Abstract
Recent studies have suggested that the tethering of viral genomes to host cell chromosomes could provide one of the ways to achieve their nuclear retention and partitioning during extrachromosomal maintenance in dividing cells. The data we present here provide firm evidence that the partitioning of the bovine papillomavirus type 1 (BPV1) genome is dependent on the chromatin attachment process mediated by viral E2 protein and its multiple binding sites. On the other hand, the attachment of E2 and the E2-mediated tethering of reporter plasmids to host chromosomes are not necessarily sufficient for efficient partitioning, suggesting that additional E2-dependent activities might be involved in the latter process. The activity of E2 protein in chromatin attachment and partitioning is more sensitive to the point mutations in the N-terminal domain than its transactivation and replication initiation functions. Therefore, at least part of the interactions of the E2 N-terminal domain with its targets during the chromatin attachment and partitioning processes are likely to involve specific receptors not involved in transactivation and replication activities of the protein. The mutational analysis also indicates that the binding of E2 to chromatin is not achieved through interaction of linear N-terminal subsequences of the E2 protein with putative receptors. Instead, the composite surface elements of the N-terminal domain build up the receptor-binding surface of E2. In this regard, the interaction of BPV1 E2 with its chromosomal targets clearly differs from the interactions of LANA1 protein from Kaposi's sarcoma-associated human herpesvirus and EBNA1 from Epstein-Barr virus with their specific receptors.
The life cycles of several double-stranded DNA tumor viruses are characterized by a distinctive latency stage, during which the viral genomes are stably maintained as nuclear extrachromosomal multicopy plasmids at an approximately constant copy number in proliferating cells. The information about the possible mechanism of the stable maintenance process has only recently started to emerge in more detail, mainly based on studies of bovine papillomavirus type 1 (BPV1) and two members of the gammaherpesvirus family, Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus. These studies have suggested that the proper partitioning and nuclear retention of the stably maintained viral genomes during mitosis might be achieved through their tethering to host mitotic chromosomes (4, 13, 14, 21, 27, 33, 51, 52). A single viral protein acts as a molecular linker between the viral genome and host chromatin. These linker proteins are viral transactivator EBNA1 in the case of Epstein-Barr virus (27), transcriptional repressor LANA1 in the case of Kaposi's sarcoma-associated herpesvirus (4, 5), and viral regulatory protein E2 in the case of BPV1 (26, 33, 52). E2 protein in trans and its multiple binding sites in cis, the latter referred to as minichromosome maintenance element (MME), are both necessary and sufficient for chromatin attachment of the extrachromosomal plasmids in proliferating cells (2, 26). MME is located in the ∼1-kb noncoding upstream regulatory region (URR) of the BPV1 genome.
The papillomavirus E2 protein is a central coordinator of the biochemical processes that operate during viral gene expression and genome replication. It is a multifunctional protein which is required throughout the papillomavirus life cycle (37, 57). The E2 protein regulates gene expression by modulating the transcription from viral promoters (24) and is essential for the initiation of papillomavirus DNA replication in vivo as a factor responsible for loading of the viral E1 helicase to the virus replication origin (37, 55). BPV1 E2 is built up as a modular protein, with its N-terminal part forming a distinct transactivation domain and its C-terminal part forming a dimerization and sequence-specific DNA-binding domain (DBD). These two domains are linked to each other through a flexible hinge region. Both domains can function as independent units under certain conditions, i.e., DBD can bind to the E2 binding site in the absence of the transactivation domain (12, 15, 39, 43, 46), and the transactivation domain is able to activate transcription and replication when linked to another DBD (8, 9, 28, 61). On the other hand, only the full-length protein can carry out certain activities, such as the cooperative binding to two neighboring binding sites on DNA (38, 39). Two main activities of the E2 protein, activation of transcription and participation in replication initiation, can be genetically separated by single-point mutations in the N-terminal transactivation domain in the case of E2 proteins from several papillomavirus types (1, 10, 11, 16, 19, 22, 49). The N-terminal transactivation domain is also the part of the E2 protein responsible for interactions with host chromatin, as it is able to localize to host mitotic chromosomes even if all other parts of the protein are removed (6). It has been shown that E1 may dislocate E2 from mitotic chromosomes (60). Thus, the presence of other viral proteins may affect E2 chromatin attachment and URR tethering.
In this study, employing a novel experimental approach, we have analyzed the activity of E2 as a central trans factor in MME-dependent chromatin attachment and partitioning processes, making use of extensive mutational analysis of the N-terminal chromatin-binding domain of the protein. The results of our study demonstrate clearly that the partitioning function of BPV1 during the host cell division can be provided by the chromatin attachment process mediated by the E2 protein and its multiple binding sites in MME. The deletion and mutation analysis of the E2 N-terminal domain indicates that composite surface elements rather than linear subsequences of this domain build up the ligand surface for interaction with the putative chromatin receptor. In this regard, the interaction of BPV1 E2 with its chromosomal targets clearly differs from the interactions of EBNA1 and LANA1 with their specific receptors.
MATERIALS AND METHODS
Plasmids.
The expression plasmids for wild-type (wt) E2, E2C, E8/E2, and dNco (58, 59) as well as for E2 proteins with point mutations (1) and deletions (30) have been described previously. All proteins are expressed from the pCG vector (56), and all but pCGE2C and pCGE2dNco contain the mutation M162I, which disrupts the initiation codon for the E2C protein. The pNeoBgl40 reporter plasmid, which contains the approximately 1-kb-long URR region from the BPV1 genome (nucleotides 6946 to 6963), is described in reference 44. The map of pRetE2 is shown in Fig. 1. This plasmid carries BPV1 MME and expresses destabilized (with a half-life of about 1 h) enhanced green fluorescent protein (EGFP) under the control of the cytomegalovirus (CMV) promoter as well as wt E2. pRetE2fr is derived from pRetE2 by introducing the frameshift mutation into the E2 sequences corresponding to the hinge region of E2 (A. Janikson, A. Männik, and M. Ustav, unpublished data). The pRetE2 vectors expressing different mutated E2 proteins were constructed by replacing the Psp1406I-StuI fragment in the wt E2 coding region with the corresponding mutated fragment from the respective pCGE2 plasmids. pRetVP16:E2 was constructed by replacing the XbaI-AgeI fragment from the wt E2 open reading frame in pRetE2 with the XbaI-AgeI fragment from the pCGVP16:E2 vector (35). The resulting constructs were verified by sequencing.
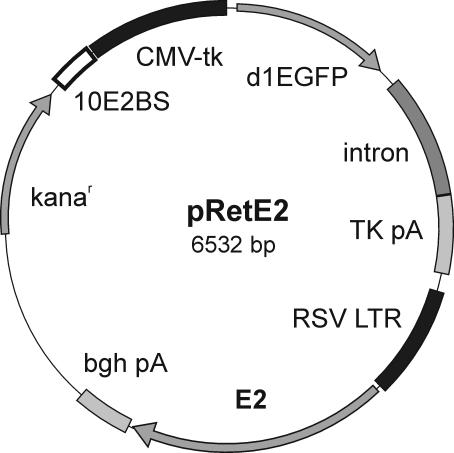
FIG. 1.
Map of plasmid pRetE2 used in partitioning assay. CMV-tk, CMV immediate early promoter with thymidine kinase leader sequence; d1EGFP, destabilized enhanced GFP coding sequence; intron, rabbit β-globin IV intron; TK pA, thymidine kinase poly(A) signal; RSV LTR, Rous sarcoma virus 5′ long terminal repeat promoter; E2, BPV1 E2 coding sequence with mutation Met162Ile; bgh pA, bovine growth hormone poly(A) signal; kanar, kanamycin resistance gene; 10E2BS, oligomerized E2BS9 in 10 copies as described in Piirsoo et al. (44).
Cell culture and transfections.
The Chinese hamster ovary (CHO) cell line and its derivatives CHO49 (expressing the BPV1 E2 protein), CHO4.15 (expressing the BPV1 E1 and E2 proteins), and CHOBgl40 (constitutively expressing the BPV1 E1 and E2 proteins and maintaining reporter plasmid pNeoBgl40) (44) were grown in Ham's F-12 medium supplemented with 10% fetal calf serum and appropriate antibiotics. Electroporation experiments were carried out as described earlier (58) with a Bio-Rad Gene Pulser II apparatus supplied with a capacitance extender. Capacitance was set to 975 μF and voltage was set to 230 V in all experiments. Five hundred nanograms of different pCGE2 plasmids and/or 500 ng of pNeoBgl40 was transfected in all experiments with CHO cell lines. Jurkat cells were grown in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum and antibiotics.
Chromatin immunofluorescence (ChiF) analysis of mitotic chromosomes.
