Abstract
Douglas Theobald recently developed an interesting test putatively capable of quantifying the evidence for a Universal Common Ancestry uniting the three domains of life (Eukarya, Archaea and Bacteria) against hypotheses of Independent Origins for some of these domains.
We review here his model, in particular in relation to the treatment of Horizontal Gene Transfer (HGT) and to the quality of sequence alignment.
Keywords: tree of life, model selection, common ancestry
Introduction
Recently Douglas Theobald developed an interesting test putatively capable of quantifying the evidence for a Universal Common Ancestry (UCA) uniting the three domains of life (Eukarya, Archaea and Bacteria) against hypotheses of Independent Origins (IO) for some of these domains.1 He imagined the UCA hypothesis as modeled by a single phylogeny connecting all three domains, while each competing IO hypothesis being represented by a partitioning of domains into independent phylogenetic trees. Thus for instance if we want to describe the hypothesis that Eukarya share a common ancestor with Bacteria but neither shares an ancestor with Archaea, we can think of one single phylogeny connecting all Eukarya and Bacteria, and another non-overlapping phylogeny describing the evolution only of the Archaea (Figure 1). This model is represented by A+BE to highlight the fact that Bacteria and Eukarya are together while Archaea is alone. Here, each extant species will coalesce into the past until the root, called the Most Recent Common Ancestor (MRCA) and which represents an individual from a population of interbreeding individuals. However, we will see that the hypotheses generalize to the ancestral populations, not just the individuals. By population here we mean groups of individuals with shared genetic material. The question being posed is actually if the ancestral/primeval populations that gave rise to the diversity of life we see today did or did not exchange genetic material between themselves -- if there were barriers to the exchange then the apparent homology we observe is in fact product of ancestral convergence.
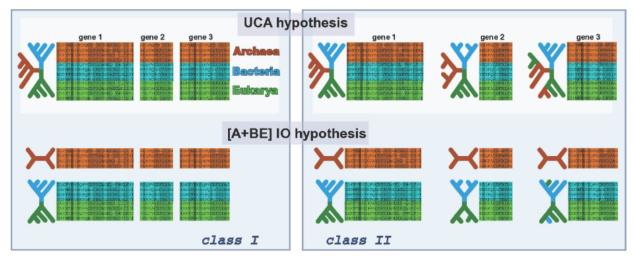
Figure 1.
Representation of the Universal Common Ancestry (UCA) and Independent Origins (IO) models, and how to compare them based on alignments. The left panel represents the class I models where all genes are concatenated into one large alignment. On the right we have the class II models where each gene is free to evolve under a distinct phylogenetic history. At the top we have the UCA hypothesis, that claims that all sequences from each gene can be traced back to a common ancestor, while at the bottom we have one example of the IO hypothesis where Bacteria and Eukarya share a common ancestor, which is not shared with Archaea. Other possible IO scenarios are not shown.
DNA or protein sequences can be used to gather data sets that are naturally amenable to test these ideas. With these type of data we can build biologically reasonable statistical models based on the phylogenetic likelihood, the probability of the data (the sequence alignment) given the hypothesis (the tree and the model of substitution). Importantly, all models used in the study are oblivious to the root position, i.e., the likelihood of the competing hypotheses are identical for any rooting and in practice we can assume unrooted trees.2 Theobald devised in this way a model selection approach based on the Likelihood Ratio Test (LRT), Akaike Information Criterion (AIC) and Bayes Factors (BF)3,4 to compare the UCA and IO hypotheses. He assumed that the ancestry of organisms can be properly represented by a set of protein-coding genes, and without modeling convergence explicitly. After analyzing 23 alignments of universally conserved proteins spanning the three domains of life (4 species for each domain), he concluded that the UCA hypothesis was strongly favored over any IO hypothesis, either after concatenating all alignments into one supermatrix or studying each alignment separately.1 Since we don’t need to know the location of the phylogenetic roots (representing the MRCAs), the test assumes that each gene has an MRCA potentially distinct from other genes - representing different individuals from the population.
