Abstract
The halogenated benzimidazoles BDCRB (2-bromo-5,6-dichloro-1-β-d-riborfuranosyl benzimidazole riboside) and TCRB (2,5,6-trichloro-1-β-d-riborfuranosyl benzimidazole riboside) were the first compounds shown to inhibit cleavage and packaging of herpesvirus genomes. Both inhibit the formation of unit length human cytomegalovirus (HCMV) genomes by a poorly understood mechanism (M. R. Underwood et al., J. Virol. 72:717-715, 1998; P. M. Krosky et al., J. Virol. 72:4721-4728, 1998). Because the simple genome structure of guinea pig cytomegalovirus (GPCMV) provides a useful model for the study of herpesvirus DNA packaging, we investigated the effects of BDCRB on GPCMV. GPCMV proved to be sensitive to BDCRB (50% inhibitory concentration = 4.7 μM), although somewhat less so than HCMV. In striking contrast to HCMV, however, a dose of BDCRB sufficient to reduce GPCMV titers by 3 logs (50 μM) had no effect on the quantity of GPCMV genomic DNA that was formed in infected cells. Electron microscopy revealed that this DNA was in fact packaged within intranuclear capsids, but these capsids failed to egress from the nucleus and failed to protect the DNA from nuclease digestion. The terminal structure of genomes formed in the presence of BDCRB was also altered. Genomes with ends lacking a terminal repeat at the right end, which normally exist in an equimolar ratio with those having one copy of the repeat at the right end, were selectively eliminated by BDCRB treatment. At the left end, BDCRB treatment appeared to induce heterogeneous truncations such that 2.7 to 4.9 kb of left-end-terminal sequences were missing. These findings suggest that BDCRB induces premature cleavage events that result in truncated genomes packaged within capsids that are permeable to nuclease. Based on these and other observations, we propose a model for duplication of herpesvirus terminal repeats during the cleavage and packaging process that is similar to one proposed for bacteriophage T7 (Y. B. Chung, C. Nardone, and D. C. Hinkle, J. Mol. Biol. 216:939-948, 1990).
The Herpesviridae family of viruses includes several significant human pathogens, including herpes simplex virus type 1 (HSV-1) and HSV-2, varicella-zoster virus, Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus, and human cytomegalovirus (HCMV). Herpesviruses have large (130 to 235 kb) double-stranded linear DNA genomes that circularize shortly after infection (24, 46, 47, 57). Viral DNA is replicated to form large concatemers of head-to-tail linked genomes that are packaged and cleaved to produce encapsidated unit length genomes (4, 6, 33, 43, 46, 57, 60, 71).
Mechanistically, herpesvirus DNA packaging is highly reminiscent of the large double-stranded DNA bacteriophage, in that packaging proceeds in one direction and cleavage occurs one genome length in from concatemer ends (43, 46, 49, 60, 71). As in many bacteriophage, cleavage of herpesvirus DNA cannot take place until two requirements have been met: (i) a near-genome length of DNA has entered the capsid and (ii) specific cis-acting DNA sequences constituting a cleavage site are encountered (11). The cis-acting sequences remain poorly defined, but two elements, designated pac1 and pac2, are highly conserved near the genomic ends of diverse herpesviruses (20, 63) and essential to the combined process of cleavage and packaging (48). More recent studies suggest that pac1 and pac2 may have distinct roles. The selective association of pac2 with concatemer ends from a variety of herpesviruses led to the proposal that pac2 may define packaging directionality by mediating initiation of concatemer packaging (49) in a manner analogous to that described for bacteriophage λ (14). Data from HSV-1 transient amplicon packaging assays support this model, suggesting distinct roles in which pac2 serves to initiate packaging while pac1 serves to terminate packaging via cleavage (28).
In bacteriophage, both translocation of the DNA into capsids and cleavage are carried out by a two-subunit enzyme complex called terminase (8). A functionally equivalent herpesvirus terminase has been postulated to be comprised of two subunits; in HSV-1, these are the products of the UL15 and UL28 genes. These proteins or their homologs in other herpesviruses have been shown to associate with one another (1, 5, 29, 34, 35), bind in vitro to packaging sequences (2, 10), and possess in vitro nucleolytic (10, 58) and ATPase activities (29, 59) (the later is a requirement for DNA translocation in bacteriophage). Additional clues to their activities have come from the study of compounds that block the cleavage and packaging process, which include the halogenated benzimidazoles BDCRB (2-bromo-5,6-dichloro-1-β-d-riborfuranosyl benzimidazole riboside) and TCRB (2,5,6-trichloro-1-β-d-riborfuranosyl benzimidazole riboside), and a structurally distinct compound, BAY38-4766. Resistance mutations to BDCRB and TCRB map to the HCMV terminase genes UL89 and UL56 (homologs of HSV-1 UL15 and UL28, respectively) (37, 66), and very similar mutations within the murine cytomegalovirus (MCMV) terminase genes m89 and m56 confer resistance to BAY38-4766 (12). How these compounds exert their inhibitory effects and the precise roles of these presumed terminase subunits in vivo remain to be elucidated.