About 36 h after transfection, Colcemid was added to culture media to a final concentration 0.1 μg/ml and cells were incubated for an additional 2 to 6 h to increase the fraction of mitotic cells. Colcemid-treated cells were harvested after treatment with phosphate-buffered saline (PBS)-EDTA (3 mM), pelleted by centrifugation (190 × g for 5 min at room temperature), and suspended in 5 ml of 0.075 M KCl solution. Suspended cells were incubated at room temperature for 15 min, collected by centrifugation as before, resuspended again in a small volume of 0.075 M KCl, and transferred to slides (at this point, before transferring to slides, an aliquot of cells was also taken for parallel fluorescent in situ hybridization [FISH] analysis that was carried out as described in the next section). The cell suspension was stored on slides for 10 min at room temperature followed by fixation in cold (−20°C) methanol for 10 min. Fixed slides were allowed to dry at room temperature and stored at −20°C. For immunostaining, the slides were warmed up to room temperature and 125 μl of primary antibody solution was added (containing a mixture of 2.5 ng [each] of monoclonal antibodies 1E2, 3F12, 1H10, 3E8, and 5H4/μl [30] and 20 μg of bovine serum albumin [BSA]/μl in PBS). After incubation for 1 h at room temperature in a humid atmosphere, the cells on slides were washed with PBS, incubated with 125 μl of fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibody (LabAs, Tartu, Estonia) diluted 1/200 in BSA-PBS for another hour at room temperature, and then washed again with PBS. The chromosomes were counterstained with propidium iodide and mounted in p-phenylenediamine antifade mounting medium. Slides were analyzed with an Olympus VANOX-S fluorescence microscope equipped with the appropriate filter set. Over 200 mitotic metaphases were analyzed in each experiment, and the percentage of metaphases carrying the E2-specific signal was calculated. The experiments were repeated at least four times. The fraction of E2-positive mitosis was calculated in the case of each mutant protein relative to the positive fraction in the case of wt E2-transfected cells, and the latter was set as 1.0 in each separate series.
FISH analysis.
FISH analysis was performed as described previously (26). At least 100 mitotic metaphases were analyzed in each experiment, and the percentage of metaphases carrying URR reporter plasmid-specific signals was calculated. The fraction of URR reporter-positive mitosis in the case of each mutant was calculated relative to the positive fraction in the case of wt E2-transfected cells, and the latter was set as 1.0 in each separate series.
Examination of the cell cycle dependence of the E2 level.
CHO cells were transfected by electroporation as described above. The cells from each transfection were split equally between two culture dishes. Approximately 24 h after transfection, Colcemid was added to one of the dishes. The cells were collected for analysis ∼36 h after transfection by PBS-EDTA treatment, pelleted by centrifugation, resuspended in 100 μl of PBS, and fixed by the addition of 1 ml of 80% ethanol (cooled to −20°) drop-wise to the cell suspension with simultaneous vortexing. Cell suspensions were kept on ice for 15 min, then 5 ml of 1% BSA in PBS-0.05% Tween 20 was added, and the cells were collected by centrifugation (5 min, 300 × g). The pellet was resuspended in 300 μl of primary antibody solution containing anti-E2 antibodies 3F12, 5H4, 3E8, and 1E4 (1 μg of each/ml in PBS-Tween 20-5% nonfat dry milk) (30) and incubated for 1 h. Five milliliters of 1% BSA in PBS-Tween 20 was added; cells were incubated for 5 min and pelleted by centrifugation as described above. The pellet was resuspended in 100 μl of anti-mouse FITC-conjugated secondary antibody solution (from Taxo A/S, Glostrup, Denmark, diluted 1:50 in PBS-Tween 20-5% nonfat dry milk supplemented with 1 mM MgCl2 and 100 μg of RNaseA/ml) and incubated in the dark for 1 h. BSA solution (1%) was added, and cells were pelleted exactly as after primary antibody incubation. Finally, the cells were resuspended in 300 μl of PBS supplemented with 15 μg of DNA dye 7-aminoactinomycin D (7-AAD)/ml, incubated for 15 min, and analyzed on a Becton-Dickinson FACSCalibur flow cytometer. One hundred thousand signals were collected from each sample. The threshold for autofluorescence was set to contain 99% of the signals from the mock-transfected control cells. In the case of E2-transfected cells, the signals with higher fluorescence levels were defined as corresponding to E2-positive cells.
Partitioning assay.
Jurkat cells were transfected by electroporation with 1 μg of the pRetE2 reporter plasmids, and control transfections were carried out with carrier DNA only. Transfections were performed essentially as described in reference 58, with 3 × 106 cells per transfection and pulse conditions of 210 V and 1 mF. At the set time points after transfection, equal aliquots of the cell suspension from all transfected samples were collected for flow cytometry analysis and the remaining cells were diluted with fresh medium, keeping the cell concentration between 0.3 × 106 to 1 × 106 cells per ml. The total number of cells was determined by counting only the signals with characteristic cell size and surface parameters. The fluorescence histogram from mock-transfected control samples was used as a reference for exclusion of the nonspecific auto-fluorescence signals inside every series. The threshold for autofluorescence inside the series was set to contain 99% of the signals from the mock-transfected control cells. All the signals that were above this threshold were considered to correspond to EGFP-positive cells. The number of EGFP-positive cells per milliliter was calculated in samples according to the formula [(number of positive cells × 1,000)/(time × flow rate)] × dilution (time is measured in minutes, and flow is measured in microliters/minute). The number of positive cells at different time points was normalized to the first time point of respective transfection. Based on the growth in the total number of cells, the cell doublings were calculated, setting the first time point (∼20 h after transfection) as zero.
Western and Southern blot analysis.
The extraction of low-molecular-weight DNA from cells and Southern blot analysis of the DNA level in plasmid were performed essentially as described previously (58). For Western blot analysis of the level of E2 proteins, the total protein from the same number of transfected cells was separated by 10% polyacrylamide gel-sodium dodecyl sulfate electrophoresis and transferred to an Immobilon-P membrane (Millipore). E2-specific monoclonal antibody 1E4 (30), peroxidase-conjugated goat-anti mouse secondary antibody, and the enhanced chemiluminescence detection kit from Amersham Pharmacia Biotech were used to detect the proteins by following the standard manual provided by the supplier.
Homology modeling of E2 transactivation domain.
The BPV1 E2 transactivation domain was modeled by the automatic Swiss-Model homology modeling server (20, 41), with the structure of the human papillomavirus type 16 (HPV16) E2 full-length N-terminal domain (Protein DataBank code 1DTO) as a template. Further analysis of the modeled structure was performed with the programs SPDBView and RasMol (20, 50).
RESULTS
Localization of the BPV1 E2 protein on mitotic chromosomes follows a speckled pattern, which is similar to the localization pattern of BPV1 URR reporter plasmids.
For more-detailed insight into the functioning of the E2 protein in the BPV1 chromatin attachment process, we first developed the ChIF assay. It is based on a standard immunofluorescence technique that was optimized for in situ detection of the E2 protein bound to the mitotic chromosomes. For initial adjustment of the ChIF method, we chose the CHOBgl40 cell line, which expresses both E2 and E1 proteins from chromosomally integrated cassettes, and extrachromosomally maintains the BPV1 URR reporter plasmid pNeoBgl40. We found that the localization of E2 protein on mitotic chromosomes follows a random speckled pattern both in the CHOBgl40 cells that were treated with Colcemid prior to fixation to enrich the mitotic fraction (Fig. 2A) and in the cells that were left untreated (Fig. 2B). At the same time, no signal was detected in the control experiments, where primary or secondary antibody was omitted (data not shown). A similar pattern has been described also in the case of E2-dependent chromatin tethering of the URR reporter plasmid in CHOBgl40 and other cell lines, as revealed by FISH (26).
FIG. 2.