Indeed, this test is not a validation of the evolutionary theory but a comparison between evolutionary models well-defined within the theory - the evidence for common ancestry in general includes astounding congruence between phylogenetic, morphological, paleontological and phylogeographic studies,5 the existence of a nearly universal genetic code6 and the ubiquitous presence of many orthologous genes.7 His test also cannot solve how many times life originated on Earth, since it already assumes one or several basal populations and even then it only looks at the successful ones (the ones still represented today). This way it is possible that several populations of independent origins were present in the past, and nonetheless all present life coalesces to a single one of these populations - in which case all life shares a UCA since all information from IO was lost. His test is restricted to gene alignments, and does not address the evolution of the genetic code, genomic content or morphological characters, for instance. In the following sections we will describe the phylogenetic theory behind his test together with his implementation, some published criticism and some further caveats.
Phylogenetic models
By assuming that a character - e.g. a nucleotide, a codon or an amino acid – evolves according to a continuous-time Markov chain along a phylogenetic tree, we can promptly calculate the probability of a set of homologous characters given the tree with branch lengths (in any arbitrary unit) and other parameters of the substitution process (the Markov chain).8 Many phylogenetic reconstruction methods will thus assume that each column of an alignment (a site) represents a distinct homologous character while each row represents a single taxon, and together with other assumptions will try to estimate the phylogenetic tree that maximizes the likelihood or the posterior distribution of trees that are compatible with the alignment and prior distributions.9 These assumptions might for instance impose equal, proportional or independent substitution processes for sites and/or branches, and usually are a simplification of the underlying unknown processes.10 We must remember that even though each site evolves independently from each other, all share the same tree topology11 and parameters like the alpha shape of the Gamma distribution for among-site rate variation12 or the stationary state frequencies. There are however exceptions (like mixture and recombination models), that we won’t discuss here.
Programs implementing these methods usually allow the user to fix the parameters at a single value or estimate them otherwise, as long as the model is fully defined. A fully defined model, as is relevant for our discussion,13,14 is one where we decide: i) if the instantaneous substitution matrix is fixed or variable; ii) if it’s fixed, which one we should use (for protein sequence there are several alternatives estimated from large data sets); iii) if the equilibrium frequencies should be estimated by Maximum Likelihood (ML) or observed proportions, or fixed at their model-derived values; iv) if all sites evolve at same rate or at distinct rates described by a discretized Gamma distribution (whose parameter should be estimated); and v) if we assume that a proportion of sites is invariable (and then estimate this proportion).
Can’t we have other, more parameter-rich models? We can indeed, like for instance a general time-reversible substitution matrix, where all instantaneous transition probabilities are estimated from the data. Or assume that each site has its own substitution rate, or substitution matrix. The problem is that as we increase the number of parameters to be estimated from the data we increase the uncertainty about the estimates in a way that might not be justified by the increase in the likelihood. And some models might lead to inconsistent estimates - like the example of one rate per site - therefore needing further constraints. We usually assume that the existing models are a good compromise, and if we want to decide objectively among the available options we must subject our data to a model selection approach.3,4
Model selection approaches aim at finding the best model that explains the data, where best is some optimum between bias and variance, or between parsimony and realism.3 The simplest case is when the models are nested (one is a particular case of the other), in which case we can approximate the difference between the log likelihoods by a chi-squared distribution - the standard LRT method. But there are many other methods like the AIC, the Bayesian nformation criterion or the BF, that don’t need the models to be nested. The AIC value of a model is simply twice the ML value under this model, substracted by twice the number of variables. The BIC is similar but takes into account also sample size, something never defined in molecular phylogenetics but usually assumed to be the number of sites in the alignment. If we do Bayesian analyses under each competing model, the BF can be calculated as the ratio between the marginal likelihoods for each model. These marginal likelihoods are usually calculated by the harmonic mean over all MCMC samples, despite more stable algorithms exist.15
We note that since model selection procedures try to find the model that best explains a set of observations, it is essential that the same data set be used when comparing different hypotheses. Indeed, when comparing the likelihood, L ~ P(D|Hi), of the different hypotheses (H1, H2, … corresponding to hypothesis 1, hypothesis 2, …), the data D should be fixed.16 Importantly, here the data observations are the columns of the alignment, as the alignment is given.
