fixed on 1%, cutoff value of the normalized relative difference was -2.00. miRs with relative difference lower than -2.00 were identified as significantly down-regulated miRs. C. FDR estimation for up-regulated miRs. When FDR was fixed on 1%, cutoff value of the normalized relative difference was 1.73. miRs with relative difference higher than 1.73 were identified as significantly up-regulated miRs. B. Selection of significantly altered miRs. Criterions above mentioned were applied to all miRs on the microarray chip, significantly up-regulated miRs were labeled with blue dots and significantly down-regulated miRs were dotted with red color. “sigma” in (A) and (C) means the relative normalized difference calculated by SAM method.
Table 4.
The most significantly altered miRs
| up-regulated | down-regulated | ||
|---|---|---|---|
|
| |||
| miR | sigma* | miR | sigma* |
| hsa-miR-31-star | 2.10 | hsa-miR-3150b-5p | -2.62 |
| hsa-miR-1264 | 2.08 | hsa-miR-210 | -2.24 |
| hsa-miR-20b-star | 2.02 | hsa-miR-3163 | -2.05 |
| hsa-miR-3168 | 1.88 | hsa-miR-2964a-3p | -2.04 |
| hsa-miR-3144-5p | 1.84 | hsa-miR-2277-3p | -1.95 |
| hsa-miR-188-5p | 1.71 | hsa-miR-214 | -1.78 |
| hsa-miR-1250 | 1.66 | hsa-miR-200b-star | -1.75 |
| hsa-miR-3074-3p | 1.60 | hsa-miR-199a-3p | -1.64 |
| hsa-miR-140-5p | 1.58 | hsa-miR-193b-star | -1.64 |
| hsa-miR-195 | 1.57 | hsa-miR-29c | -1.63 |
| hsa-miR-302b-star | 1.54 | hsa-miR-3065-3p | -1.63 |
| hsa-miR-1265 | 1.50 | hsa-miR-3161 | -1.57 |
| hsa-miR-1197 | 1.46 | hsa-miR-1277 | -1.56 |
| hsa-miR-3173-3p | 1.41 | hsa-miR-200a | -1.51 |
| hsa-miR-28-5p | 1.36 | hsa-miR-2114 | -1.43 |
| hsa-miR-1200 | 1.35 | hsa-miR-221 | -1.42 |
| hsa-miR-3153 | 1.34 | hsa-miR-124-star | -1.41 |
| hsa-miR-296-3p | 1.29 | hsa-miR-10b-star | -1.39 |
| hsa-miR-130a | 1.29 | hsa-miR-23c | -1.38 |
| hsa-miR-3164 | 1.27 | hsa-miR-15a | -1.38 |
| hsa-miR-3167 | 1.27 | hsa-miR-2278 | -1.38 |
| hsa-miR-1268 | 1.25 | hsa-miR-30a | -1.37 |
| hsa-miR-130b-star | 1.25 | ||
| hsa-miR-27b | 1.24 | ||
| hsa-miR-219-2-3p | 1.24 | ||
| hsa-miR-1291 | 1.21 | ||
sigma means the relative normalized difference calculated by SAM method.
MiRs-genes network construction
To construct the network between significantly expression-altered miRs and genes, 8 genes whose expression altered the most markedly and was validated by qPCR were selected to perform miR target prediction and miRNA target correlation. For example, CCL4 gene was predicted to be putatively mediated by 32 miRs (predicted by ≥3 programs) with the aid of miRWalk online software. Then this 32 miRs set was done intersection with the down-regulated 22 miRs and the only overlapped miR was miR-214. Thus the up-regulated expression of CCL4 was putatively mediated by the down-regulated expression of miR-214. For the other 7 genes, we did the prediction and target correlation according to the same procedure and found that there were 8 miRs that putatively mediated the expression of 6 of those 8 genes, the network was constructed with the aid of Cystoscope 2.8 software (Figure 4). Expression-increased miR-27b putatively mediated the decreased expression of FASN and INSIG1, while the down-regulated INSIG1 was putatively mediated by expression-increased miR-27b, miR-130a and miR-1264.
Figure 4.