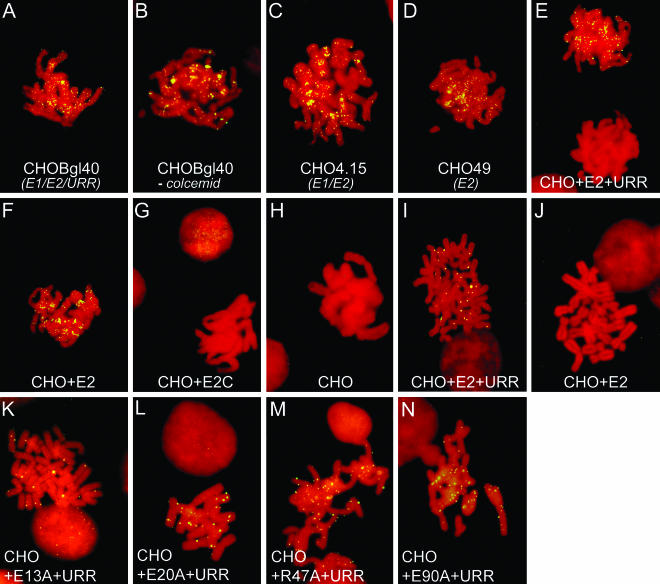
In situ analysis of the attachment of E2 protein, or URR-containing reporter plasmid, to mitotic chromosomes, as revealed by ChIF or FISH assays, respectively. The FITC signals corresponding to E2 protein or URR reporter appear as yellow dots on the red background of propidium iodide-counterstained chromosomes. (A to H) ChIF analysis of E2 protein; (I to N) FISH analysis of URR reporter. (A) CHOBgl40 cells (express E1 and E2, stably maintain URR reporter plasmid); (B) CHOBgl40 cells without Colcemid treatment; (C) CHO4.15 cells (express E1 and E2); (D) CHO49 cells (express E2); (E and I) CHO cells transfected with E2 expression vector pCGE2 and URR reporter pNeoBgl40; (F and J) CHO cells transfected with pCGE2 alone; (G) CHO cells transfected with pCGE2C and pNeoBgl40; (H) mock-transfected control cells. (K to N) FISH analysis of CHO cells transfected with mutant pCGE2 (E13A, E20A, R47A, and E90A, respectively) and URR reporter plasmid.
We then analyzed the chromosomal localization of E2 protein by ChIF in the following cell-lines: CHO4.15, which expresses E1 and E2 but lacks stably maintained URR-carrying reporter plasmid (Fig. 2C); CHO49, which expresses only E2 (Fig. 2D); and CHO cells that were transiently transfected either with the E2 expression construct alone (Fig. 2F) or together with BPV1 URR reporter plasmid (Fig. 2E). In all these experiments, the random speckled pattern of bound E2 on mitotic chromosomes was observed, which was indistinguishable from the pattern we saw in the case of CHOBgl40 cells. Thus, the pattern of E2 association with mitotic chromosomes is not affected by the presence of URR reporter plasmid with multiple E2 binding sites or by moderate E1 coexpression. In addition, the ChIF analysis of transiently transfected CHO cells demonstrates that the E2 localization on chromosomes is not specific to established cell lines and can be studied in transiently transfected cells. Only very few interphase nuclei, but no metaphase chromosomes, showed E2-specific staining if E2C protein lacking the N-terminal domain was expressed in CHO cells (Fig. 2G), and no specific signal was observed in mock-transfected cells (Fig. 2H). This is in agreement with the data from previous studies that have pointed to the importance of the N-terminal part of the E2 protein in interactions with chromatin (6, 52) and demonstrates that our method specifically analyzes the chromatin-bound proteins.
We also examined the localization of BPV1 URR reporter plasmid on metaphase chromosomes in the CHO cells that were transiently cotransfected with the respective reporter and E2 expression vector. FISH analysis revealed that the localization of the URR reporter in these cells (Fig. 2I) follows the speckled pattern, which is similar to that observed in CHOBgl40 cells that stably maintain the same reporter (26). As a negative control, the cells transfected with either the URR-containing plasmid or E2 expression vector alone did not show any signal on mitotic chromosomes by FISH analysis (Fig. 2J and data not shown). These data show that the E2-dependent localization of the extrachromosomal BPV1 URR reporter plasmid, like that of the E2 protein, follows a similar pattern on the mitotic chromosomes of both transiently transfected and established cell lines.
Chromatin attachment of E2 protein is abolished by deletions in the transactivation domain.
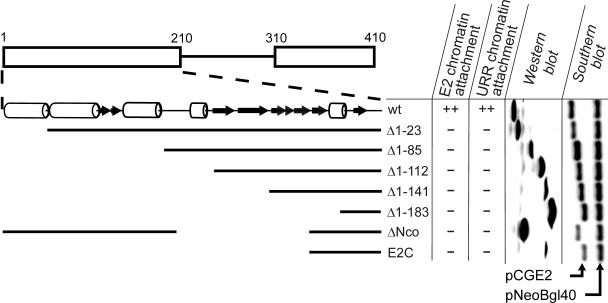
It has been shown previously that the N-terminal transactivation domain of the E2 protein itself can associate with mitotic chromosomes (6). In the case of EBNA1 and LANA1, the chromatin attachment activity has been mapped to specific linear subsequences of the protein (36, 45). To test whether similar linear sequences with chromatin attachment activity are also present in the E2 protein, we performed a deletion mapping of the E2 N-terminal domain. A panel of E2 deletion constructs was transiently expressed in CHO cells, and the attachment of the respective proteins to metaphase chromosomes was studied by ChIF. As shown in Fig. 2E, we can discriminate between E2-positive and E2-negative chromosome spreads from different metaphase cells without merging images or changing filters on the microscope. Therefore, it is possible to count the number of total metaphase cells as well as the number of E2-positive metaphase cells, which enables us to determine the percentage of E2-positive metaphase cells as a quantitative evaluation of the activity of different E2 proteins in our chromosome attachment assay. The fraction of positive metaphase cells in the case of wt E2 varied between 10 to 20% from the total population, depending on the transfection efficiency in the respective experimental series. To enable the comparative analysis of the data from different experimental series, the fraction of E2-positive metaphase cells in the parallel control transfections with wt E2 protein was used as a basis for normalization of the data from transfections with mutant proteins. The results from at least four independent experiments with E2 deletion mutants are summarized in Fig. 3 (E2 chromatin attachment column). In accordance with previously published results (6, 52), we could not detect any E2-specific signal on the metaphase chromosomes from the cells expressing the E2C protein that lacks the majority of the N-terminal domain (Fig. 3, lane E2C). The same results were obtained even if much higher levels of the protein were expressed in the cells after transfection of higher amounts of the respective expression construct (data not shown). The analysis of E2 proteins with shorter truncations showed that even the removal of the first 23 amino acids from the N-terminal domain (Fig. 3, lane Δ1-23) as well as longer N-terminal deletions (Fig. 3, lanes Δ1-85, Δ1-112, Δ1-141, and Δ1-183) and internal deletions (Fig. 3, lane ΔNco) leads to the complete loss of chromosome attachment activity of the protein. The expression level of E2 protein and the amount of URR plasmid in analyzed cells were found to be essentially on similar levels in all experiments (Fig. 3). All E2 deletion mutants also localized to cell nucleus (data not shown).
FIG. 3.
Summary of data from ChIF and FISH analyses of E2 deletion mutants. Over 100 individual mitotic metaphases in the case of the FISH assay and over 200 individual mitotic metaphases in the case of ChIF assay were examined in each separate experiment. The average data from at least four independent series are summarized on the figure. Activity less than 5% from the wt is indicated by −, and more than 80% activity from the wt is indicated by ++. As a control, the data from parallel Western and Southern blot analyses are shown, to indicate the presence of E2 protein and URR reporter plasmid in transfected cells. Arrowheads indicate the approximate positions of signals corresponding to different E2 expression plasmids (pCGE2) and the URR reporter plasmid (pNeoBgl40) on the Southern blot. At the top of the figure, the schematic representation of the E2 N-terminal domain is indicated, with alpha-helical regions of the protein marked as open barrels and beta-sheets marked as dark arrows.
In the ChIF experiments described above, the E2 expression constructs were cotransfected into the cells together with a BPV1 URR reporter plasmid. That enabled us to perform the parallel FISH analysis of the transfected cells to determine the ability of E2 proteins to mediate the chromatin tethering of extrachromosomal DNA constructs with BPV1 MME. The corresponding results are summarized in Fig. 3 (URR chromatin attachment column). As expected, on the basis of results from ChIF studies, none of the E2 deletion mutants was able to mediate the tethering of URR reporter to the mitotic chromosomes. All the deletion mutants used in our studies can bind to the single E2BS (30) and are thus expected to bind to BPV1 URR thorough the E2 DBD. Therefore, the lack of chromatin attachment reflects the defects in the interactions between the E2 N-terminal domain and the host chromosomes. These data indicate that the attachment of E2 protein to host chromosomes is required for URR-dependent tethering of the extrachromosomal DNA molecules to the cell chromosomes in the absence of other viral proteins. Most importantly, the ChIF and FISH analyses of E2 deletion mutants suggest that the E2 transactivation domain, in contrast to two known attachment proteins from herpesviruses, EBNA1 and LANA1, is unlikely to contain simple linear sequences or subdomains responsible for the chromatin attachment activity.
E2 chromatin attachment activity can be modulated by single-point mutations in the N-terminal transactivation domain of the protein.
