Abstract
Adeno-associated virus serotype 2 (AAV2) uses heparan sulfate proteoglycan as a cell surface-attachment receptor. In this study the structures of AAV2 alone and complexed with heparin were determined to ~18 Å resolution using cryo-electron microscopy and three-dimensional image reconstruction. A difference map showed positive density, modeled as heparin, close to the icosahedral twofold axes and between the protrusions that surround the threefold axes of the capsid. Regions of the model near the threefold place the receptor in close proximity to basic residues previously identified as part of the heparin binding site. The region of the model near the twofold axes identifies a second contact site, not previously characterized but which is also possibly configured by heparin binding. The difference map also revealed two significant conformational changes: I) at the tops of the threefold protrusions, which have become flattened in the complex, and II) at the fivefold axes where the top of the channel is widened possibly in response to movement of the HI loops in the capsid proteins. Ordered density in the interior of the capsid in the AAV2-heparin complex was interpreted as nucleic acid, consistent with the presence of non-viral DNA in the expressed capsids.
Keywords: Adeno-associated virus, receptor attachment, parvovirus, virus structure, electron cryo-microscopy, 3D image reconstruction, atomic modeling, heparin sulfate
INTRODUCTION
Adeno-associated virus (AAV) is a non-pathogenic member of the Parvoviridae family belonging to the Dependovirus genus (Muzyczka and Berns, 2001). A large number of AAV serotypes have been described and studied for their use in gene-delivery applications. They differ in receptor binding, host-cell tropism, and antigenic properties. Most of the gene therapy applications to date have made use of AAV serotype 2 (AAV2), which is currently the subject of clinical trials to correct hemophilia, cystic fibrosis, and other diseases (e.g. Flotte et al., 2004; Kay et al., 2000; Maguire et al., 2008). Detailed understanding of the molecular mechanisms by which the virus particle can recognize and enter host cells is necessary to develop efficacious, targeted vectors.
AAV capsid shells are constructed with T=1 icosahedral symmetry, are ~260 Å in diameter, and encapsidate a 4.7 kb ssDNA genome. The genome has two open reading frames (ORFs), rep and cap, that encode the replication and capsid proteins, respectively. The 60-subunit viral capsid consists of three viral proteins (VPs), with the 62 kDa VP3 protein comprising approximately 90% of the structure. The minor capsid proteins, VP1 (87 kDa) and VP2 (73 kDa), share a common C-terminal amino acid sequence with VP3 but have N-terminal extensions. The N-terminal sequence unique to VP1 has a phospholipase A2 (PLA2) activity that is essential for infectivity at a step between endocytosis and nuclear entry (Girod et al., 2002) as well as nuclear localization signals. The AAV capsid, in addition to protecting the ssDNA genome, mediates several steps in the viral lifecycle, including primary and secondary receptor recognition as well as antigenicity.
Cryo-electron microscopy and three-dimensional (3D) image reconstruction (“cryo-reconstruction”) and X-ray crystallography have been used to determine the structures of several AAV serotypes, including AAV1, AAV2, AAV4, AAV5, AAV7, and AAV8 (Govindasamy et al., 2003; Kronenberg et al., 2001; Miller et al. 2006; Nam et al., 2007; Padron et al., 2005; Quesada et al., 2007; Walters et al., 2004; Xie et al., 2002). The AAV capsids contain a highly homologous core subunit structure, consisting of an eight-stranded, antiparallel β-barrel motif similar to that observed for members of other parvovirus genera. The β-barrel strands are denoted as letters from B-I and the variable, inter-strand loops are named for the letter of the β-strands that flank the loops (reviewed in Chapman and Agbandje-McKenna, 2006; Kontou et al., 2005). Depressions are located on the capsid surface at the icosahedral twofold (“twofold depressions”) and around the fivefold (fivefold depressions) symmetry axes. Three prominent protrusions encircle the icosahedral threefold axes and are separated by “valleys”. A channel at each fivefold axis (channel) is implicated in packaging of the AAV2 genome and extrusion of the PLA2 activity of the VP1 unique region (Bleker et al., 2005; Sonntag et al., 2006).
Heparan sulfate proteoglycan, the first receptor identified for an AAV virus (Summerford and Samulski, 1998), functions during infection to attach AAV2 and AAV3 to the cell surface. Soluble heparin has been shown to compete with permissive cells for AAV2 binding and de-sulfating heparin reduces binding to AAV2 (Qiu et al., 2000). AAV2 residues R484, R487, K527, K532, R585, and R588 have been shown to affect heparin binding in mutagenesis studies (Kern et al., 2003; Opie et al., 2003; Wu et al., 2000). These residues form a basic patch between neighboring protrusions and are contributed by two, threefold-related VP monomers. R585 and R588 form the wall of the valleys, R484, R487, and K527 are on the floor, and K532 is also on the floor but closer to the twofold depression (Kern et al., 2003; Opie et al., 2003). AAV2 residues G512 and R729 lie near the basic patch and are also reported to have minor effects on heparin binding (Lochrie et al., 2006).
Efficient AAV2 infection also requires a co-receptor, such as human fibroblast growth factor receptor 1 (FGFR1), integrins αvβ5 and α5β1, or hepatocyte growth factor receptor (Asokan et al., 2006; Kashiwakura et al., 2005; Summerford et al., 1999; Qing et al., 1999). These co-receptors mediate endocytosis of the virus via clathrin-coated pits (Bartlett et al., 2000; Duan et al., 1999). The region of the capsid surface responsible for α5β1 binding was mapped to a moderate affinity asparagine-glycine-arginine (NGR) integrin binding motif at residues 511-NGR-513 on the AAV2 capsid by mutagenesis (Akokan et al., 2006). The binding site(s) for the other co-receptors are unknown.
Regions of the capsid surface that bind to host-cell receptors have been identified for three autonomous parvoviruses: human parvovirus B19 (B19), canine parvovirus (CPV), and minute virus of mice (MVM) (Chipman et al., 1996; Hafenstein et al., 2007; Lopez-Bueno et al., 2006). Cryo-reconstruction was used to show that globoside, the receptor for B19, binds in the depression at the threefold axis of the capsid (Chipman et al., 1996). The binding site for the CPV transferrin receptor was mapped by mutagenesis and cryo-reconstruction to the raised portion of the capsid that lies between the icosahedral two- and fivefold axes (Hueffer et al., 2003a; Hueffer et al., 2003b; Govindasamy et al., 2003; Hafenstein et al., 2007). For MVM, the binding site for its sialic acid receptor was mapped by X-ray crystallography to the twofold depression (Cotmore et al., 1987; Lopez-Bueno et al., 2006). These three examples illustrate diversity in the regions utilized by parvovirus capsids for host recognition.