Theobald’s implementation
Having succeeded in writing the UCA and IO hypotheses in phylogenetic terms, Theobald could then extend existing model selection methodologies to compare one phylogeny (UCA) against more than one (IO).1 Under the UCA hypothesis it suffices to find the model and its corresponding parameters that best fit the alignment, while for each IO hypothesis he splitted the alignment into taxa belonging to each independent group and then found the best model for each group independently. This way it could be found that e.g. under the hypothesis AB+E (that is, Archaea and Bacteria having ancestry independent from Eukarya) the model that best explains AB is not the same as the optimal one for Eukarya. Under IO, one can simply multiply the likelihoods of each ML model since they describe independent events.
The difference in the number of parameters among hypotheses due solely to the number of trees - that is, neglecting other model parameters like presence/absence of rate heterogeneity, which nonetheless must be taken into account when comparing UCA and IO models - is 3 per IO assumption, since every time we split a tree in 2 we lose 3 branches with their corresponding lengths. It can be shown that both hypotheses can be accommodated by the same general model, where each IO hypothesis is equivalent to an arbitrarily long branch (it is a consequence of the property that ergodic chains converge to their equilibrium distributions). In frequentist terms, the IO would be the null hypothesis - of a branch length fixed at infinity - nested in the alternative, more complex UCA hypothesis. This general model could, in principle, incorporate a variable substitution process along the tree such that each branch has not only a distinct length but also a potentially different substitution matrix, proportion of invariant sites, equilibrium frequencies and rate heterogeneity process. This way we could ideally isolate the effect of one branch length while keeping all other parameters at the same values under both hypotheses, leading to the previously described difference of 3 in the number of parameters. But as we mentioned above it is hard to work with such overparameterized models, and most programs simply assume that the substitution matrix is constant along the phylogeny. Therefore in practice the substitution model can vary between independent trees while it is assumed constant under UCA, because of a limitation of available software to consider only homogeneous models. That is why the model representing the IO hypothesis has more parameters that the model representing UCA even though IO is a particular case of UCA in general: the software can infer the best, distinct models for each domain under IO, but cannot use all these models (i.e., heterogeneous models changing in different parts of the tree) under the single phylogeny of the UCA.
This may be a problem whenever the test favors the IO hypothesis, since we cannot know if the improvement of IO over the UCA model stands from the independent trees assumption (with fewer branches) or from better substitution models for each tree. The former indeed provides evidence for IO, but the later suggests that the true evolutionary process cannot be assumed to be homogeneous along the tree. As we will see in the analyses presented by Theobald the IO hypotheses were always rejected, therefore he didn’t need to consider this issue. A provisional solution would be to constrain all sequences under each IO hypothesis to follow the same evolutionary model - estimated e.g. from the UCA model. A better solution would be to employ a program that allows heterogeneous models along the phylogeny, ideally under a Bayesian framework so that the marginal distribution of the branch lengths of interest can be estimated.
Theobald used the crude LRT based on the ML values from Prottest,17 the AIC as given by the same program, and also the Bayes factor between the marginal likelihoods given by MrBayes13 - which as before can be simply multiplied under the IO hypothesis. Under MrBayes he chose a broad, fixed parametrization and for the substitution matrices he used a mixture model that samples from all the matrices. If we are then interested in inferring the best ones we can simply look at their posterior distribution, but here to calculate the marginal likelihood they are integrated out.
His data set comprised 23 proteins and his first analysis (called class I models) assumed that all proteins shared the same phylogenetic tree and same parameters (under a given hypothesis). In other words he concatenated all proteins into a single alignment with 6591 sites, and then compared the UCA model with models where one or all domains of life were separated. Such scenarios are represented at the left panel of Figure 1. Under all model selection criteria the UCA hypothesis was very strongly favored, and in general there was strong agreement between them.