Because the deletions in the E2 transactivation domain had strong effects on chromatin attachment, we next decided to test the E2 mutants with less drastic changes in the transactivation domain in these assays. In the following experiments, we used E2 proteins that carry single amino acid changes of the conserved charged residues to alanine in N-terminal domain. These mutant proteins were previously characterized in transient replication, transactivation, and sequence-specific DNA-binding assays (1). Every mutant protein has significant activity at least in one of these assays, and all are localized to the cell nucleus (1). Therefore, it is unlikely that any of these mutations could cause major inactivating structural changes in the E2 protein.
We transfected the mutant E2 expression constructs and URR reporter plasmid into the CHO cells. The expression of E2 protein and the amount of URR plasmid in transfected cells were found to be on similar levels in all experiments (Fig. 4B). The attachment of E2 proteins to metaphase chromosomes was studied by ChIF, and the ability of E2 proteins to tether the URR reporter plasmid to host mitotic chromosomes was studied by FISH essentially as described above in the case of truncated E2 constructs (examples of images of FISH analysis of some mutants are shown in Fig. 2K to N). The fraction of E2- or URR reporter-positive mitosis in the respective assays was estimated relative to the positive fraction in the case of wt E2-transfected cells in each separate series. The results from at least four independent experiments for each mutant are summarized in Fig. 4A.
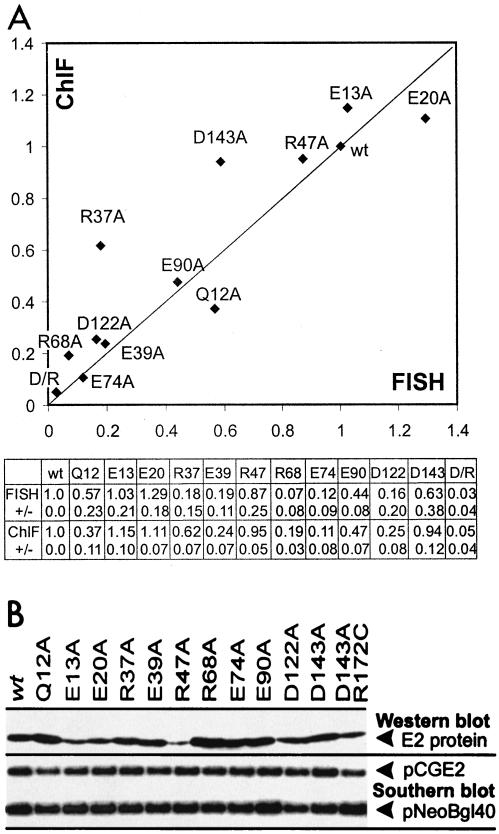
FIG. 4.
Summary of data from ChIF and FISH analyses of E2 point mutants. (A) Comparison of the results from ChIF (chromosome attachment of E2 proteins, on y axis) and FISH (tethering of URR reporter to chromosomes, on x axis) analyses. All mutations are as indicated on the figure; D/R corresponds to the double mutant D143A/R172C. Over 100 individual mitotic metaphases in the case of the FISH assay and over 200 individual mitotic metaphases in the case of ChIF assay were examined in each separate experiment. The average data from at least four independent series are summarized on the figure. The relative activity in the case of each mutant protein was estimated as a percentage of positive metaphases relative to percentage of positive metaphases in the wt E2-transfected cells, with the latter set as 1.0 in each separate series. For a better overview, the error bars are omitted from the graph. The numerical values as well as average deviations (±) are shown at the bottom of the figure. (B) As a control, the data from parallel Western and Southern blot analyses are shown to indicate the presence of E2 protein and URR reporter plasmid in transfected cells in the ChIF and FISH assays. Arrowheads indicate approximate positions of signals corresponding to different E2 expression plasmids (pCGE2) and the URR reporter plasmid (pNeoBgl40) on the Southern blot.
According to ChIF analysis (Fig. 4A), five of the tested mutant proteins, E39A, R68A, E74A, D122A, and double mutant D143A/R172C (D/R), were defective in chromosome attachment. Four mutants, E13A, E20A, R47A, and D143A, were attached to the mitotic chromosomes as efficiently as wt E2. The Q12A, R37A, and E90A proteins formed an intermediate activity group, with diminished, but still clearly displayed activity in the ChIF assay. These data show that E2 chromatin attachment can be affected to various extents by single-point mutations in the transactivation domain. The expression level of E2 varies on a cell-to-cell basis in the transiently transfected cell population (more than 2 orders of magnitude) (Fig. 5 and data not shown). If the mutations in E2 weaken its interaction with mitotic chromosomes but do not abolish it completely, the binding could still occur in the cells expressing higher E2 levels. Consequently, in the conditions where the average expression level is the same for different mutant E2 proteins, the decreased interaction leads to a percentage of E2-positive metaphase cells that is lower than in the case of wt protein. The results from parallel FISH analysis of cells from the same transfection series demonstrate that the ability of mutant E2 proteins to tether the URR reporter to mitotic chromosomes correlates well with the activity of respective mutants in chromatin attachment (Fig. 4A). Good correlation between activities of E2 mutants in FISH and ChIF assays provides an additional indication that chromatin attachment of the E2 protein is required for the tethering of the URR to mitotic chromosomes in the absence of other viral proteins. On the other hand, even the adequate interaction of E2 protein with both mitotic chromatin and its binding sites in the URR could be insufficient for the tethering of URR to chromosomes under certain conditions. This is suggested by the behavior of R37A and D143A proteins, which seem to be more active in protein attachment than URR tethering. At the same time, both proteins, as well as all the other mutant proteins tested in the series, have been shown to effectively bind to E2-specific binding sites on DNA. Therefore, the possibility of point mutations leading to changes in the URR binding activity of the protein could be excluded. It should be noted, however, that for unknown reasons the results of the FISH assay in the case of the D143A protein were more diverse in different series than in the case of other mutant proteins (the average deviation was ∼0.4 instead of the usual ∼0.2).
FIG. 5.
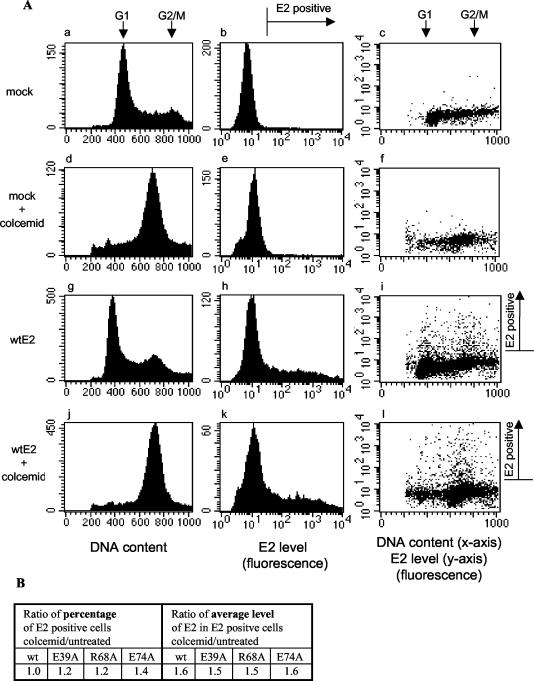
Analysis of cell cycle dependence of E2 expression level in transfected CHO cells. The transfected cells were fixed, the E2 protein was visualized by mixture of anti-E2 monoclonal antibodies and FITC-conjugated secondary antibody, and DNA content was visualized with 7-AAD, as described in Materials and Methods. The intensities of FITC and 7-AAD signals were measured in cells by flow cytometry. (A) On the dot plots in the right column, the relative DNA content (x axis) and the E2 level in respective cells (y axis) are analyzed for mock- or wt E2-transfected cells. The cells from the same transfections were split to separately analyze Colcemid-treated and untreated populations, exactly as indicated on the left of the respective rows. The histograms in the left column illustrate the distribution of cells according to their relative DNA content, and those in the middle column illustrate the distribution of cells according to relative E2 expression level in the same analysis. Note that in the case of E2 expression analysis, only the signals above the autofluorescence threshold represent the E2-expressing cells (indicated at the top of the middle column and to the right of the right column). (B) The percentage of E2-positive cells and the average E2 level in E2-positive cells were measured in the case of CHO cells transfected with wt or partitioning-deficient mutant E2 expression constructs. The ratio of respective parameters in the case of Colcemid-treated cells relative to untreated cells is shown. Values shown are the averages of the results from two separate experiments.
There is no specific degradation of partitioning-deficient E2 proteins during mitosis.