Structural studies of a number of parvovirus virions have demonstrated the presence of icosahedrally-ordered, genomic ssDNA inside the capsid. The crystal structures of AAV4 and AAV8 showed the ordering of a single nucleotide (NT) under each threefold axis (Govindasamy et al., 2006; Nam et al., 2007). This NT is stabilized by two highly conserved prolines in a pocket formed by conserved residues. The crystal structures of CPV and the immunosuppressive strain of MVM (MVMi) showed the presence of at least 11 ordered ssDNA NTs inside the capsids (Agbandje-Mckenna et al., 1998; Kontou et al., 2005; Xie et al., 1996). The strongest DNA-protein associations inside CPV and MVMi involved residues that form grooves bounded by β-strand regions at the inner capsid surface near the two- and threefold axes. Some of the ordered NTs observed in MVM are located close to those seen in AAV4 and AAV8, suggesting the presence of a common binding pocket for DNA in parvoviruses. Though parvoviruses are currently believed to not require genomic DNA for capsid assembly (e.g. (Hernando et al., 2000)), presence of an ordered NT in wild type (wt) AAV4 and in the same conserved capsid region in AAV8, determined from “empty”, baculovirus-expressed capsids, suggests that NTs may at least play a role in AAV assembly.
In this study cryo-reconstruction methods were used to determine the 3D structures of AAV2 virus-like particles (VLPs) alone and complexed to heparin oligosaccharides (~6 kDa average molecular mass) to ~18 Å resolution. As predicted by mutagenesis, density interpretable as heparin occurs proximal to basic residues between the threefold protrusions in the AAV2-heparin complex. Additional density was also unexpectedly observed close to the twofold depressions, which was modeled as heparin oligosaccharides, suggesting that new binding sites are exposed upon initial heparin binding. Density differences were observed at the fivefold axes and at the top of the protrusions in the AAV2-heparin complex, suggesting that heparin binding induces conformational changes in the capsid at these regions. Density associated with the interior surface of the capsid in the AAV2-heparin complex was interpreted as icosahedrally-ordered DNA.
MATERIALS AND METHODS
Cells
Human embryonic kidney cells (HEK293) were grown in Dulbecco Modified Media, supplemented with 10% bovine calf serum and 100 U/ml penicillin and streptomycin. Cells were incubated in 5% CO2 at 37°C.
Virus-like particle (VLP) expression and purification
To express AAV2 VLPs containing VP1, VP2, and VP3, ten 15cm plates of low passage 293 cells, at ~80% confluence, were infected with an adenovirus expression vector containing the AAV2 cap ORF under a cytomegalovirus promoter. Infections were at an MOI of 5, carried out in serum free conditions for 1 hour. The infected cells were supplemented with 10% bovine calf serum and incubated for 48–72 hours until cytopathic effect was detected. The cells were harvested by centrifugation at 1000 × g for 10 minutes for purification.
The cell pellets were resuspended in 30 ml lysis buffer (150 mM NaCl, 50 mM Tris pH 8.4) and lysed by three freeze/thaw cycles. Benzonase (Sigma) was added to the lysate (50 U/ml) after the final thaw followed by clarification through centrifugation at 4000 × g for 20 minutes. The supernatant was purified using a discontinuous iodixanol (OptiPrep Nycomed) step gradients. Samples were centrifuged in a 70 Ti rotor at 350,333 × g for 1 hour at 18 °C as previously described (Zolotukhin et al., 2002). VLPs were removed from the 40%:25% interface of the gradient. The sample was further purified by loading onto an heparin agarose Type I column (Sigma), eluted with TD buffer (1 X PBS, 1 mM MgCl2, 2.5 mM KCl) containing 500mM NaCl, and concentrated using Amicon Ultra Centrifugal filters (100,000 MW cut off).
The purity of the VLPs was confirmed by silver-stained SDS-PAGE, western blot analysis with the AAV2 B1 antibody that recognizes a linear peptide at the C-terminus of the VPs, and transmission electron microscopy of particles negatively stained with 2% uranyl acetate. Protein concentration was determined by UV absorbance (Sommer et al., 2003).
Extraction of DNA from VLPs
Purified VLPs, at ~1mg, were treated with 20 μg/ml of RNAse A and DNAse (Sigma) at 37°C to degrade non-encapsidated nucleic acid. Nuclease-treated samples were purified on a heparin column as described above and then treated with Proteinase K (Sigma) to release any nucleic acid that might be packaged within the VLPs. Phenol-chloroform extraction was used to remove protein and the remaining sample was precipitated with ethanol. To determine whether the extracted nucleic acid was RNA or DNA, a portion of the precipitated sample was treated with 20 μg/ml of either RNAse-free DNAse I or DNAse-free RNAse A (Roche). Treated and untreated samples were visualized by agarose gel (0.5%) electrophoresis and ethidium bromide staining. UV absorbance was used to estimate the protein to nucleic acid ratio in the original sample.
Cryo-reconstructions of VLPs and AAV2-Heparin complexes
AAV2 VLP-heparin (AAV2-Hep) complexes were produced by mixing purified VLPs with heparin oligosaccharides (Sigma) with an average molecular mass of 6 kDa (~9 disaccharides per oligosaccharide) at a ratio of 120 heparin molecules per VLP. Small aliquots (~3.5 μl) of samples at ~0.5 mg/ml were vitrified on carbon-coated copper EM grids in liquid ethane as described (Baker et al., 1999). Grids with vitrified samples were loaded into a pre-cooled Gatan 626 cryotransfer holder, maintained at a constant temperature of −176°C, and were examined in a Philips CM200 FEG electron microscope. The diameter of the electron beam was adjusted to not exceed the limits of the Gatan 2k2 slow-scan CCD detector (0.4 μm on the specimen) to allow images to be recorded without pre-irradiating adjacent regions. A low-magnification (4,560X) survey image was captured on the CCD, and areas for high-magnification data collection were identified and selected. Images were recorded at 0.5–3.0 μm under-focus at 200kV, at a nominal magnification of 38,000X, and on Kodak SO-163 film under low-dose conditions (~19e-/Å2). Film micrographs were digitized on a Zeiss SCAI scanner at 7 μm steps and the pixels were then twofold bin-averaged to yield pixels equivalent to 3.68 Å at the specimen.
Individual images of the VLPs and AAV2-Hep complexes were manually selected from the digitized micrographs and extracted within equivalent size boxes (1212 pixels) using the interactive graphics program, RobEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt). A total of 2335 particle images was boxed from ten VLP micrographs and 1523 particle images were boxed from eight AAV2-Hep micrographs as previously described (Bowman et al., 2002; Walters et al., 2004). RobEM was used to remove linear density gradients from the images and to normalize the means and variances in the image intensities. The center and orientation of each boxed particle were initially estimated by means of the model-based polar Fourier transform routine (Baker and Cheng, 1996), with the AAV5 structure (Walters et al., 2004) serving as the starting search model for the procedure. A low resolution 3D map was generated from a selected subset for each set of images using the programs EMSEL and EM3DR (Baker et al., 1999; Fuller et al., 1996). Those particle images that exhibited a correlation coefficient greater than 0.3 in comparison with the corresponding projected image of the current model were included in the reconstruction. The resultant reconstructions served as models for refinement of particle origins and orientations for the respective full sets of images using the program EMPFT (Baker et el., 1999). The quality of each reconstructed density map was assessed by visual inspection using tools like RobEM and by quantitative Fourier Shell Correlation (FSC) analysis (Baker et el., 1999). The final reconstructions contained 2045 (from ten micrographs) and 1466 (from eight micrographs) particle images, respectively, for the VLPs and AAV2-Hep complex. The estimated resolution of each of the final 3D density maps was computed by subdividing each set of particle images into two subsets to generate two different reconstructions, which were compared by FSC and phase residual analyses (Baker et el., 1999). The estimated resolution achieved with each reconstruction was defined by the lowest spatial frequency at which the FSC first dropped below 0.5 and the average phase difference exceeded 45°.