To take into account the possibility that some gene suffered horizontal gene transfer (HGT), that is, genetic material was laterally exchanged between unrelated organisms in the past,18 Theobald devised the class II models. Under these models each one of the 23 genes can have a phylogenetic history independent from the others, with its own model, tree and parameters. This stands from the phylogenetic effect of HGT: possible existence of incongruent phylogenies between genes, with some HGTs being undetectable.
As before, the test is between models where all taxa are connected against models where the taxa are partitioned into independent clusters (Figure 1). Because each gene will lead to an independent unrooted phylogeny, class II models can effectively accommodate distinct common ancestors for each gene - the exact position of the root is unknown for each tree - that nonetheless belong to a single population. Under class II models, again, the UCA hypothesis was strongly favored against any IO scenario, using any model selection criterion. Furthermore, the UCA hypothesis using the class II model was preferred over its class I equivalent. This means that indeed the evolutionary history of the analyzed data set could be a reticulate one, and one should not force all genes to follow the same phylogenetic tree.
There have been so far two relevant criticisms to Theobald’s work, and both are somehow related to the fact that the homology of each site is assumed for phylogenetic inference methods and therefore might not be easily refuted. The first criticism was based on the need to explicitly model convergent and parallel evolution, such that even taking those into account to explain sequence similarity, UCA would still be preferred over IO.19 To emphasize their point the authors showed that UCA was preferred even for a data set of unaligned DNA sequences known to not be homologous. Theobald replied that UCA was properly rejected once codon or amino acid models were used, and that his data set with over 55% average sequence identity could not be explained by convergent evolution alone.20
The second criticism came from a simulation study where each alignment column was simulated by sampling amino acids from a discrete distribution such that columns did not follow any phylogenetic tree structure, and each column was generated by a distinct distribution.21 Still, the UCA hypothesis was favored for all 100 such simulations due to the similarity between sequences alone, according to the authors. Theobald contested that this model is equivalent to a star tree, which indeed represents common ancestry, and showed that a modification of his test to include such a model would in fact distinguish his data set from these simulations.22 And he also showed that his test would favor IO for an engineered data set composed of a phylogenetic mixture. Unfortunately he used the observed significant pairwise similarity between the sequences to conclude that the sequences are highly similar, while the average identity was only 0.33 for every column of the resulting multiple sequence alignment.
Discussion
While indeed we believe (in a Bayesian sense) that the UCA hypothesis is correct, there are, however, two caveats with Theobald’s analysis that are worth mentioning. The first relates to the theoretical model and its treatment of HGT within Class II hypotheses. The Class II models, despite being an elegant solution to within-domain lateral transfers, do not consider transfers between domains of independent origins. This general scenario would be equivalent to an IO model where any division of the taxa into independent phylogenies is possible (while the UCA model does not need correction). A formal test that fully takes into account HGT should allow for one gene favoring AE+B, another favoring AB+E, and even another favoring a single origin ABE - if this particular gene is the product of an ancestral sweeping HGT. This reminds us that a really informative test of the UCA hypothesis that accounts for HGT should be actually one about the existence or not of at least one gene for which it can be rejected in favor of an IO.
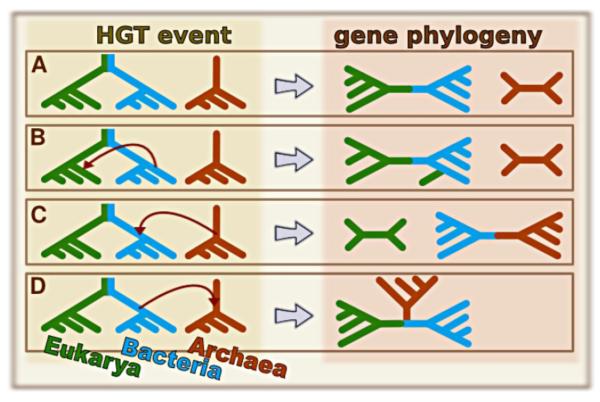
This is represented in Figure 2, where we have several possible HGT scenarios under the EB+A hypothesis, but only scenarios represented by panels A and B maintain the topologies EB and A - and therefore would be detected by Theobald’s test under EB+A hypothesis. The HGT scenario from panel C generates topologies that resemble an AB+E model, since the ancestral bacterial gene was replaced by one of archaean origin. In panel D the archaean gene from independent origin was replaced by the bacterial version, such that the signal from independent ancestry is lost. For all effects, genes like these where their ancestral version was completely replaced by a foreign one will support a UCA nonetheless. It then becomes essential to consider as many genes as possible, since even if the organisms conform to IO, some genes may have lost this information and support UCA.