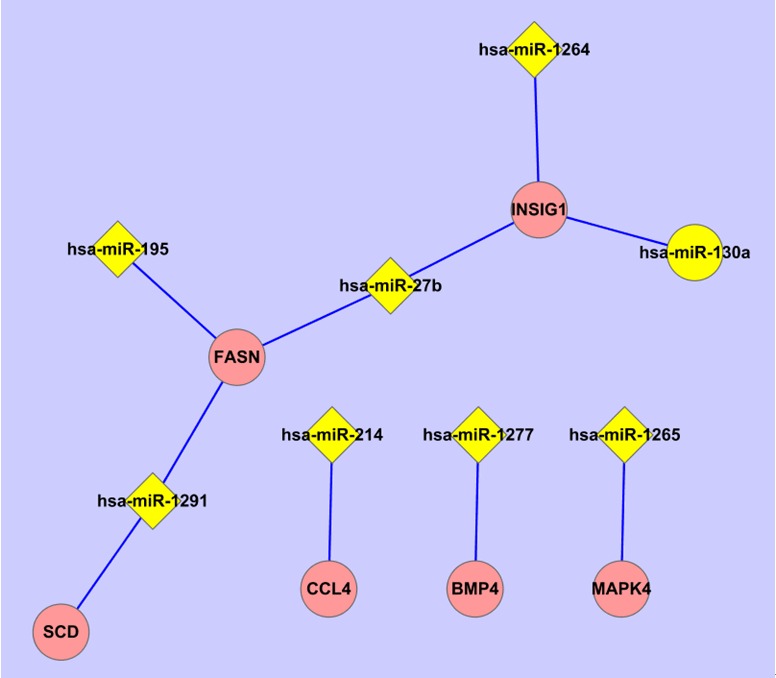
miRs-genes network. The most significantly expression-altered 8 genes validated by qPCR were respectively applied to miRWalk online software to predict the upstream miR targets putatively mediated the gene expression. And then the miR targets were done intersection with the expression-altered miRs (FDR<10%) determined by microarray. The overlapped miRs were considered as potential upstream targets and used to construct the miRs-genes network with the aid of Cytoscape 2.8 software.
Discussion
Paclitaxel is a widely used chemotherapy drug for advanced laryngeal cancer patients, whether in induction chemotherapy or in concurrent chemoradiotherapy. However, the fact that 20-40% of advanced laryngeal cancer patients do not response to paclitaxel makes it necessary to figure out biomarkers for paclitaxel sensitivity prediction. In this work, Hep2 cells, untreated or treated by lower dose of paclitaxel for 24 h, were applied to DNA microarray chips for gene and miR expression profile analysis. The differentially expressed genes and miRs were identified and the relationships between miRs and genes were examined.
It was proposed that treating cancer cells with lower dose of antitumor drug in vitro can mimic the physiological status in vivo when patients were treated with the drug, and thus this method is suitable for study on cancer cells sensitivity to some drug in vitro [20]. In present study, 2 nmol/L of paclitaxel, far lower than the corresponding IC50 dose (143 nmol/L), was administrated to Hep2 cells for 24 h. Under this condition, the cell viability was reduced by 15% or so. It is assumed that when cancer cells growth was slightly inhibited, expression of genes involved sensitivity or resistance to some drug may be altered more severely than those linked to cell apoptosis or cell cycle. Subsequent gene expression profile analysis showed that expression of p53, Bax, CCND1 or CCNB1 did not change markedly. And this result strongly supports the above hypothesis.
Expression of ID2, CCL4, BMP4, ACTG2, FASN, INSIG1, MAPK4 and SCD was the most greatly altered following lower dose of paclitaxel treatment, which was further confirmed with qPCR. The former four genes were up-regulated, while the latter four genes were down-regulated following paclitaxel treatment. Within the up-regulated genes, ID2 and BMP4 are intriguing. ID2, encoding a nuclear-localized protein, is associated to DNA-damage response and found to be induced by Ecteinascidin 743 in human primary sarcoma cells [21]. Furthermore, ID2 is proposed to one of the crucial genes linked to chemosensitivity of glioblastoma to Semustine (Me-CCNU) [22]. BMP4 protein is a member of the bone morphogenetic protein family belonging to the transforming growth factor-beta superfamily. Previous study showed that BMP4 inhibits proliferation and induces the apoptosis by p53-dependent endoplasmic reticulum (ER) dysfunction in myeloma and B-cell hybridoma cells [23]. Moreover, BMP4 was highly expressed in cisplatin-resistant gastric cancer lines and targeted genetic inhibition of BMP4 caused significant sensitization of gastric cancer cells to cisplatin [24]. Interestingly, BMP4 induces ID2 expression in prostate cancer cell line [25]. This may be used to explain the co-elevated expression of BMP4 and ID2 in our data. These reports together with our data suggest that ID2 and BMP4 might be potential markers for paclitaxel sensitivity in laryngeal cancer.