Reconstructed map comparisons, difference density map calculation, and pseudo-atomic model building
The known crystal structure of wt AAV2 (Xie et al., 2002) (PDB accession No. 1LP3) provided the structural framework for interpreting the reconstructed density maps of the VLP and the AAV2-Hep complex. The atomic coordinates of the VP3 monomer of AAV2 were used to generate a complete capsid shell through matrix multiplication using icosahedral symmetry operators for comparing the low resolution reconstructions of the VLP and AAV2-Hep complex. Structure factors were calculated using the Cα atom coordinates of the 60 VP3 monomers and used to calculate a density map computationally filtered to 18-Å resolution using the CCP4 package (CCP4, 1994). The Chimera program (Petterson et al., 2004) was used to superimpose and compare the VLP and AAV2-Hep maps to each other and to the AAV2 crystal structure model and map.
Difference maps computed by two separate procedures were used to locate density associated with bound heparin molecules and to identify heparin-induced conformational changes. In the first method, RobEM was used to subtract the VLP map from an appropriately scaled (density range and magnification) AAV2-Hep map as previously described (Baker et el., 1999). A second difference map was calculated with the CCP4 package (CCP4, 1994) using structure factors generated by inverse Fourier transformation of the two density maps (Baker et el., 1999). The two resultant, essentially identical, difference density maps were inspected using Chimera (Petterson et al., 2004) and O (Jones et al., 1991).
The difference map was superimposed onto the AAV2 Cα atom model map as well as VLP and AAV2-Hep reconstructed density maps for further analysis. A trimer and a pentamer of AAV2 VP3 generated from the crystal structure coordinates using icosahedral threefold and fivefold symmetry operators, respectively, were docked into the maps. This facilitated identification of capsid residues that are proximal to positive difference density interpreted as heparin and to enable model building at difference density map regions interpreted as being due to structural re-arrangements. Heparin molecules, extracted from the atomic coordinates of the structure of a heparin hexasaccharide complexed with fibroblast growth factor (FGF-heparin) (Faham et al., 1996) (PDB accession No. 1BFC) were modeled into positive density close to the twofold axes and at the valleys between the threefold protrusions by means of rigid body rotations and translations. One heparin disaccharide was built into density at one threefold valley and the symmetry-related disaccharides were generated. Five disaccharides were built into the positive density near the twofold axis and the twofold-related decasaccharide molecules were generated.
Positive and negative difference density surrounding the channel at the icosahedral fivefold was interpreted as movement of the HI loop (region between βH and βI, residues T651 to Y673) which lies on top of adjacent fivefold-related VP monomers and forms the majority of fivefold-related interactions on the parvovirus capsid. This movement was modeled by extracting the coordinates of residues 651 to 673 from a VP3 monomer of AAV2 (PDB accession No. 1LP3) docked into the reconstructed maps and manually rotating and translating this peptide into positive difference density followed by the use of the fit model to map function in the program Chimera (Petterson et al., 2004). The terminal peptide residues T651 and Y673 were constrained to remain near residues 650 and 674 of VP3, respectively, during model building. The coordinates of the modeled loop were then re-inserted into the VP3 coordinate file and used to generate fivefold-related monomers for further analysis of the re-arrangement. Additional negative and positive difference density inside and close to the channel, respectively, was interpreted as movement of the loop at the top of the βDE ribbon (between β-strands βD and βE which form the walls of the channel), but this re-arrangement was not modeled.
Positive difference density observed inside the capsid was interpreted as ordered nucleic acid and N-terminal VP residues. The modeling of nucleic acid density was guided by the crystal structures of MVMi (Agbandje-McKenna et al., 1998) (PDB accession No. 1MVM) and AAV4 (Govindasamy et al., 2006) (PDB accession No. 2G8G). The atomic coordinates of MVM VP2 along with the observed ordered NTs were superimposed onto the VP3 monomer of AAV2 in its fitted position in the VLP and AAV2-Hep reconstructed maps. The AAV2 coordinates were then examined with respect to their proximity to the MVMi NTs that superimpose within the positive difference density inside the capsid. A similar procedure was used to dock the NT observed in the crystal structure of AAV4 within this positive difference density. In addition to the density onto which the MVMi and AAV4 NTs could be docked, further positive density was observed in the capsid interior proximal to the last N-terminal residue, G217, built into the AAV2 crystal structure. This density was not modeled.
RESULTS AND DISCUSSION
Heparin binding induces several changes in the AAV2 capsid
Cryo-reconstruction methods were used to determine the structures of VLPs and an AAV2-Hep complex from 2045 and 1466 particle images, respectively (Fig. 1A and E). The VLP data set was refined to ~12-Å resolution, but the AAV2-Hep data set could not be refined beyond ~18 Å resolution. Hence, to make side-by-side comparisons and analysis valid, density maps presented here were computed only to 18-Å resolution. The difference in refined map resolution was likely attributable to heterogeneity of the heparin oligosaccharides in the AAV2-Hep complex sample, which introduced background noise and possibly introduced some asymmetry in the complex.
Figure 1.
Cryo-reconstructions of AAV2 VLPs and AAV2-Hep complex. (A–D) Shaded-surface representations of VLP reconstruction showing an entire particle viewed along an icosahedral twofold axis (A) and close-up views along two- (B), three- (C), and five-fold (D) axes. (E–H) Same as (A–D) for AAV2-Hep complex. The top of one βDE ribbon is marked with a black dot in D and H to visualize the ~36° difference in the position of this loop in the two maps. Broken circles (B, C, F, and G) highlight regions of obvious difference in the two reconstructions. These images were generated in RobEM (Baker et al., 1999).
The VLP and AAV2-Hep reconstructed density maps both exhibited surface features typical for AAV2, including the twofold depressions, threefold protrusions, and fivefold depressions and the fivefold channel (Fig. 1B-D, F-H). The VLP map was very similar to an 18-Å resolution density map generated from the Cα-atomic coordinates of the crystal structure of wt AAV2 (Xie et al., 2002) with the exception that the channel appears more open in the crystal structure map (Fig. 2A). This difference was previously recognized (Kronenberg et al., 2005) and is likely due to the reported flexibility of the loop at the top of the βDE ribbons (between the βD and βE strands of the core β-barrel) that form this channel in parvoviruses (Kontou et al., 2005, Govindasamy et al., 2006).
Figure 2.