Figure 2.
Effect of horizontal gene transfers (HGTs) on the observable gene phylogenies. Here we show 4 examples where Eukarya and Bacteria do not share a common ancestor with Archaea – each domain is represented by a color but individual species are not identified. In the left column we have the transfers represented by red arrows defining donor and recipient, while in the right we have the resulting phylogenetic trees. The first scenario (A) is one with no apparent HGT (no change in topology), while B) represents a gene transfer from a Bacteria to an Eukarya, such that for this gene the eukaryotic species will have an homolog resembling a bacterial one (such homologs are called xenologs actually). C) shows a transfer from an archaean ancestor to a bacterial one, and D) describes a transfer from Bacteria to Archaea. Notice that under scenario D the archaean version of the gene is lost and all species share the bacterial one. For scenarios B, C and D we assume that the recipients of the gene transfer lose their original copies, which are replaced by the foreign ones.
Our second caveat is about the practical implementation of the test, that unintentionally neglects the contribution of alignment properties.23 Theobald later clarified that his test can only be applied without corrections for highly similar sequences, mentioning that the alignment optimization can result in bias toward UCA for data sets with lower similarity or alignment uncertainty.22 To have an idea of how the alignment properties are related to the evolutionary scenarios, we simulated 8-sequence data sets under the UCA hypothesis and under an IO scenario with two independent quartets. We evolved these sequences under trees and parameters that resembled the original BE data set, but with the total sum of branch lengths randomly assigned between 0.01 and 10. We then aligned the resulting sequences using ProbCons,24 a program that can also provide a measure of alignment quality - the expected percentage of correct pairwise matches per column. Furthermore we calculated the average pairwise identity before and after optimizing the alignment. The alignment optimization step (the process of aligning the sequences) is a standard procedure in phylogenetics since the primary data are sequences likely to be homologous as a whole but still with homology status unknown for each site.25 This optimization aims at finding the scenario of indels (insertions and deletions) such that each column could be optimally assigned as an homologous character, generally in practice trying to maximize the columns’ similarity while not increasing too much the length of the sequences.26
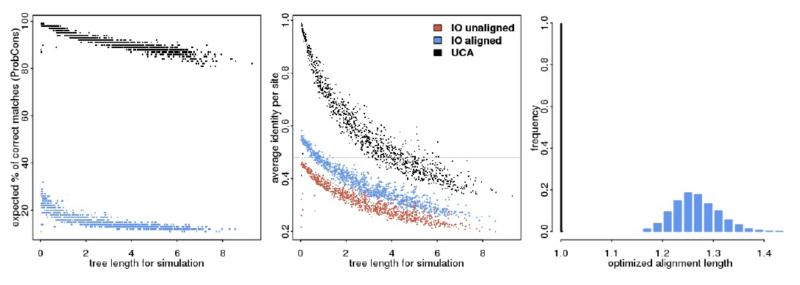
These results are summarized in Figure 3, where we can see that alignments of sequences we simulated under UCA have very different properties from those simulated under IO - for data simulated under a UCA the optimized alignment was essentially the same as the unaligned. The quality values given by ProbCons, for instance, are a good predictor of common ancestry since even large trees under UCA present an average expected accuracy much higher than very short trees under an IO model. We can also see how the alignment optimization improved the average pairwise identity (by definition, the average identity per site) for the sequences we simulated under IO, values which are nonetheless much lower than those under UCA with similar divergence levels.