According to the Western blot analysis (Fig. 4B), the total expression levels of different E2 proteins are on approximately similar levels. Therefore, the deficient partitioning activity of some E2 mutant proteins was not a result of the general destabilization and degradation of the respective proteins. However, if the destabilization occurred only during a short period of the cell cycle, such an effect could have been left unnoticed on the background of the protein from cells in the remaining cell cycle phases. The ChIF and FISH analyses were performed on mitotic cells. Therefore, the deficient partitioning activity of certain E2 mutant proteins could have also been a result of the mitosis-specific degradation of the respective proteins. To examine this possibility, we performed a simultaneous analysis of the E2 level and DNA content of transfected cells by measuring these parameters on a cell-by-cell basis by flow cytometry (see Materials and Methods for details). As shown in Fig. 5A, treatment with the efficient mitotic blocker Colcemid for 12 h causes the vast majority of cells to gather at the G2/M peak (compare panels a and g to panels d and j). The homogenous auto-fluorescence peak, which is not affected by Colcemid treatment, appears in the case of mock-transfected cells (compare panel b to panel e). The transfection of cells with E2 expression constructs leads to the appearance of signals with higher fluorescence intensities. These signals, corresponding to E2-expressing cells, reveal a very heterogeneous distribution of the E2 expression level in both Colcemid-treated and control cells (panels h, i, k, and l). E2-expressing cells are efficiently present in the G2/M subpopulation, and the number of E2-expressing cells in this cell cycle period rises significantly as a result Colcemid-induced mitotic enrichment (compare Colcemid-treated cells on panel l to untreated cells on panel i). The results of quantitative analysis of these data and the data from experiments with several E2 mutant proteins inactive for partitioning (E39A, R68A, and E74A) are shown in Fig. 5B. The percentage of wt E2-positive cells in the Colcemid-treated population relative to the untreated transfected population remains unaltered (the ratio of respective percentages equals 1.0). If the mitosis-specific E2 degradation took place, the Colcemid-induced mitotic enrichment should have led to a diminished number of cells with detectable E2 levels, reflected by the lower percentage of these cells in the total population. There is also no apparent mitotic degradation in the case of partitioning-deficient mutant proteins, as the percentage of E2-expressing cells was certainly not lower but even seemed to rise (1.2 to 1.4 times higher) as a result of mitotic enrichment. The average level of E2 in E2-positive cells was also somewhat increased in the mitosis-enriched population (1.5 to 1.6 times higher in Colcemid-treated cells), this time equally for wt and mutant E2 proteins. In conclusion, there is certainly no specific degradation of partitioning-deficient mutant E2 proteins during mitosis.
Analysis of partitioning function of wt E2 protein.
A good correlation between chromatin attachment and efficient long-term maintenance of extrachromosomal URR plasmids (26) or BPV1 genomes (33) suggests that E2-dependent attachment of the MME-containing plasmids or virus genomes is necessary for their successful nuclear retention and partitioning in dividing host cells. However, this correlation does not prove that the E2- and MME-dependent chromatin attachment is sufficient for partitioning of the episomal MME-containing plasmids.
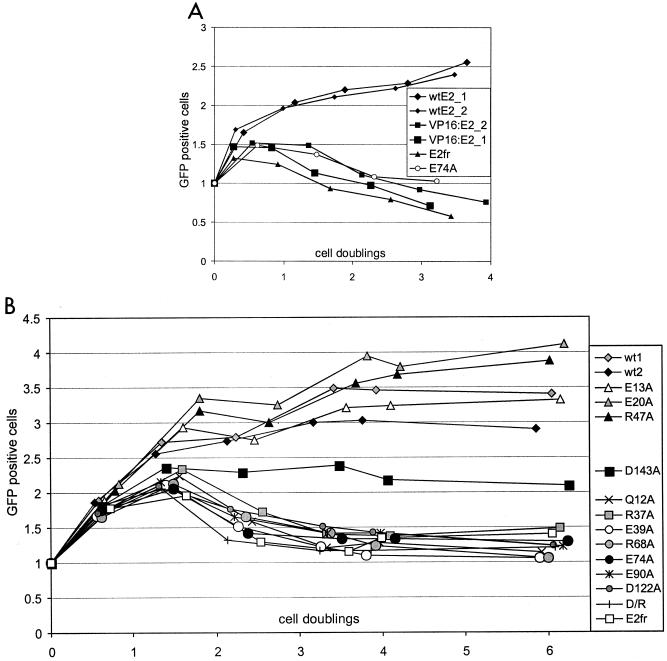
To prove this, we characterized the E2 protein in a functional partitioning assay developed in our laboratory (A. Männik, K. Janikson, and M. Ustav, unpublished data). For these experiments, the suspension culture of the Jurkat cell line was transfected with pRetE2 reporter plasmid that contains an expression cassette for destabilized EGFP under the strong constitutive CMV promoter, an expression cassette for E2 (or its derivatives), and MME (oligomerized E2 binding sites) (Fig. 1). The number of GFP-positive cells per milliliter of medium and the total number of cells per milliliter were determined by fluorescence-activated cell sorter analysis over the course of several days (up to 1 week) after transfection. The constructs used in the partitioning assay did not contain the BPV1 origin of replication and the E1 protein was not expressed in the cells; thus, the transfected reporter plasmid was unable to replicate. In this way, we could examine the nuclear maintenance and partitioning of the transcriptionally active plasmids on a cell-by-cell basis in proliferating cells. The use of the destabilized version of EGFP (with a half-life of ∼1 h) ensures that the steady-state EGFP level in the cells responds quickly to the changes in reporter plasmid copy number. The number of GFP-positive cells in this assay depends on several aspects. If both nuclear retention and partitioning of the reporter plasmid take place in dividing cells and if the copy number of the reporter in host cells is sufficient, the increase in the number of GFP-positive cells should occur over the course of the first cell divisions. On the other hand, in the case of the nuclear retention of plasmids that lack any partitioning mechanism, the number of GFP-positive cells should remain constant. The transfection results in a cell population that is heterogeneous in regards to the plasmid copy number and, consequently, GFP expression level per cell. After every cell division, the copy number of the nonreplicating reporter plasmid decreases even if active partitioning takes place. As a result, the GFP signal in the cells with a lower copy number will fall below the threshold level sufficient for the detection under the assay conditions. As shown in Fig. 6A, where the number of GFP-positive cells is plotted against cell doublings, the number of positive cells in the case of reporter plasmid expressing wt E2 increases over the course of at least four doublings, which indicates that, in addition to nuclear retention, the active partitioning has taken place. On the other hand, in the case of reporter expressing E2fr protein inactive in specific DNA binding as a result of a frameshift in the hinge region, or E74A mutant inactive in chromatin attachment, the number of GFP-positive cells starts to decrease after the first doubling. The reporter plasmids, which express the E2fr or E74A mutant, behave in the partitioning assay very similarly to those that do not express E2 or lack MME (data not shown). This indicates that the nuclear retention and partitioning of the MME reporters expressing attachment-defective E2 proteins is essentially on the basal residual level. The apparent E2-MME-dependent partitioning in our assay is not a primary effect of the transactivating activity of the E2 protein, as the reporter expressing a fusion protein VP16-E2, which is a potent transactivator, was lost from the cells at the rate comparable to E2fr and the transactivation-defective mutant E74A. In the experiments that are shown in Fig. 6A, the wt E2 and VP16-E2 reporters are transfected in duplicate to show the reproducibility of the assay. These data indicate that E2 has a specific effect on MME-containing plasmid partitioning. This effect also requires, in addition to the intact C-terminal DBD, the N-terminal transactivation domain from the E2 protein, which cannot be replaced by the heterologous transactivating domain.
FIG. 6.
Plasmid partitioning assay. Jurkat cells were transfected with pRetE2 plasmids containing MME and expression cassettes for E2 (or its derivatives) and destabilized EGFP protein. The total number of cells per milliliter and the number of EGFP-positive cells per milliliter were determined by fluorescence-activated cell sorter analysis at different time points. From these data, the number of doublings and the number of EGFP-positive cells per milliliter relative to the first time point of each separate transfection were calculated. Zero doublings corresponds actually to first time point, ∼20 h posttransfection. (A) Partitioning properties of plasmids expressing E2 or its derivatives. The number of EGFP-positive cells per milliliter on the y axis is plotted against cell doublings on the x axis. Note that the constructs expressing wt E2 or VP16-E2 were transfected in duplicate to show the reproducibility of the assay. (B) A similar partitioning assay was performed with plasmids expressing different point-mutated E2 proteins; D/R corresponds to the double mutation D143A/R172C. The wt E2 construct was transfected in duplicate.
Effective chromatin attachment is required for partitioning function of E2 protein.