AAV2 crystal structure with superimposed AAV2-Hep minus AAV2 VLP positive difference map and heparin models. A) Shaded-surface electron density map of AAV2 (PDB accession No. 1LP3, Xie et al., 2002) (grey) with the positive density (red mesh, contoured at 1σ) close to the two-, three-, and fivefold axes, 2f pos, 3f pos and 5f pos1, respectively, superimposed. (B) Close-up of shaded-surface representation of VLP reconstruction (grey) and AAV2-Hep (pink, density not in present VLPs), with the pseudo-atomic coordinates of heparin oligosaccharides (solid red) modeled into positive density (red mesh). hep2f = decasaccahride (twofold symmetry related decasaccahride is also shown); hep3f = single disaccharide (threefold symmetry related models are also shown). (C) Ribbon diagram of an AAV2 VP3 trimer and modeled heparin molecules. Surface heparin binding residues, R484, K527, K532, R585, and R588, are shown in blue (Arginines) or light blue (lysines) with terminal amino groups depicted as black balls. Residues from a reference monomer are denoted with A after the residue number; B indicates a threefold related residue. Heparin models are as labeled in (B). (D) Close up of basic (blue), acidic (orange) residues flanking the heparin binding residues. Basic residues are colored as in (C) and carboxyl groups are white for acidic residues. The broken lines indicate potential ionic interactions. The location of the icosahedral symmetry axes are labeled with numbers in (A–C). (A) is viewed along a twofold axis, and B, C, and D along a threefold axis. The figure was generated using Chimera (Pettersen et al., 2004).
The VLP and AAV2-Hep cryo-reconstructions show many distinct features (Fig. 1A–H). In AAV2-Hep, additional densities surround the twofold depressions (compare Fig. 1B and F) and in the valleys that separate the protrusions (compare Fig. 1C and G). Differences occur on the floor of the fivefold depressions (compare Fig. 1D and H) and the protrusions are more pointed in the VLP compared to the flattened peaks in the complex (compare Fig. 1C and G). Finally, more density occurs inside the channel in the VLP compared to the AAV2-Hep structure (compare Fig. 1D and H). Comparison of equatorial cross-sections from the two cryo-reconstructions highlights the above observations and a general structural re-arrangement of the capsid shell (Fig. 3A and B).
Figure 3.
Capsid conformational transitions at the fivefold axis. Equatorial cross-sections of the (A) VLP and (B) AAV2-Hep reconstructions. (C) Difference map (AAV2-Hep minus AAV2 VLP), with positive (red) and negative (cyan) differences superimposed onto a shaded-surface density map (grey mesh) of the AAV2 crystal structure close to the fivefold axis. Positive and negative density differences are labeled (e.g. 2f pos = positive density close twofold axis). (D) Close-up of fivefold channel and HI loop difference densities. HI loop is shown as observed in the crystal structure (cyan coil) close to the negative (cyan) density and modeled (magenta coil) into positive (red) density. βDE ribbons (cyan coil, as in the crystal structure) are indicated with arrows. (E) Top-down view of HI loop model. Positions of residues K321 and E322 at the base of the βDE ribbon, and Y673 at the base of the HI loop are indicated by arrows for some of the monomers. The cyan loops and residues indicate the position of these regions in the crystal structure. The magenta coil indicates proposed new position of HI loop. (F) Side-view of HI loop model. The positions of the HI loops when heparin is not bound (cyan) and when heparin is bound (magenta). Residue D327 at the tip of the βDE ribbon, and the HI loop are indicated with arrows for one monomer. A two-, three-, and fivefold axis are indicated on each panel where appropriate. Figure (A) was generated using RobEM (Baker et al., 1999) and (C–F) by Chimera (Pettersen et al., 2004).
An AAV2-Hep minus VLP difference map pinpointed regions of change between the two structures (Fig. 2A, B, and 3C and D). Positive difference densities close to the twofold axes and in the valleys between the protrusions were interpreted as bound heparin (Fig. 2A–C), and those at and surrounding the channels were interpreted as resulting from movement of the loops at the top of the βDE ribbon and between the βHI strands (the HI loop) (Fig. 3C and D). Negative difference densities were observed at the top of the protrusions, inside the channel, and close to the position of the HI loop as modeled in the AAV2 crystal structure (Fig. 3C and D). These differences are interpreted as structural re-arrangements at the surface resulting from heparin binding. A dodecahedral cage of positive difference density also occurred inside the AAV2-Hep capsid (Fig. 4A). This density was interpreted as ordered DNA (Fig. 4B and C).
Figure 4.
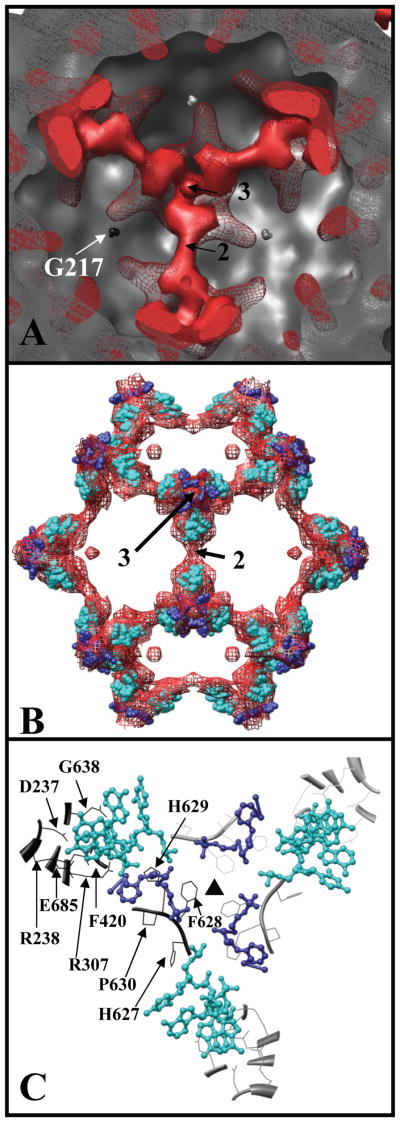
Difference density associated with the interior of the AAV2 capsid. (A) The positive difference density at 1σ (solid red) and 0.5σ (red mesh) superimposed onto a shaded-surface representation of the AAV2 crystal structure (solid grey). (B) The dodecahedral cage at 1σ (red mesh) superimposed with the nucleotides (NTs) ordered in the crystal structures of AAV4 (blue) and MVMi (cyan). (C) Close-up of Cα chains (ribbons and coil) and the side-chains (stick) of AV2 residues (in a trimer) that closely associate with the AAV4 (blue) and the MVMi (cyan) NTs. The amino acids at a radial distance of <3.5 Å (D237, R238, R307, F420, G638, E685) and <5.0 Å (H627, F628, H629, P630) from the modeled NTs are indicated with black arrows for one VP monomer. The two- and threefold axes are indicated with numbers in (A and B) and the threefold axis is indicated with a filled triangle in (C). (A and C) are viewed down a threefold axis, and (B) down a twofold axis. The figure was generated using Chimera (Pettersen et al., 2004).
Heparin binds close to the residues in the AAV2-Heparin complex structure mapped by mutagenesis
Positive density, rendered at a low threshold (0.5σ) in the difference map, extends from the wall of the twofold axes along the valley between the threefold protrusions and traverses the residues expected to bind heparin based on mutagenesis studies (data not shown). Preliminary modeling showed that nine heparin disaccharides (~6 kDa oligosaccharide used for complexing) could be fitted into this low threshold difference map (data not shown). However, only six heparin disaccharides were modeled into positive density (at ~1.0σ), labeled 2f pos and 3f pos (Fig. 2A), lying close to twofold and threefold axes, respectively. These densities coincided with regions of the AAV2-Hep map that include density (pink surface regions in Fig. 2B) not represented in the VLP map. Five heparin disaccharides were built into the 2f pos density (hep2f in Fig. 2B and C), with K527 and K532 being the closest residues predicted by mutagenesis to affect heparin binding. One heparin disaccharide was modeled into the 3f pos density (hep3f in Fig. 2B and C) close to three residues: R585 and R588, which are those most critical for heparin binding in AAV2, and R484, another heparin binding residue from a symmetry-related VP. The hep2f model is within 7Å of K532 and hep3f is within 5Å of R588. These mark the closest points to the heparin binding residues identified by mutagenesis (Kern et al., 2003; Opie et al., 2003) in the crystal structure of the AAV2 capsid (Xie et al., 2002).