As for the four down-regulated genes by paclitaxel, three genes are involved in carboxylic acid biosynthetic process (SCD and FASN) and ER (SCD, INSIG1), the fourth one, ACTG2, is linked to actin cytoskeleton. Member of actin cytoskeleton has been proposed to be involved in paclitaxel sensitivity [10,11]. Growing evidences suggest that ER-associated genes are critically linked to paclitaxel sensitivity [26-28]. In 2007, Shajahan et al showed that activation of Caveolin-1, an ER-localized protein, sensitizes breast cancer cells to paclitaxel [26]. Bortezomib, an approved drug for the treatment of certain hematological neoplasms, was shown to efficiently induce apoptosis by an upregulation of the ER stress sensor ATF3 in ovarian cancer cells and to enhance the sensitivity of ovarian cancer cells to TRAIL [27]. However, bortezomib attenuated the efficacy of paclitaxel treatment [27], suggesting that the cytotoxic effect of paclitaxel on cancer cells might partially be dependent on the down-regulation of ER stress, which is in line with our data in some ways. Recently, it was proposed that targeted inhibition of GRP78, a chaperone protein mainly located in the ER of normal cells, significantly increases paclitaxel sensitivity of castrate-resistant prostate cancer cell line expressing GRP78 at its surface [28]. All the above reports show the roles of ER-associated genes on modulating cancer cells sensitivity to paclitaxel. The possible mechanism by which ER-associated genes mediate paclitaxel sensitivity is in that ER is associated with the microtubule cytoskeleton, exactly site where paclitaxel exerts its cytotoxic effect in cancer cells. Specific to the SCD, a gene involved in ER and fatty acid biosynthesis, was suggested to be critical for cancer cell survival and thus to be a potential target for cancer therapy [29]. It was proposed that the docetaxel-resistance of progesterone receptor-positive breast cancer cells induced by progestin treatment is in part caused by the increased expression of SCD [30], indicating that SCD overexpression may be putative marker for docetaxel resistance. Considering that Hep2 cells are sensitive to paclitaxel, the repressed expression of SCD following paclitaxel treatment is reasonable.
MiRs have been implicated to progression of human cancers and drug sensitivity. In this work, the majority of the most significantly expression-altered miRs (49 miRs with FDR<10%) have not putative targets in the 8 genes whose expression altered the most markedly, except for 8 miRs (Figure 4). Because most miRs are typically thought to regulate gene expression post-transcriptionally, this phenomenon is not surprising. However, recent studies concluded that degradation of miRNA targets is a widespread effect of miRNA-based regulation, which alone accounts for most of the repression mediated by miRNAs in mammalian cell cultures [31]. Up-regulated miR-130a putatively mediated decreased expression of INSIG1 in our data, while previous report showed that miR-130a was up-regulated in cisplatin-resistant SKOV3 cells and silencing of miR-130a resulted in MDR mRNA inhibition and hence help SKOV3 cells overcome cisplatin resistance [32]. Increased expression of miR-27b putatively repressed expression of INSG1 and FASN, while Pan et al suggested that ectopic expression of miR-27b suppressed CYP3A4 via posttranscriptional and transcriptional manner and thus led to a lower sensitivity of PANC1 cells to cyclophosphamide [33]. Up-re-gulated miR-195 putatively inhibited expression of FASN, while it showed that miRNA-195 sensitizes hu-man hepatocellular carcinoma cells to 5-FU by targeting BCL-w [34]. Down-regulated miR-214 putatively mediated the increased expression of CCL4, while Wang et al demonstrated that miR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells [35]. In spite that some of above reports are in accordance with our data, the relationship between altered miR and mRNA expression need to be further studied by functional validation in more laryngeal cancer cell lines. Taken together, we provide here a new clue for looking for paclitaxel sensitive biomarkers and the candidates from our results are worth of further study, which might do better to the clinical application of paclitaxel.
Acknowledgments
We gratefully acknowledge Feng-qing Li, Yi-hui Lin, and Lei Xiong for helpful discussion, Qing-zhou Zhang and Gang Niu for the technical supports of microarray data analysis, Tao Zhou and Zhi-Qiang Zeng for the technical supports of qPCR. This work is supported by grants of Shanghai Science and Technology Development Fund (No. 09411951000 & No. 054119550), China.