To look for a possible connection between E2-dependent chromatin attachment and partitioning processes, we tested the panel of E2 point mutants with known attachment and plasmid-tethering activities in the partitioning assay described in the previous section. As shown in Fig. 6B, where the relative number of GFP-positive cells is plotted against cell doublings, the graphs corresponding to different point-mutated E2 constructs are clustering into two groups, those similar to the wt E2 and those similar to E2fr carrying a frameshift in the hinge region. The only exception is the graph that corresponds to D143A, which is located between these two clusters. Similar clustering was apparent in at least four independent experiments and was preserved even if the number of GFP-positive cells rather than the normalized relative value was plotted against time or cell doublings. All the mutant E2 proteins were expressed at comparable levels when examined by Western blotting (data not shown). The wt-like cluster consists of graphs corresponding to the data from experiments with E2 proteins that carry point mutations E13A, E20A, or R47A. The number of GFP-positive cells in the case of these E2 mutations increases over the course of at least 4 doublings, exactly as in the case of wt E2. All the members of this group also behave like wt E2 in chromatin attachment assays. The E2fr-like cluster includes graphs corresponding to experiments with E2 mutants Q12A, R37A, E39A, R68A, E74A, E90A, D122A, and D/R. In the case of these mutations, the number of GFP-positive cells already starts to decrease after the first cell doubling and continues to do so over the examined time period. All the mutants in this cluster are also at least partially defective in chromatin attachment assays. These data show that effective chromatin attachment is required for E2-dependent partitioning function.
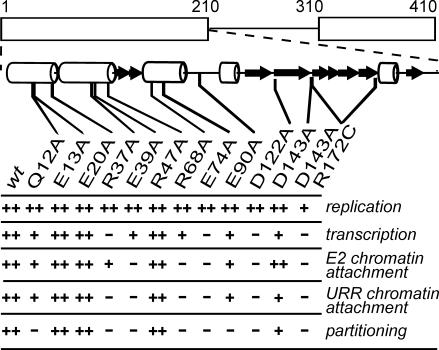
In Fig. 7, the data from the partitioning assay are compared to the activity of the respective mutant proteins in chromatin binding, tethering of the URR reporter plasmid to mitotic chromatin, transcription activation, and initiation of transient replication. The mutants have been divided into three groups in each respective assay, according to their activity relative to that of wt E2. The comparison shows that the group of mutants, which had wt-like activity in both chromatin attachment assays and in partitioning (E13A, E20A, and R47A), also demonstrates a wt-like activity in transcription and replication assays. On the other hand, none of the remaining E2 mutants behaves similarly in all five assays. From the eight point mutants inactive in the partitioning assay, seven perform like the wt protein in the transient replication assay and D/R retains at least partial activity. R37A, E74A, D122A, and D143A/R172C (D/R) from the same group are inactive both in partitioning and transactivation, but Q12A, E39A, R68A, and E90A retain at least some activity in transactivation assays. Therefore, the activity of the E2 protein in the partitioning assay, if compared to its activities in the chromatin attachment, transcription activation, and replication initiation, is the most sensitive to point mutations in the conserved charged positions of the N-terminal domain.
FIG. 7.
The data from ChIF (E2 chromatin attachment), FISH (URR chromatin attachment), and partitioning assays are compared to the activity of respective mutant E2 proteins in transcription activation (with simple artificial reporter pSV3BSCAT) and transient replication assays according to the method of Abroi et al. (1). Activity less than 30% from the wt is indicated by −, activity more than 80% from the wt is indicated by ++, and activity between 30 and 80% from the wt is indicated by +.
Analysis of the location of structural determinants on the homology-modeled E2 activation domain.
Publication of the crystal structure of the full-length HPV16 E2 N-terminal domain (3) enables us to use the homology modeling programs to compose an atomic model for the full N-terminal domain from BPV1 E2. Despite the low sequence similarity between BPV1 and HPV16 E2 C-terminal DBDs (∼33% identity), the resolved structures of those domains are very similar, with a root mean square of 0.9 Å for backbone atoms according to FSSP data for PDB entries 2BOP (BPV1 E2 DBD) and 1BY9 (HPV16 E2 DBD) (23). It is, for example, less than the differences between the different monomers in the structure of the HPV18 E2 DBD (PDB code 1F9F, containing 4 E2 chains per crystallographic asymmetric unit; root mean square for most different chains, 1.1 Å) according to the FSSP database. Therefore, it is reasonable to assume that the transactivation domains of BPV1 E2 and HPV16 E2, which have the same sequence identity (33% in the modeled region) as the corresponding DBDs, could also have very similar structures. The designed model for the BPV1 E2 N terminus is certainly acceptable for studying the localization of mutated amino acids.
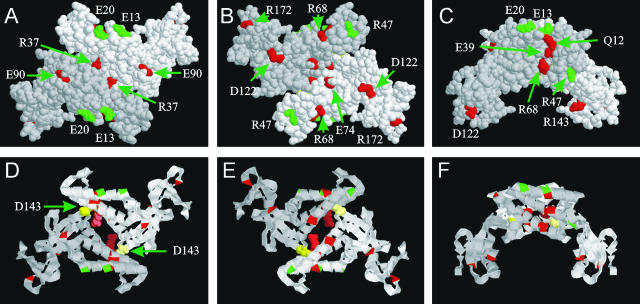
We constructed the model structure for the BPV1 E2 N-terminal domain between amino acids 1 and 199, by using the automatic homology modeling program Swiss-Model (20, 41). As shown in Fig. 8A and B, the mutated amino acids localized quite randomly over the surface of the N-terminal domain of E2. However, a single continuous patch of mutated amino acids can be observed (Fig. 8C). This consists of amino acids Q12, E13, E39, and R68, which are located on three neighboring alpha-helices forming the subdomain N1 (according to nomenclature by Antson et al.) (3).
FIG. 8.
Localization of the mutated amino acids on the homology modeled structure of dimerized N-terminal domains of the BPV1 E2. The model is viewed from the angle which is rotated 180° over the x axis in panels B and E and 90° in panels C and F relative to panels A and D. The respective models are presented in a space-fill mode in panels A to C and in ribbon mode in the corresponding panels D to F in the bottom row. The amino acids, the substitution of which does not affect the activity of E2 in partitioning assay, are shown in green; those inactivating the protein are shown in red; and those leading to medium activity are shown in yellow. One monomer (chain A) is shown in darker gray than another (chain B). Note that amino acids R37 and D143 on the putative dimerization surface are shown in space-fill mode on all panels and in darker red and yellow, respectively, in chain A than in chain B (see also supplemental material at http://www.ebc.ee/∼aabroi/art3).
Only two of the analyzed E2 point mutations, R37 and D143, are located directly on the N-terminal dimerization interface. These are the same mutations which led to differing activities of E2 by FISH and ChIF analysis (Fig. 4A and 7). The conserved amino acid R37 seems to be a very important player in N-terminal dimerization, as it is localized on the central region of the dimerization interface and forms 1 intrachain and 3 interchain hydrogen bonds on the proposed structure of BPV1 E2 and on the established structure of HPV16 E2 (3). One intra- and one interchain bond are conserved between these two structures.
DISCUSSION
The importance of the E2 protein and its binding sites on viral DNA in the process of BPV1 stable extrachromosomal maintenance in dividing cells was suggested initially by Piirsoo et al. (44). This suggestion was later supported by the findings that both the E2 protein and BPV1 genomes are attached to the mitotic chromosomes in the host cells and that the mutations in the E2 open reading frame may lead to defects in the chromatin attachment of both the E2 protein and the BPV1 genome (6, 33, 52). The E2 protein in trans as well as a sufficient number of E2 binding sites that form an MME in cis are prerequisites for chromatin attachment and long-term extrachromosomal maintenance of the BPV1 (26, 44). In the present study, we have performed analysis of the E2 protein as a central trans factor required for MME-dependent chromatin attachment and partitioning processes. We have used extensive mutational analysis of the N-terminal chromatin-binding domain of the E2 protein and relatively traditional approaches for analyzing the chromosome attachment of E2-MME as well as a novel assay for testing the E2-dependent nuclear maintenance and partitioning of nonreplicating MME reporters. Our analysis shows that the chromatin-bound E2 protein seems to localize into discrete centers on the mitotic chromosomes, appearing as speckles of bound E2. This pattern was altered neither by simultaneous moderate E1 expression nor by the presence of reporter plasmid carrying the URR portion of the BPV1 genome containing intact MME. The speckled pattern of the E2 protein localization on chromosomes is very similar to that in the case of E2-mediated tethering of URR reporters. In addition, the number of E2-specific dots on metaphase chromosomes seems to be roughly in the same range of that of the URR-specific dots, both in the established CHOBgl40 cell line and in CHO cells transiently cotransfected with E2 and the URR reporter. Therefore, it is tempting to speculate that the number of E2 localization centers on host chromosomes could be one of the parameters which determines the potential virus genome copy number during the latent maintenance stage in infected cells. However, additional studies are clearly required to support this speculation.