The AAV2 heparin binding residues and the heparin oligosaccharide models built into the positive densities are too far apart for direct contact. This suggests that some heparin binding residues may change conformation to permit closer contacts with the receptor or, alternatively, solvent molecules (not visible at this resolution) may mediate the interactions. Structural re-arrangements would be consistent with the observed conformational change at the top of the threefold protrusions, which become flattened in the AAV2-Hep complex map, and with the negative density observed in this region in the AAV2-Hep minus VLP difference map (cyan density, 3f neg, in Fig. 3C). Residues R585 and R588 are located on the inner face (towards threefold axis) of these protrusions where negative difference density is observed. In addition, the 2f pos density (Fig. 2A and B) modeled as hep2f (Fig. 2B and C) is difficult to explain on the basis of the heparin binding residues, R484, R487, K527, K532, R585, and R588 (Kern et al., 2003; Opie et al., 2003). The closest mapped heparin binding residues, K527 and K532, are ~7 Å away from the first disaccharide of the decasaccharide model. Thus, this positive density may result from a conformational change in the capsid triggered by heparin binding possibly at the threefold axes (hep3f in Fig. 2C), and is not likely due to heparin binding alone.
The resolution of the VLP and AAV2-Hep cryo-reconstructions does not allow analysis of the atomic details of interactions between the heparin models and the heparin binding residues mapped by mutagenesis. However, the juxtaposition of the amino acids close to these residues in the AAV2 crystal structure points to stabilizing interactions that may re-arrange to enable receptor binding. The basic, heparin-binding patch is flanked by a highly conserved acidic loop containing residues 528-DDEE-531, D494, and D574, which are involved in electrostatic interactions with the heparin binding residues (Fig. 2D, D494 is not shown) and close to the heparin models. Of these, residues D529 and E530 are closest to hep2f, and lie within 6Å and 5Å, respectively. Another acidic loop (561-DEEE-564), which is not surface accessible, is located at the base of the protrusions next to 528-DDEE-531 and does not contact any heparin-binding residues. However, alanine substitution for this stretch of four acidic residues (561-DEEE-564) produces a mutant virus that does not bind heparin and is non-infectious (Wu et al., 2000). Furthermore, other acidic (D269, D514, and D594) and basic (R471, K507, and H509) residues decorate the capsid surface near the heparin binding residues and extend the charged patch on the valley floor (not shown). Hence, salt bridges (such as the ones illustrated in figure 2D) might serve to maintain a specific conformation of the heparin binding region that favors virus-receptor interactions. Heparin binding might disrupt these interactions and result in side-chain conformations for the binding residues that are closer to the modeled heparin molecules than their positions in the uncomplexed crystal structure used in this analysis.
Conformational change induced by heparin binding is also consistent with a model proposed by Lochrie et al. who suggested that primary receptor binding by AAV2 might lead to structural re-arrangements close to the twofold axis to facilitate co-receptor attachment (Lochrie et al., 2006). Residues G512 and R729 were identified in this study as exhibiting minor effects on heparin binding in HepG2 cells (Lochrie et al., 2006). These residues are located on the capsid surface, positioned below the hep2f model with G512 immediately adjacent to R527 in the valley and R729 at the floor/wall junction near the twofold axis (not shown). In addition, AAV2 residue N511, which together with G512 and R513 has been implicated in the binding of integrin α5β1 (Asokan et al., 2006), is shielded from the capsid surface by K527 and suggests that either G512 or R513 would have to shift position in order for N511 to directly contact a receptor. Furthermore, a conformational change at the twofold axis resulting from heparin binding that “raised” residues in the twofold depression could bring additional basic residues, including R389, K490, K544, R566, R724, and R729 close to the hep2f model. Thus, a second binding event may be facilitated by structural re-arrangements that expose additional, basic residues within contact distance. This may help explain how heparin can bind at the twofold axes away from K532. The lack of negative difference density at the original position of these residues may be due to subtle movements or the low resolution of the maps. It is possible that this second interaction might normally be with a co-receptor, or that heparin binding at this site may facilitate capsid recognition by co-receptors such as the integrins or FGFR1, which also interact with heparin.
Heparin binding induces conformational changes at and around the fivefold axes of the AAV2 capsid
The region at and near the channel at the fivefold axis of the AAV2 capsid differs in the VLP and AAV2-Hep structures (Fig. 1D and H, 3A and B). Comparison of the capsid surface and equatorial sections of the reconstructions shows a widening and opening at the top of the channel in the AAV2-Hep structure compared to the narrower, more closed channel in the VLP. Also, the floor of the fivefold depression appears to bulge upwards in the AAV2-Hep structure compared to the VLP floor (Fig. 1D and H). These conformational changes in AAV2-Hep were unexpected and could not be predicted solely by mutagenesis studies. A review of the literature showed that a similar re-arrangement could be seen in human parvovirus B19 capsids bound to its globoside receptor (Chipman et al., 1996).
A superimposition of the AAV2-Hep minus VLP difference map onto a Cα atom map generated at 18 Å resolution from the AAV2 crystal structure showed positive and negative difference density on the floor of the fivefold depression and negative difference density inside the channel (Fig. 3C, D). The positive (5f pos1) and negative (5f neg1) difference densities (figure 3C and D) appear to result from movement of the HI loop (residues T651 to Y673). This loop interacts with neighboring VP monomers in the fivefold depression and with the base of the βDE ribbons that form the wall of the channels (Xie et al., 2002). The shapes of the difference densities were consistent with the structure of the HI loop. The observed change in density suggests that upon heparin binding, the HI loop rotates ~90° from a position tangential to the capsid surface to running radially about the fivefold axis. This repositions the HI loop away from the negative density and into the observed positive density (5f neg1 and 5f pos1, respectively, in Fig. 3D). The HI loop is present in the VP of all the parvovirus structures determined to date, and is positioned tangential to the capsid surface in the AAVs and members of the parvovirus genera (reviewed in Chapman and Agbandje-McKenna, 2006). However, the HI loop in B19 adopts a radial orientation relative to the capsid surface (Kaufmann et al., 2004), similar to the AAV2-Hep HI loop conformation described above.