Figure 3.
Alignment properties for data sets simulated under Universal Common Ancestry (UCA) and Independent Origins (IO). The left panel shows the relation between quality values from ProbCons (averaged over all sites) and tree length used in simulation. The middle panel shows the average pairwise identity under UCA and IO as a function of tree length. For IO simulations we have the identity values before and after optimizing the alignment, and the gray horizontal line at 0.47 represents the observed value for the BE data set. The panel at the right show the histogram of optimal alignment lengths divided by the unaligned values, for simulations under UCA and IO. Here the distribution is pooled over several tree lengths. For all panels we have the UCA simulations in black (after alignment optimization, which is essentially the same as before optimization), the unaligned IO data sets in red and after alignment optimization in blue.
These simulations were inspired by the Bacteria and Eukarya data set, and therefore we wanted to compare the real data set with our simulations. Douglas Theobald kindly provided to us the complete data set, and in Figure 4 we show some column-wise statistics for the observed BE data set compared to simulations under UCA or IO hypotheses. This data set has average pairwise identity of 0.47 (shown as well in Figure 3 – the gray horizontal line – for comparison), and the ML tree under the best model had a total tree length of 3.3, which was used in simulations as well. To be more conservative, under UCA all simulated trees had total length larger than 6, while under the IO simulation scenario the sum of branch lengths from both trees did not exceed 3. It is worth noticing that our simulation scenarios under the IO hypothesis are similar to the doppelgänger sequences described in Pollock DD et al.:27 these sequences were simulated under a tree and parameters of interest and then analyzed together with the original sequences from which the tree was inferred. Their objective was to increase the signal for rate heterogeneity without providing information about internal branches. They also recognized that these resulting data sets are unlikely to provide alignable sequences in nature.27
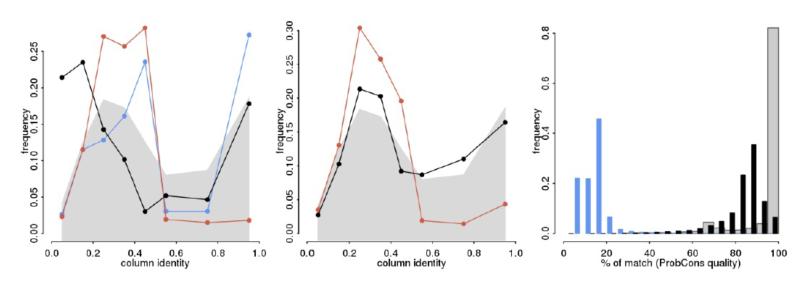
Figure 4.
Distribution of pairwise identities and percentage of correct matches (as given by ProbCons) per alignment column for simulated and real data sets. On the left we have the distribution of column-wise identities over all simulations - that is, under short Independent Origins (IO) trees and under large Universal Common Ancestry (UCA) scenarios. The gray background indicates the equivalent frequencies for Theobald’s BE data set. The middle panel shows the same information as the left one, but here all trees were simulated under branch lengths compatible to those for Theobald’s BE data set. The right panel is the histogram of quality values per column as given by ProbCons for all simulations, together with the values for the observed BE data set. The colors are the same as Figure 3 (blue for aligned IO and black for UCA), with gray columns indicating Theobald’s BE data set.
Notice that we did not conduct the model selection test proposed by Theobald and nonetheless we already have strong evidence that the BE data set resembles UCA much more than IO: at overall and column-wise sequence identities, distribution of correct pairwise matches per column, alignment size after optimization, and probably at many other alignment quality estimates. As a side note, this kind of procedure is formally applied on approximate Bayesian computation (ABC) analyses, where we cannot calculate the likelihood of arbitrary models and instead look at the parameter sets that best approximate the observed data.28
The simulations presented here constitute just a few idealized situations, and we recognize that we didn’t explore here other alignment statistics that might serve as relevant indicators (e.g. congruence of alignment optimization methods and other measures of robustness of the alignment, as well as homology detection algorithms). But the few statistics we analyzed here can already distinguish UCA from IO data sets, even before doing the formal test proposed by Theobald. It is not only a mistake but a misdirection to neglect the correlation between these alignment measures and the ancestrality of the sequences. In this sense if we follow Theobald’s approach of restricting the analysis to robust alignments, the test might be giving us the right answer (UCA hypothesis) for the wrong reason: we would have selected beforehand sequences that would maximize our chances of favoring the UCA hypothesis.