References
- 1.Myers EN. Cancer of the head and neck. Philadelphia, Pa.: Saunders; 2003. [Google Scholar]
- 2.Schrijvers D, Vermorken JB. Role of taxoids in head and neck cancer. Oncologist. 2000;5:199–208. doi: 10.1634/theoncologist.5-3-199. [DOI] [PubMed] [Google Scholar]
- 3.Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, Peters G, Lee DJ, Leaf A, Ensley J, Cooper J. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003;349:2091–2098. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- 4.Pfreundner L, Hoppe F, Willner J, Preisler V, Bratengeier K, Hagen R, Helms J, Flentje M. Induction chemotherapy with paclitaxel and cisplatin and CT-based 3D radiotherapy in patients with advanced laryngeal and hypopharyngeal carcinomas--a possibility for organ preservation. Radiother Oncol. 2003;68:163–170. doi: 10.1016/s0167-8140(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 5.Machtay M, Rosenthal DI, Hershock D, Jones H, Williamson S, Greenberg MJ, Weinstein GS, Aviles VM, Chalian AA, Weber RS Penn Cancer Center Clinical Trials Group. Organ preservation therapy using induction plus concurrent chemoradiation for advanced resectable oropharyngeal carcinoma: a University of Pennsylvania Phase II Trial. J. Clin. Oncol. 2002;20:3964–3971. doi: 10.1200/JCO.2002.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Schrijvers D, Vermorken JB. Taxanes in the treatment of head and neck cancer. Curr Opin Oncol. 2005;17:218–224. doi: 10.1097/01.cco.0000158735.91723.0e. [DOI] [PubMed] [Google Scholar]
- 7.Cmelak AJ, Murphy BA, Burkey B, Douglas S, Shyr Y, Netterville J. Taxane-based chemoirradiation for organ preservation with locally advanced head and neck cancer: results of a phase II multi-institutional trial. Head Neck. 2007;29:315–324. doi: 10.1002/hed.20522. [DOI] [PubMed] [Google Scholar]
- 8.Cmelak AJ, Li S, Goldwasser MA, Murphy B, Cannon M, Pinto H, Rosenthal DI, Gillison M, Forastiere AA. Phase II trial of chemoradiation for organ preservation in resectable stage III or IV squamous cell carcinomas of the larynx or oropharynx: results of Eastern Cooperative Oncology Group Study E2399. J. Clin. Oncol. 2007;25:3971–3977. doi: 10.1200/JCO.2007.10.8951. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Scagliotti G, Danenberg KD, Lord RV, Bepler G, Novello S, Cooc J, Crino L, Sanchez JJ, Taron M, Boni C, De Marinis F, Tonato M, Marangolo M, Gozzelino F, Di Costanzo F, Rinaldi M, Salonga D, Stephens C. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene. 2003;22:3548–3553. doi: 10.1038/sj.onc.1206419. [DOI] [PubMed] [Google Scholar]
- 10.Verrills NM, Po’uha ST, Liu ML, Liaw TY, Larsen MR, Ivery MT, Marshall GM, Gunning PW, Kavallaris M. Alterations in gamma-actin and tubulin-targeted drug resistance in childhood leukemia. J Natl Cancer Inst. 2006;98:1363–1374. doi: 10.1093/jnci/djj372. [DOI] [PubMed] [Google Scholar]
- 11.Dan S, Tsunoda T, Kitahara O, Yanagawa R, Zembutsu H, Katagiri T, Yamazaki K, Nakamura Y, Yamori T. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002;62:1139–1147. [PubMed] [Google Scholar]
- 12.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, Nicke B, Downward J, Swanton C, Bell SD, Earl HM, Laskey RA, Caldas C, Brenton JD. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agen-ts. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita Y, Kojima K, Ohhashi R, Hamada N, Nozawa Y, Kitamoto A, Sato A, Kondo S, Kojima T, Deguchi T, Ito M. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285:19076–19084. doi: 10.1074/jbc.M109.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O, Riker AI, Tan M. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren Y, Zhou X, Mei M, Yuan XB, Han L, Wang GX, Jia ZF, Xu P, Pu PY, Kang CS. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M, Wistuba II, Gazdar AF, Minna JD, Pertsemlidis A. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh S, Chitkara D, Mehrazin R, Behrman SW, Wake RW, Mahato RI. Chemoresistance in prostate cancer cells is regulated by miRNAs and Hedgehog pathway. PLoS One. 2012;7:e40021. doi: 10.1371/journal.pone.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci U S A. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez N, Sanchez-Beato M, Carnero A, Moneo V, Tercero JC, Fernandez I, Navarrete M, Jimeno J, Piris MA. Transcriptional signature of Ecteinascidin 743 (Yondelis, Trabectedin) in human sarcoma cells explanted from chemo-naive patients. Mol Cancer Ther. 2005;4:814–823. doi: 10.1158/1535-7163.MCT-04-0316. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z, Liu Y, He H, Chen X, Chen J, Lu YC. Candidate genes influencing sensitivity and resistance of human glioblastoma to Semustine. Brain Res Bull. 2011;86:189–194. doi: 10.1016/j.brainresbull.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda N, Saitoh M, Kobayashi N, Miyazono K. Execution of BMP-4-induced apoptosis by p53-dependent ER dysfunction in myeloma and B-cell hybridoma cells. Oncogene. 2006;25:3509–3517. doi: 10.1038/sj.onc.1209393. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova T, Zouridis H, Wu Y, Cheng LL, Tan IB, Gopalakrishnan V, Ooi CH, Lee J, Qin L, Wu J, Lee M, Rha SY, Huang D, Liem N, Yeoh KG, Yong WP, Teh BT, Tan P. Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer. Gut. 2013;62:22–33. doi: 10.1136/gutjnl-2011-301113. [DOI] [PubMed] [Google Scholar]
- 25.Wahdan-Alaswad RS, Song K, Krebs TL, Shola DT, Gomez JA, Matsuyama S, Danielpour D. Insulin-like growth factor I suppresses bone morphogenetic protein signaling in prostate cancer cells by activating mTOR signaling. Cancer Res. 2010;70:9106–9117. doi: 10.1158/0008-5472.CAN-10-1119. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Shajahan AN, Wang A, Decker M, Minshall RD, Liu MC, Clarke R. Caveolin-1 tyrosine phosphorylation enhances paclitaxel-mediated cytotoxicity. J Biol Chem. 2007;282:5934–5943. doi: 10.1074/jbc.M608857200. [DOI] [PubMed] [Google Scholar]
- 27.Bruning A, Burger P, Vogel M, Rahmeh M, Friese K, Lenhard M, Burges A. Bortezomib treatment of ovarian cancer cells mediates endoplasmic reticulum stress, cell cycle arrest, and apoptosis. Invest New Drugs. 2009;27:543–551. doi: 10.1007/s10637-008-9206-4. [DOI] [PubMed] [Google Scholar]
- 28.Delie F, Petignat P, Cohen M. GRP78-targeted nanotherapy against castrate-resistant prostate cancer cells expressing membrane GRP78. Target Oncol. 2012 doi: 10.1007/s11523-012-0234-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Roongta UV, Pabalan JG, Wang X, Ryseck RP, Fargnoli J, Henley BJ, Yang WP, Zhu J, Madireddi MT, Lawrence RM, Wong TW, Rupnow BA. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol Cancer Res. 2011;9:1551–1561. doi: 10.1158/1541-7786.MCR-11-0126. [DOI] [PubMed] [Google Scholar]
- 30.Schlaepfer IR, Hitz CA, Gijon MA, Bergman BC, Eckel RH, Jacobsen BM. Progestin modulates the lipid profile and sensitivity of breast cancer cells to docetaxel. Mol Cell Endocrinol. 2012;363:111–121. doi: 10.1016/j.mce.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Li N, Wang H, Jia X, Wang X, Luo J. Altered microRNA expression in cisplatin-resistant ovarian cancer cells and upregulation of miR-130a associated with MDR1/P-glycopro- tein-mediated drug resistance. Oncol Rep. 2012;28:592–600. doi: 10.3892/or.2012.1823. [DOI] [PubMed] [Google Scholar]
- 33.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Yin J, Yu J, Xiang Q, Liu Y, Tang S, Liao D, Zhu B, Zu X, Tang H, Lei X. miRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol Rep. 2012;27:250–257. doi: 10.3892/or.2011.1472. [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Liu M, Li X, Tang H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013;587:488–495. doi: 10.1016/j.febslet.2013.01.016. [DOI] [PubMed] [Google Scholar]