The loops at the top of the βDE ribbons which form the opening of the channel moved radially outwards from the fivefold axes and rotate by ~36° clockwise following heparin binding (compare Fig. 1D and H). The difference densities 5f neg2 and 5f pos2 (Fig. 3C and D) were interpreted as representing the original and new positions, respectively, of this loop. This movement is consistent with widening of the channel seen in the equatorial section of the AAV2-Hep cryo-reconstruction (Fig. 3B). K321 and E322 in AAV2 form a ring of five pairs of alternating basic/acidic residues at the base of the βDE ribbon (base of the channel), close to the bottom of the HI loop where it contacts the βDE ribbon (Fig. 3E and F). Heparin binding and HI loop re-arrangements may disrupt this ring of electrostatic interactions or lead to formation of new contacts between the relocated HI loop and the βDE ribbon residues as modeled (Fig. 3F). Such movements might result in an iris-like rotation of the ring of residues leading to the widening of the top of the channel observed in the AAV2-Hep complex structure (Fig. 1H).
Specific roles for the βDE ribbon and HI loop transitions proposed here are yet to be verified, but previous mutagenesis work does support their importance in AAV capsid assembly, genome packaging, and infectivity. An AAV2 K321-E322 double-mutant produces a temperature sensitive capsid phenotype (Wu et al., 2000), and mutation of E322 alone produces a capsid assembly defect (Bleker et al., 2005). In addition, a nine residue insertion in the HI loop (at position 664) affects capsid formation and infectivity (Wu et al., 2000). Recently, mutagenesis, confocal microscopy, and VP structural alignment studies reported a role for the HI loop in genome packaging (DiPrimio et al., 2008). Furthermore, an AAV2 point mutation (F661G) within the HI loop reduces viral infectivity at the nuclear entry step (DiPrimio et al., 2008). Significantly, the fivefold channels of parvovirus capsids have been implicated as conduits for extrusion of the N-terminal portions of VP1 and VP2 (both of which are reported to normally lie inside the capsid and for genome packaging (Bleker et al., 2005; Farr et al., 2004; Farr et al., 2006). Extrusion and activity of the VP1-N terminal PLA2 domain is essential for efficient infection and is believed to occur during or shortly after virus entry into the cell (Bleker et al., 2005; Farr et al., 2006; Mani et al., 2006; Suikkanen et al., 2003; Vihinen-Ranta et al., 2002). Nuclear localization signal sequences present at the N-terminal regions of VP1 and VP2 are also reported to be required for nuclear entry. Thus, the structural arrangements proposed above for the βDE ribbon and HI loop may represent structural capsid transitions/dynamics, following AAV2 heparin receptor attachment, required for subsequent steps in infection, such as the externalization of internally disposed VP regions through the channel.
AAV2-Hep encapsidates an ordered, dodecahedral cage of DNA
Heparin binding, in addition to altering the AAV2 surface structure, leads to an ordering of density inside the capsid, resulting in a dodecahedral cage (Fig. 4A and B). Density in the cage runs beneath the interfaces between two- and threefold-related VP monomers and is particularly pronounced close to the threefold axes. NT models fitted into ordered, internal density in the crystal structures of AAV4 (dark blue model in Fig. 4B and C) (Govindasamy et al., 2006) and MVMi (cyan model in Fig. 4B and C) (Agbandje-McKenna et al., 1998) superimpose onto this cage in the AAV2-Hep cryo-reconstruction and naturally led to the conclusion that the cage was nucleic acid. Notably, this cage-like density appears very similar to ordered, duplex RNA structures seen in some ssRNA viruses (e.g. (Bink et al., 2002; Prasad et al., 1994; Speir et al., 1995; Tang et al., 2001)).
The presence of oligonucleotides in purified AAV2 VLPs in a form inaccessible to DNAse was confirmed by means of a nuclease protection assay (Fig. 5). The presence of DNA, but not RNA, in the VLPs was confirmed by noting that extracted nucleic acid was digested by DNAse but not RNase (Fig. 5, lanes 4 and 3, respectively). The DNA found was present as short oligonucleotides, 100 to 700 bp long.
Figure 5.
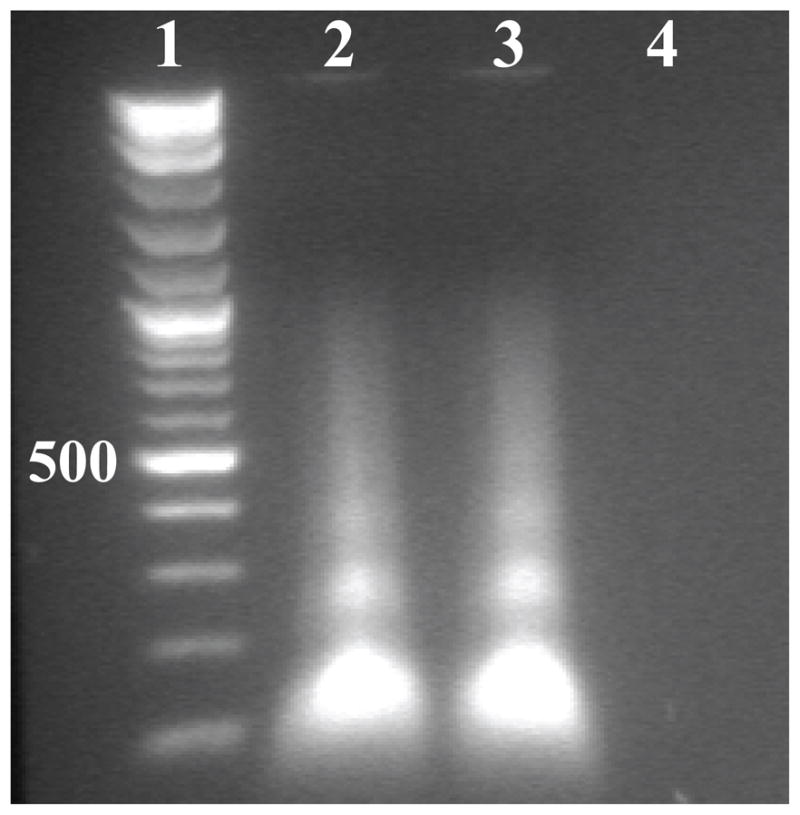
Agarose gel showing the presence of DNA in VLPs. Lane 1, molecular weight marker (500 bp band labeled); Lane 2 untreated (no DNAse or RNAse); Lane 3 treated with RNAse; and Lane 4 treated with DNAse.
AAV terminal repeats (TRs) rather than heterologous DNA sequences are known to form the signal used to package AAV (McLaughlin et al., 1988). Several investigators have shown that AAV DNA and heterologous, chromosomal DNA that contains a Rep binding element and a terminal resolution site, but no TRs, can also be packaged into AAV virions (Huser et al., 2003; Nony et al., 2003). This suggests that a Rep molecule, covalently attached to the 5′ end of a DNA molecule, is sufficient for packaging. In the present study, VLPs were assembled in cells infected with an adenovirus vector that expressed only the AAV capsid gene and no rep gene. Thus, the observation that DNA is present in AAV2 VLPs suggests that random pieces of DNA, perhaps generated by apoptosis during the course of adenovirus infection, get “packaged” into particles or become “trapped” within particles during assembly. This observation challenges the paradigm that AAV does not require nucleic acid for assembly, since non-viral DNA is packaged during assembly in the absence of viral genome. In addition, recent observation of NTs in baculovirus expressed AAV8 (Nam et al., 2007) provides further evidence contrary to the existence of truly “empty” AAV particles. Hence, since Rep-facilitated DNA packaging is proposed to occur with preformed empty capsids through the channel, these observations suggest that small portions of viral genome are pre-packaged during assembly before Rep completes the packaging (Myers and Carter, 1980; Wistuba et al., 1995; King et al., 2001).