The N-terminal domain of the BPV1 E2 protein carries the determinants both necessary and sufficient for the localization of the protein to host chromosomes (6). This domain has also been shown to incorporate major determinants required for transactivation and replication initiator activities of the E2 protein. E2 is the only viral protein required for tethering of the BPV1 URR to host mitotic chromosomes (2, 26). We demonstrate here that in the same cell population, the E2 protein and the BPV1 URR are attached to chromosomes in the absence of other BPV1 proteins and genomic sequences. The analysis of the behavior of E2 proteins with N-terminal deletions and point mutations confirms that chromatin attachment of the E2 protein is necessary for the tethering of URR (containing intact MME) to mitotic chromosomes. Perhaps the most important conclusion based on our present study is that successful nuclear maintenance and partitioning of nonreplicating BPV1 MME reporters in proliferating cells is dependent on efficient E2-mediated chromatin attachment. On the other hand, the attachment of E2 protein to the mitotic chromosomes does not guarantee the mediation of equally efficient tethering of URR to mitotic chromosomes. It is demonstrated by the effect of mutations R37A and D143A, which cause the E2 protein to be more active in its chromatin attachment than in its mediation of the URR tethering, even though the respective mutant proteins bind to a single E2 binding site on the DNA with essentially the same efficiency as wt protein (1). Moreover, the E2-mediated tethering of the URR reporter does not necessarily lead to efficient partitioning, as the proteins with Q12A and E90A mutations are at least partial active in URR tethering but are essentially inactive in the partitioning assay. These data suggest that in addition to binding to the chromatin components required for URR tethering, binding to some additional target(s) of E2 is required for efficient BPV1 partitioning process. However, we cannot entirely exclude the alternative explanation that even relatively modest defects in chromatin attachment could perhaps have cumulative effects on the long-term extrachromosomal DNA retention over the course of multiple cell divisions. On the other hand, a still relatively good correlation of the results from the partitioning assay with the data from ChIF and FISH analysis shows that the information about chromatin attachment activity obtained from the latter two assays predicts well the behavior of E2 protein in the partitioning and long-term extrachromosomal maintenance of DNA constructs carrying BPV1 MME.
The defects in any of the E2 activities that we observed as a result of point mutations in the conserved surface amino acids of the N-terminal domain can be explained in two ways. First, the mutations that we introduced might have directly disrupted the interactions of the E2 N-terminal domain with the target proteins (or other molecular structures) essential for carrying out respective activity of E2. Second, the mutations might have led to certain structural changes in the protein, thus indirectly disrupting vital interactions with specific targets. In both cases, as a consequence, the interaction pattern of E2 with its molecular targets is altered. Previous analysis has shown that E2-dependent transient replication and chromosome tethering of BPV1 use clearly separated mechanisms, as indicated by the requirement for separate cis sequences from the URR (26, 44). The data presented in the present study show that from the mutants with wt-like activity in transient replication, four (E39A, R68A, E74A, and D122A) are essentially inactive, and four (Q12A, R37A, E90A, and D143A) have defects in the tethering of URR to chromosomes and in the partitioning assay. Therefore, BPV1 transient replication is separated from the chromosome tethering and partitioning also on the level of E2 as a trans factor. All three viral proteins that are known to mediate the tethering of genomes of the respective viruses to host cell chromosomes, E2, EBNA1, and LANA1, can also function as transcriptional modulators (17, 24, 29, 34, 48). Therefore, one might speculate about the possible overlap between transcriptional modulation and chromatin attachment (and consequently, partitioning) activities. However, differences in the sensitivity to point mutations exist not only in the case of E2-dependent replication initiation and partitioning but also in the case of transcription activation and partitioning, even though in the latter case they are somewhat less pronounced. This assumption is based on the effect of mutations Q12A, E39A, R68A, and E90A, which render E2 inactive in partitioning but retain at least partial activity in its transcription activation. These data suggest that at least some of the targets and interactions of the E2 N-terminal domain that participate in chromatin attachment and partitioning are not involved in transcription activation or replication initiation.
The structure of the N-terminal domain of the HPV16 E2 protein (3) and part of that of the HPV18 E2 protein have been resolved (22). As expected, the homology of different E2 proteins on the amino acid sequence level is reflected well in the striking similarity of these proteins on the structural level. As no structural information is available yet for EBNA1 or LANA1 chromatin attachment domains, the E2 activation domain is the only chromatin attachment domain with available structural information. Four amino acids that were analyzed in our studies, Q12, E13, E39, and R68, form a continuous patch on the surface of the homology-based structure model of the BPV1 E2 protein. This patch is situated on the opposite sides of the homodimer of the BPV1 E2 N-terminal domains, modeled on the basis of a similar dimer that has been previously demonstrated to exist in the case of HPV16 E2 (3). The point mutations of three amino acids from this group, Q12A, E39A, and R68A, inactivate the partitioning function but retain full activity in replication initiation and significant activity in transactivation. As only one of the other mutations analyzed, E90A, has a similar specific inactivating effect on partitioning function, three of the four mutants from our panel specifically inactive in partitioning are located in the same continuous surface region of the protein (see supplemental material at http://www.ebc.ee/∼aabroi/art3). Thus, we can speculate that this surface may represent one of the targets, the interaction of which with cellular factors is responsible for E2 mediated partitioning. This surface region of the protein is also the most-conserved surface in the family of E2 transactivation domains according to the ConSurf server analysis (18) (see supplemental material at http://www.ebc.ee/∼aabroi/art3). However, we cannot entirely exclude the possibility that respective residues may allosterically control some other E2 N-terminal surfaces directly involved in chromatin attachment. Homology modeling of the N-terminal homodimer of BPV1 E2 reveals also that the amino acids R37 and D143, located on the proposed N-terminal dimerization interface, are responsible for multiple predicted hydrogen bonds that stabilize the dimeric structure. As discussed above, the mutation of these amino acids causes the BPV1 E2 protein to be more active in its own chromatin attachment than in mediation of the URR tethering. This might suggest that the dimerization of N-terminal domains, even though it is not required for the efficient attachment of BPV1 E2 protein to mitotic chromosomes, could somehow affect the efficiency of URR tethering to mitotic chromosomes. The cooperative binding of E2 protein to two neighboring binding sites on DNA has been defined as a function of the E2 transactivation domain (38, 39). Therefore, the fact that the R37A mutation leads to defects in the cooperative DNA binding of BPV1 E2 and that the complex of the respective protein with DNA displays diminished mobility in gel shift assays (A. Abroi and M. Ustav, unpublished data) also supports the possibility of N-terminal dimerization in the case of BPV1 E2.
Deletion analysis of EBNA1 and LANA proteins has shown that chromatin attachment is achievable by short, probably relatively loosely structured protein fragments (25, 27, 45). In this aspect, BPV1 E2 is likely to differ from these two proteins. It has been shown earlier that internal deletions between amino acids 41 to 120 and 51 to 120 abolish the attachment of E2 protein to chromosomes. However, these data do not exclude the possibility that the region responsible for chromatin binding may lie between amino acids 50 and 120. In this study, we show that the deletion of as few as the first 23 amino acids of the E2 protein and longer truncations and an internal deletion that retains the first 91 amino acids from the N-terminal domain (dNco) all completely abolish the chromosome attachment activity. These data strongly suggest that E2 has no linear subsequence responsible for chromatin attachment. The truncations that we have introduced in the N-terminal domain led to the exposure of an epitope which is poorly accessible in the wt E2 protein (30). Therefore, these truncations are likely to considerably affect the structural integrity of the E2 transactivation domain. In addition, the amino acids, point mutations of which lead to inactivation of E2 chromatin attachment and partitioning functions, are located all over the N-terminal sequence. In conclusion, the overall structural integrity of the N-terminal domain rather than the intactness of some short linear sequences is most likely required for the chromatin attachment and partitioning functions of the E2 protein. Therefore, component(s) of the host chromatin, which are targeted by respective activities of the E2, can be expected to be different from those used by EBNA1 and LANA1.