We must also mention an even larger problem with such data selection: sequences simulated under IO are unlikely to be detected by BLAST as potential homologs, given their low similarity scores. In our simulations above, all sequences simulated under the same phylogeny displayed a significant similarity according to the e-value from BLAST, while no significant hits were found between sequences simulated under distinct trees (results not shown). This suggests that a data set produced under the IO hypothesis not only is unlikely to fit the requirements needed by Theobald’s test, but it wouldn’t even be considered to start with. And even sequences with significant similarity based on pairwise comparisons routinely produce uncertain alignments, with overall low sequence identities, as shown for instance by the BaliBASE data sets.29 We must not forget that no matter how imperfect and biased a database homology search might be, it is still an inference that we cannot ignore. Far from replacing it, the test developed by Theobald actually relies on a set of candidate homologs. The puzzle then is to develop a model that can systematically produce sequences under the IO hypothesis that also conforms to the alignment properties needed for the test to be valid without corrections, according to Theobald.22
Conclusions
Although we agree that high similarity does not imply homology, they are certainly not independent. A formal test capable of distinguishing common ancestry from independent origins should not be valid only on data sets where we already have strong evidence favoring one of the outcomes - the data selection must be considered as part of the test.30 Theoretically such a test should consider arbitrary sequences for which we do not have prior indication of homology - that is, it should not rely on blast-like database searches and should rather supplant it.
And since with the presence of HGT we must consider all sequences potentially present in the ancestral pool, we cannot conclude for the UCA hypothesis based only on the most particularly similar sequences. The signal for IO will most likely be preserved only in the more divergent genes, even if we find most other genes supporting UCA - in a manner similar to an HGT sweep.31 In a nutshell, a test that is not biased towards UCA should be capable of handling sequences with low similarity, from the database search to the alignment optimization. Sequences with low similarity may not only contain the (lack of) signal characteristic of IO, but are also informative about deep branches and will be essential in estimating realistic evolutionary rates and/or times.32 This test should also contemplate the inclusion of paralogs: what we classify nowadays as different genes due to limitations of homology detection algorithms might be in fact product of an ancient duplication and therefore support a UCA of these gene families.
Notice that this criticism refers mostly to the implementation of the test and not to the models, that we do believe are appropriate phylogenetic representations of UCA and IO hypotheses, as developed in Sober E et al.33 Here we always assume that the UCA hypothesis can be resolved by looking at the set of gene phylogenies, which may be insufficient to describe the organismal evolution. There are further complications when we consider the limitations of the tree of genes when compared to the tree of cells,34 to the evolution of the genomes taking into account noncoding regions, or to the evolution of the genetic code. For example, the history of the cells may point to a common ancestry while some gene may have appeared more than once. Theobald did not explore all possible scenarios, neither we tried to do it here. But Theobald was clear, from the beginning, that his test was not the last word on the subject, but a first step in trying to solve it.
Acknowledgements
We would like to thank Douglas Theobald for fruitful discussions and for sharing with us his data sets. Two anonymous reviewers and David Liberles provided insightful comments that improved this manuscript. The programs with our analyses can be found at http://darwin.uvigo.es/common_origin/.
References
- 1.Theobald DL. A formal test of the theory of universal common ancestry. Nature. 2010;465:219–22. doi: 10.1038/nature09014. [DOI] [PubMed] [Google Scholar]
- 2.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 3.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of AIC and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan J, Joyce P. Model Selection in Phylogenetics. Annl Rev Ecol Evol and Syst. 2005;36:445–66. [Google Scholar]
- 5.Theobald DL. 29+ Evidences for Macroevolution: the scientific case for common descent. The Talk. Origins Archive. :c1999–2011. Available from: http://www.talkorigins.org/faqs/comdesc.