Regardless of how DNA is encapsidated, the dodecahedral cage in the AAV2-Hep complex suggests that heparin binding triggers conformational changes at the inner surface of capsid. These, in turn, likely create more favorable interactions with the packaged DNA. A re-organization of the capsid interior following heparin binding is evident in the equatorial cross-sections (Fig. 3A and B). However, the significance, if any, of the ordering of DNA following heparin binding during the AAV2 life cycle remains unknown. This ordering perhaps represents a means to prepare the capsid for genome uncoating or DNA re-organization/stabilization prior to transfer into the nucleus.
AAV2 residues closest to the cage density were identified by superimposing the VP3 and NT coordinates of AAV4 and MVMi (Govindasamy et al., 2006; Agbandje-McKenna et al., 1998) onto the VP3 of AAV2 (Xie et al., 2002) docked into the difference map (AAV2-Hep minus VLP). Residues of AAV2 that lie 3.5 Å or closer to the NTs at the twofold interface include D237, R238, R307, F420, G638, and E685 (Fig. 4C). These are highly conserved residues in AAV serotypes 1–8, with substitutions only in AAV4 (D237G, R238H) (Padron et al., 2005). Notably, mutations of charged residues to alanine, which include substitutions of AAV2 D237, R238, and R307, lead to defective capsid assembly (Wu et al., 2000). In addition, mutations in residues E681-E683 and E689-R693, all of which lie close to E685, create defects in capsid assembly (Wu et al., 2000). The vertex of the dodecahedron cage (at the threefold axis) most closely associates with loop 627-HFHP-630 (Fig. 4C). These residues constrict a small pore in the capsid crystal structure. Negative difference density (in the AAV2-Hep minus VLP map) was observed above the vertex of dodecahedron cage at the interior surface of the capsid (not shown), which overlaps with the 627-HFHP-630 region, suggesting a re-arrangement. This is a highly conserved region in AAVs (Padron et al., 2005) and is positioned adjacent to the NTs ordered in both the AAV4 and AAV8 crystal structures (Govindasamy et al., 2006, Nam et al., 2007). Thus, re-arrangements of this loop may influence DNA organization in AAV2-Hep and suggests the loop plays a role in DNA stabilization or possibly capsid uncoating.
As discussed above for AAV2 residues, mutations in MVMi residues that contact ordered DNA in the crystal structure affect capsid assembly and lead to the production of empty or unstable capsids (Reguera et al., 2005). Such observations strongly support a role for DNA-VP contacts in capsid assembly. A DNA cage may serve to stabilize interactions between capsid assembly intermediates by tethering proteins at subunit interfaces and knitting pentameric intermediates together in an icosahedron. A similar mechanism has been proposed for assembly of ssRNA viruses (Rudnick and Bruinsma, 2005; Zlotnick, 1994, 2004). For the AAVs, Rep binding to a stabilized capsid stabilized by the DNA cage could complete genome packaging through a fivefold channel.
Additional positive density in the AAV2-Hep minus VLP difference map appears inside the capsid adjacent to the last N-terminal residue (G217) modeled in the AAV2 crystal structure (Fig. 4A, in red mesh). This density extends from the dodecahedron cage toward the fivefold axis and may be part of the missing VP N-terminus. The rationale for improved ordering of this structural region following heparin binding is not yet known, but may be associated with capsid transitions following receptor attachment required for the externalization of the VP1/VP2 N-termini.
Summary
We have identified and interpreted several distinct changes that occur at the surface and in the interior of AAV2 triggered by heparin binding. In addition to the expected added density in the valleys between the threefold protrusions attributed to heparin, significant changes occur close to the twofold and five depressions and at the channels. These changes are proposed to reflect structural re-arrangements conferred by heparin binding. Additional density beneath the inner wall of the capsid at the two- and threefold axes and in the form of a dodecahedral cage was interpreted as short DNA oligonucleotides packaged in the absence of replicating AAV DNA and Rep protein. The presence of such highly ordered density suggests that DNA may play a role in AAV capsid assembly. Structural changes that occurred after heparin binding are believed to represent transitions required for steps subsequent to primary receptor attachment, such as co-receptor binding and the externalization of the DNA and N-termini of VP1 and VP2 through the fivefold channel. This study further highlights the versatile and dynamic nature of the parvovirus VP and capsid as is required for receptor recognition and downstream events in the infection pathway.
Acknowledgments
We thank Dr. Sergei Zolotukhin (University of Florida) for providing the adenovirus expression vector for AAV2 and Irena Zolotukhin (University of Florida) for help with virus purification. The project was supported by NIH NHLBI grants P01 HL59412 and P01 HL51811 to MA-M and NM, and NIH GM R37-033050 to TSB. N.M. is an inventor on patents related to recombinant AAV technology and owns equity in a gene therapy company that is commercializing AAV for gene therapy applications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 2.Asokan A, Hamra JB, Agbandje-McKenna M, Samulski RJ. Adeno-associated virus type 2 (AAV2) contains a putative integrin alpha5beta1 recognition sequence for viral cell entry. J Virol. 2006;80:8961–8969. doi: 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 4.Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bink HH, Pleij CW. RNA-protein interactions in spherical viruses. Arch Virol. 2002;147:2261–2279. doi: 10.1007/s00705-002-0891-6. [DOI] [PubMed] [Google Scholar]
- 7.Bleker S, Sonntag F, Kleinschmidt JA. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol. 2005;79:2528–2540. doi: 10.1128/JVI.79.4.2528-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol. 2002;76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 10.Chapman MS, Agbandje-McKenna M. Atomic structure of viral particles. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish, Parrish CR, editors. Parvoviruses. Edward Arnold, Ltd; New York: 2006. pp. 107–123. [Google Scholar]
- 11.Chipman PR, Agbandje-McKenna M, Kajigaya S, Brown KE, Young NS, Baker TS, Rossmann MG. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc Natl Acad Sci U S A. 1996;93:7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotmore SF, Tattersall PP. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 13.DiPrimio N, Asokan A, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Surface loop dynamics in adeno-associated virus capsid assembly. J Virol. 2008;82:5178–5189. doi: 10.1128/JVI.02721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 16.Farr GA, Cotmore SF, Tattersall P. VP2 Cleavage and the Leucine Ring at the Base of the Fivefold Cylinder Control pH-Dependent Externalization of both the VP1 N Terminus and the Genome of Minute Virus of Mice. J Virol. 2006;80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farr GA, Tattersall P. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology. 2004;323:243–256. doi: 10.1016/j.virol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Flotte TR, Brantly ML, Spencer LT, Byrne BJ, Spencer CT, Baker DJ, Humphries M. Phase I trial of intramuscular injection of a recombinant adeno-associated virus alpha 1-antitrypsin (rAAV2-CB-hAAT) gene vector to AAT-deficient adults. Hum Gene Ther. 2004;15:93–128. doi: 10.1089/10430340460732490. [DOI] [PubMed] [Google Scholar]