The partitioning activity of E2, if compared to other activities tested, is the most sensitive to the point mutations in the N-terminal domain of the protein. In the initiation of replication, which is relatively insensitive to point mutations (10, 37), the primary function of E2 is to load E1 helicase to the origin of replication (54). On the other hand, the process of transcription activation by papillomavirus E2 is much more complex, involving interactions with numerous components of the transcription initiation complex (7, 47, 53, 62, 63) as well as with coactivators (31, 32, 40, 42). It could explain why the transactivation function of BPV1 E2 is more sensitive to different point mutations than the replication function is. The observations that the partitioning function is even more affected by point mutations than the transactivation function and that in the case of both activities the inactivating mutations are located diffusely over the surface of the E2 N-terminal domain suggest that the BPV1 E2-dependent partitioning represents a very complex process involving multiple molecular targets on the host chromatin. Alternatively, we can perhaps expect the complex interaction with the single chromatin receptor to involve composite rather than linear surface elements of E2 N-terminal domain, with several interactions acting additively.
Several previously published studies have provided us with extensive information about the influence of N-terminal point mutations on the activities of the BPV1 E2 protein. In light of our present study, the data by Brokaw et al. describing the behavior of E2-mutated BPV1 genomes in focus formation assay are especially interesting (10). In that study, all but 2 (E20D and Y159F) of 18 BPV1 genomes that carry mutations in E2 N-terminal domain show wt level of focus formation. Eight mutations that did not affect E2 transactivation, replication initiation, and cooperative origin binding with E1 led to defective transformation. These eight mutants include the mutations at positions 12, 37, and 122. In our present study, the mutations Q12A, R37A, and D122A in the same positions were inactive in the partitioning assay, which could explain the described transformation defects by the deficient partitioning and consequently deficient stable episomal maintenance of BPV1 genomes. In addition, the mutation E20A is active in our partitioning assay, and the mutation of the respective residue E20D in the context of the full-length genome does not cause any transformation defects.
One of the most important unanswered questions about BPV1 chromatin attachment and stable extrachromosomal maintenance is, of course, what the actual molecular targets are from the host chromatin side. Different mutant E2 proteins, whose behavior in chromatin attachment and partitioning compared to other activities of the protein have been extensively analyzed in this paper, should provide a useful tool for future studies that try to address this question.
Acknowledgments
We thank Andres Männik and Kadri Janikson for help with the partitioning assay, Anne Kalling for technical assistance, and Reet Kurg and Arvi Jõers for critical reading of the manuscript.
This study was supported by grants 4475 and 4476 from the Estonian Science Foundation, grant INTNL55000339 from the Howard Hughes Medical Institute, and grant CT96-0918 from the European Union.
REFERENCES
- 1.Abroi, A., R. Kurg, and M. Ustav. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allikas, A., D. Ord, R. Kurg, S. Kivi, and M. Ustav. 2001. Roles of the hinge region and the DNA binding domain of the bovine papillomavirus type 1 E2 protein in initiation of DNA replication. Virus Res. 75:95-106. [DOI] [PubMed] [Google Scholar]
- 3.Antson, A. A., J. E. Burns, O. V. Moroz, D. J. Scott, C. M. Sanders, I. B. Bronstein, G. G. Dodson, K. S. Wilson, and N. J. Maitland. 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403:805-809. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 7.Benson, J. D., R. Lawande, and P. M. Howley. 1997. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J. Virol. 71:8041-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg, M., and A. Stenlund. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breiding, D. E., M. J. Grossel, and E. J. Androphy. 1996. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology 221:34-43. [DOI] [PubMed] [Google Scholar]
- 10.Brokaw, J., M. Blanco, and A. McBride. 1996. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J. Virol. 70:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, C., S. Upmeyer, and P. Winokur. 1998. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions. Virology 241:312-322. [DOI] [PubMed] [Google Scholar]
- 12.Corina, K., S. Grossman, J. Barsoum, S. Prakash, E. Androphy, and R. Pepinsky. 1993. The tryptophan bridge is a critical feature of the papillomavirus E2 DNA binding domain. Virology 197:391-396. [DOI] [PubMed] [Google Scholar]
- 13.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 14.Delecluse, H. J., S. Bartnizke, W. Hammerschmidt, J. Bullerdiek, and G. W. Bornkamm. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J. Virol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dostatni, N., F. Thierry, and M. Yaniv. 1988. A dimer of BPV-1 E2 containing a protease resistant core interacts with its DNA target. EMBO J. 7:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson, M., and M. Botchan. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser, F., T. Pupko, I. Paz, R. E. Bell, D. Bechor-Shental, E. Martz, and N. Ben-Tal. 2003. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19:163-164. [DOI] [PubMed] [Google Scholar]
- 19.Grossel, M. J., F. Sverdrup, D. E. Breiding, and E. J. Androphy. 1996. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J. Virol. 70:7264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 21.Harris, A., B. D. Young, and B. E. Griffin. 1985. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J. Virol. 56:328-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris, S. F., and M. R. Botchan. 1999. Crystal structure of the human papillomavirus type 18 E2 activation domain. Science 284:1673-1677. [DOI] [PubMed] [Google Scholar]
- 23.Holm, L., and C. Sander. 1996. Mapping the protein universe. Science 273:595-603. [DOI] [PubMed] [Google Scholar]
- 24.Howley, P. 2001. Papillomaviruses and their replication, p. 2197-2230. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kivimae, S., A. Allikas, R. Kurg, and M. Ustav. 2001. Replication of a chimeric origin containing elements from Epstein-Barr virus ori P and bovine papillomavirus minimal origin. Virus Res. 75:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurg, R., J. Parik, E. Juronen, T. Sedman, A. Abroi, I. Liiv, U. Langel, and M. Ustav. 1999. Effect of bovine papillomavirus E2 protein-specific monoclonal antibodies on papillomavirus DNA replication. J. Virol. 73:4670-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, D., S. G. Hwang, J. Kim, and J. Choe. 2002. Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem. 277:6483-6489. [DOI] [PubMed] [Google Scholar]
- 32.Lee, D., B. Lee, J. Kim, D. W. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 275:7045-7051. [DOI] [PubMed] [Google Scholar]
- 33.Lehman, C., and M. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 35.Li, R., and M. R. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 36.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride, A., and G. Myers. 1997. The E2 proteins, p. 54-73. In G. Myers, C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses 1997. Los Alamos National Laboratory, Los Alamos, N.M.
- 38.Monini, P., I. Blitz, and E. Cassai. 1993. Cooperative DNA binding of the bovine papillomavirus E2 transcriptional activator is antagonized by truncated E2 polypeptides. J. Virol. 67:5668-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monini, P., S. Grossman, B. Pepinsky, E. Androphy, and L. Laimins. 1991. Cooperative binding of the E2 protein of bovine papillomavirus to adjacent E2-responsive sequences. J. Virol. 65:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76:11042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peitsch, M. C. 1996. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 42.Peng, Y. C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74:5872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pepinsky, R., S. Prakash, K. Corina, M. Grossel, J. Barsoum, and E. Androphy. 1997. Sequences flanking the core DNA-binding domain of bovine papillomavirus type 1 E2 contribute to DNA-binding function. J. Virol. 71:828-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 45.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prakash, S., S. Grossman, R. Pepinsky, L. Laimins, and E. Androphy. 1992. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 6:105-116. [DOI] [PubMed] [Google Scholar]
- 47.Rank, N. M., and P. F. Lambert. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakai, H., T. Yasugi, J. Benson, J. Dowhanick, and P. Howley. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 70:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 51.Simpson, K., A. McGuigan, and C. Huxley. 1996. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol. 16:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steger, G., J. Ham, O. Lefebvre, and M. Yaniv. 1995. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 14:329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stenlund, A. 2003. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 22:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sverdrup, F., and G. Myers. 1997. The E1 proteins, p. 37-53. In G. Myers, C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses 1997. Los Alamos National Laboratory, Los Alamos, N.M.
- 56.Tanaka, M., and W. Herr. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375-386. [DOI] [PubMed] [Google Scholar]
- 57.Ustav, E., and M. Ustav. 1998. E2 protein as the master regulator of extrachromosomal replication of the papillomaviruses. Papillomavirus report. 9:145-152. [Google Scholar]
- 58.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voitenleitner, C., and M. Botchan. 2002. E1 protein of bovine papillomavirus type 1 interferes with E2 protein-mediated tethering of the viral DNA to mitotic chromosomes. J. Virol. 76:3440-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winokur, P., and A. McBride. 1996. The transactivation and DNA binding domains of the BPV-1 E2 protein have different roles in cooperative origin binding with the E1 protein. Virology 221:44-53. [DOI] [PubMed] [Google Scholar]
- 62.Wu, S. Y., and C. M. Chiang. 2001. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276:34235-34243. [DOI] [PubMed] [Google Scholar]
- 63.Yao, J. M., D. E. Breiding, and E. J. Androphy. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 72:1013. [DOI] [PMC free article] [PubMed] [Google Scholar]