- 6.Koonin EV. Novozhilov AS. Origin and evolution of the genetic code: the universal enigma. IUBMB life. 2009;61:99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liò P, Goldman N. Models of molecular evolution and phylogeny. Genome Res. 1998;8:1233–44. doi: 10.1101/gr.8.12.1233. [DOI] [PubMed] [Google Scholar]
- 9.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–4. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 10.Huelsenbeck JP, Alfaro ME, Suchard MA. Biologically inspired phylogenetic models strongly outperform the no common mechanism model. Syst Biol. 2011;60:225–32. doi: 10.1093/sysbio/syq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Statistical inference of phylogenies. J R Stat Soc A. 1983;146:246–72. [Google Scholar]
- 12.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–14. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 13.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–74. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 14.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 15.Xie W, Lewis PO, Fan Y, et al. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol. 2011;60:150–60. doi: 10.1093/sysbio/syq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suchard MA, Redelings BD. BAli-Phy: simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics. 2006;22:2047–8. doi: 10.1093/bioinformatics/btl175. [DOI] [PubMed] [Google Scholar]
- 17.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagan T. Phylogenomic networks. Trends Microbiol. 2011;19:483–91. doi: 10.1016/j.tim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Yonezawa T, Hasegawa M. Was the universal common ancestry proved? Nature. 2010;468:E9. doi: 10.1038/nature09482. [DOI] [PubMed] [Google Scholar]
- 20.Theobald DL. Theobald reply. Nature. 2010;468:E10. [Google Scholar]
- 21.Koonin EV, Wolf YI. The common ancestry of life. Biol Direct. 2010;5:64. doi: 10.1186/1745-6150-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theobald DL. On universal common ancestry, sequence similarity, and phylogenetic structure: The sins of P-values and the virtues of Bayesian evidence. Biol Direct. 2011;6:60. doi: 10.1186/1745-6150-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anisimova M, Cannarozzi G, Liberles DA. Finding the balance between the mathematical and biological optima in multiple sequence alignment. Trends Evol Biol. 2010;2:e7. [Google Scholar]
- 24.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–40. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dessimoz C, Gil M. Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biol. 2010;11:R37. doi: 10.1186/gb-2010-11-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips A, Janies D, Wheeler W. Multiple sequence alignment in phylogenetic analysis. Mol Phylog Evol. 2000;16:317–30. doi: 10.1006/mpev.2000.0785. [DOI] [PubMed] [Google Scholar]
- 27.Pollock DD, Bruno WJ. Assessing an unknown evolutionary process: effect of increasing site-specific knowledge through taxon addition. Mol Biol Evol. 2000;17:1854–8. doi: 10.1093/oxfordjournals.molbev.a026286. [DOI] [PubMed] [Google Scholar]
- 28.Didelot X, Everitt RG, Johansen AM, Lawson DJ. Likelihood-free estimation of model evidence. Bayesian Anal. 2011;6:49–76. [Google Scholar]
- 29.Landan G, Graur D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol. 2007;24:1380–3. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- 30.Vul E, Pashler H. Voodoo and circularity errors. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.01.027. [Epub 2012 Jan 8] [DOI] [PubMed] [Google Scholar]
- 31.Koonin EV. On the origin of cells and viruses: primordial virus world scenario. Ann New York Acad Sci. 2009;1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Filipski AJ, Battistuzzi FU, et al. Statistics and truth in phylogenomics. Mol Biol Evol. 2011;29:457–72. doi: 10.1093/molbev/msr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sober E, Steel M. Testing the hypothesis of common ancestry. J Theor Biol. 2002;218:395–408. [PubMed] [Google Scholar]
- 34.O’Malley MA, Koonin EV. How stands the Tree of Life a century and a half after The Origin? Biol Direct. 2011;6:32. doi: 10.1186/1745-6150-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]