- 19.Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles--the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 20.Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 21.Govindasamy L, Hueffer K, Parrish CR, Agbandje-McKenna M. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J Virol. 2003;77:12211–12221. doi: 10.1128/JVI.77.22.12211-12221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini J, Agbandje-McKenna M. Structually mapping the diverse phenotype of Adeno-associated virus serotype 4. J Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafenstein S, Palermo LM, Kostyuchenko VA, Xiao C, Morais MC, Nelson CD, Bowman VD, Battisti AJ, Chipman PR, Parrish CR, Rossmann MG. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc Natl Acad Sci U S A. 2007;104:6585–6589. doi: 10.1073/pnas.0701574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernando E, Llamas-Saiz AL, Foces-Foces C, McKenna R, Portman I, Agbandje-McKenna M, Almendral JM. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology. 2000;267:299–309. doi: 10.1006/viro.1999.0123. [DOI] [PubMed] [Google Scholar]
- 25.Hueffer K, Govindasamy L, Agbandje-McKenna M, Parrish CR. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J Virol. 2003;77:10099–10105. doi: 10.1128/JVI.77.18.10099-10105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. 2003;77:1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huser D, Weger S, Heilbronn R. Packaging of human chromosome 19- specific adeno-associated virus (AAV) integration sites in AAV virions during AAV wild-type and recombinant AAV vector production. J Virol. 2003;77:4881–4887. doi: 10.1128/JVI.77.8.4881-4887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol. 2005;79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufmann B, Simpson AA, Rossmann MG. The structure of human parvovirus B19. Proc Natl Acad Sci U S A. 2004;101:11628–11633. doi: 10.1073/pnas.0402992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay MA, Manno CS, Ragni MV, Larson PJ, Couto LB, McClelland A, Glader B, Chew AJ, Tai SJ, Herzog RW, Arruda V, Johnson F, Scallan C, Skarsgard E, Flake AW, High KA. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 2000;24:257–261. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 32.Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, Agbandje-McKenna M. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronenberg S, Kleinschmidt JA, Bottcher B. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2001;2:997–1002. doi: 10.1093/embo-reports/kve234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kronenberg S, Böttcher B, von der Lieth CW, Bleker S, Kleinschmidt JA. A Conformational Change in the Adeno-Associated Virus Type 2 Capsid Leads to the Exposure of Hidden VP1 N Termini. J Virol. 2005;79:5296–5303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, Pierce GF, Colosi P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol. 2006;80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Bueno A, Rubio MP, Bryant N, McKenna R, Agbandje-McKenna M, Almendral JM. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J Virol. 2006;80:1563–1573. doi: 10.1128/JVI.80.3.1563-1573.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell’Osso L, Hertle R, Ma JX, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mani B, Baltzer C, Valle N, Almendral JM, Kempf C, Ros C. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J Virol. 2006;80:1015–1024. doi: 10.1128/JVI.80.2.1015-1024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLaughlin SK, Collis P, Hermonat PL, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller EB, Gurda-Whitaker B, Govindasamy L, McKenna R, Zolotukhin S, Muzyczka N, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 1. Acta Cryst. 2006;F62:1271–1274. doi: 10.1107/S1744309106048184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muzyczka N, Berns KI. Parvoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4. Lippincott Williams and Wilkins; New York: 2001. pp. 2327–2360. [Google Scholar]
- 44.Myers MW, Carter BJ. Assembly of adeno-associated virus. Virology. 1980;102:71–82. doi: 10.1016/0042-6822(80)90071-9. [DOI] [PubMed] [Google Scholar]
- 45.Nony P, Chadeuf G, Tessier J, Moullier P, Salvetti A. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J Virol. 2003;77:776–781. doi: 10.1128/JVI.77.1.776-781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. Identification of amino Acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, Aslanidi G, Byrne B, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padron E, Bowman V, Kaludov N, Govindasamy L, Levy H, Nick P, McKenna R, Muzyczka N, Chiorini JA, Baker TS, Agbandje-McKenna M. Structure of adeno-associated virus type 4. J Virol. 2005;79:5047–5058. doi: 10.1128/JVI.79.8.5047-5058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Prasad BV, Matson DO, Smith AW. Three-dimensional structure of calicivirus. J Mol Biol. 1994;240:256–264. doi: 10.1006/jmbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- 51.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 52.Qiu J, Handa A, Kirby M, Brown K. The interaction of heparin sulfate and adeno-associated Virus 2. Virol. 2000;269:137–147. doi: 10.1006/viro.2000.0205. [DOI] [PubMed] [Google Scholar]
- 53.Quesada O, Gurda B, Govindasamy L, McKenna R, Kohlbrenner E, Aslanidi G, Zolotukhin S, Muzyczka N, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 7. Acta Cryst. 2007;F63:1073–1076. doi: 10.1107/S1744309107060289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reguera J, Grueso E, Carreira A, Sanchez-Martinez C, Almendral JM, Mateu MG. Functional relevance of amino acid residues involved in interactions with ordered nucleic acid in a spherical virus. J Biol Chem. 2005;280:17969–17977. doi: 10.1074/jbc.M500867200. [DOI] [PubMed] [Google Scholar]
- 55.Rudnick J, Bruinsma R. Icosahedral packing of RNA viral genomes. Phys Rev Lett. 2005;94:038101. doi: 10.1103/PhysRevLett.94.038101. [DOI] [PubMed] [Google Scholar]
- 56.Sommer JM, Smith PH, Parthasarathy S, Isaacs J, Vijay S, Kieran J, Powell SK, McClelland A, Wright JF. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 57.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol. 2006;80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suikkanen S, Antila M, Jaatinen A, Vihinen-Ranta M, Vuento M. Release of canine parvovirus from endocytic vesicles. Virology. 2003;316:267–280. doi: 10.1016/j.virol.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 60.Summerford C, Bartlett JS, Samulski RJ. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 61.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang L, Johnson KN, Ball LA, Lin T, Yeager M, Johnson JE. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat Struct Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 63.Vihinen-Ranta M, Wang D, Weichert WS, Parrish CR. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J Virol. 2002;76:1884–1891. doi: 10.1128/JVI.76.4.1884-1891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walters RW, Agbandje-McKenna M, Bowman VD, Moninger TO, Olson NH, Seiler M, Chiorini JA, Baker TS, Zabner J. Structure of adeno-associated virus serotype 5. J Virol. 2004;78:3361–3371. doi: 10.1128/JVI.78.7.3361-3371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wistuba A, Weger S, Kern A, Kleinschmidt JA. Intermediates of adeno-associated virus type 2 assembly: Identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu P, Xiao W, Conlon T, Hughes J, Agbandje-McKenna M, Ferkol T, Flotte T, Muzyczka N. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J Virol. 2000;74:8635–8647. doi: 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie Q, Chapman MS. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J Mol Biol. 1996;264:497–520. doi: 10.1006/jmbi.1996.0657. [DOI] [PubMed] [Google Scholar]
- 69.Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J Mol Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 70.Zlotnick A. Viruses and the physics of soft condensed matter. Proc Natl Acad Sci U S A. 2004;101:15549–15550. doi: 10.1073/pnas.0406935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ, Jr, Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, Flotte TR, Byrne BJ, Snyder RO. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]