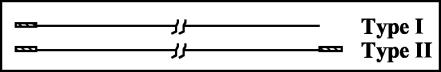
We have used guinea pig cytomegalovirus (GPCMV) to study the cleavage and packaging process because of the simplicity of its genome structure. Two genomic isomers are formed by GPCMV in roughly equimolar amounts: type I genomes have a single copy of a 1-kb terminal repeat at the left terminus but none at the right terminus, whereas type II genomes have a single copy of this repeat at each end (Fig. 1).
FIG. 1.
GPCMV genomic structures. The two possible genome structures of GPCMV are illustrated with a line representing unique sequences and hatched boxes representing the terminal repeats. The genome structure designated as type II has two directly repeated copies of the 1-kb terminal repeat, one at each end of the genome, whereas the structure designated as type I has only one copy of the repeat at the left end and lacks repeat sequences at the right end, terminating instead with unique sequences.
Analysis of a recombinant GPCMV that produced only type II genomes provided strong evidence that duplication of the terminal repeat must occur with each cleavage event (53), a phenomenon characteristic of bacteriophages T3 and T7 (22, 23). But in addition to duplicative cleavage, GPCMV must also be capable of simple, nonduplicative cleavage to produce type I genomes which lack the right-terminal repeat. Thus, the GPCMV genome structure, in which each type of cleavage is represented by a distinct terminal restriction fragment, lends itself to the study of genetic, pharmacological, and cellular factors that might alter the cleavage and packaging processes.
In this study, we examined the effects of BDCRB on the DNA cleavage, packaging, and maturation of GPCMV. Its effects on GPCMV proved to be profoundly different from its effects on HCMV, yet they provide valuable insights into the mechanistics that underlie herpesvirus cleavage and packaging.
MATERIALS AND METHODS
Cell and viral culture.
Wild-type GPCMV strain 22122 (ATCC VR-682) was propagated with guinea pig embryo or lung fibroblasts prepared as previously described (47) and cultured with Eagle's minimal essential medium (EMEM) supplemented with 10% fetal bovine serum, 50 U of penicillin ml−1, and 50 mg of streptomycin ml−1. BDCRB was a gift from John Drach and Leroy Townsend (University of Michigan) and was dissolved in dimethyl sulfoxide at a stock concentration of 25 mM.
FIGE.
Cells (2 × 106) were infected with GPCMV at a multiplicity of infection (MOI) of 3 for 4 days and then washed with phosphate-buffered saline (PBS) and 0.53 mM EDTA, trypsinized, resuspended in 10 ml of PBS-EDTA, and pelleted by centrifugation (5 min at 800 × g). Pelleted cells were either fixed for electron microscopy (described below) or resuspended in 50 μl of molten (52°C) 1% SeaPlaque agarose in TE (10 mM Tris [pH 8.0], 1 mM EDTA), cast into molds, and then prepared and analyzed by field-inversion gel electrophoresis (FIGE) as previously described (46, 47). Restriction analyses of FIGE-separated DNAs were performed as previously described (47).
Southern hybridization.
Extracellular virion DNA was prepared from infected cell culture supernatants, restricted, and separated by agarose gel electrophoresis as previously described (47). Agarose-separated DNAs were transferred to Nytran nylon membranes (Schleicher & Schuell) and hybridized as previously described (46) with the following probes: the O probe was derived from pGP14, which contains unique sequences common to HindIII O and M (47); the R probe was derived from pGP48, which contains the HindIII R terminal fragment (47); and the N probe was derived from pKTS398 (kindly provided by Mark Schleiss, University of Cincinnati), which contains the GPCMV HindIII N fragment (44).
[32P]orthophosphate uptake.
Cells (2 × 106) were infected with GPCMV at an MOI of 3 and incubated for 3 d. The medium was then removed and replaced with fresh EMEM containing either no drug or 50 μM BDCRB. After 8 h of incubation, the medium was removed and replaced with low-phosphate EMEM (3) containing 5 μCi of [32P]orthophosphate (Amersham) ml−1 with or without 50 μM BDCRB. After 16 h of incubation at 37°C, agarose plugs containing total cell DNA were prepared and separated by FIGE as described above. The 230-kb DNA was excised from the FIGE gel and digested with HindIII. The resulting fragments were separated by agarose electrophoresis and visualized by autoradiography of the dried gel.
Nuclease treatment.
Nuclease digestion was performed by a modification of the method of Lai and Chu (38). Cells (106) were infected with GPCMV at an MOI of 3 in the absence or presence of 50 μM BDCRB and incubated for 4 days. Infected cell monolayers were washed with PBS, scraped, pelleted by centrifugation (5 min at 800 × g), and then resuspended in 450 μl of ice-cold hypotonic buffer (10 mM Tris [pH 8.0], 12 mM KCl). After incubation on ice for 60 min, the lysate was homogenized 20 times in a Dounce homogenizer and complete lysis was confirmed by trypan blue staining of a small aliquot. The lysate was divided into two 150-μl aliquots, and 3.3 μl of 100 mM CaCl2 and 16 μl of 10× nuclease buffer (600 mM KCl, 150 mM NaCl, 20 mM Tris [pH 8.0]) were added to each aliquot. Digestion of one aliquot was carried out by the addition of 4 μl (600 U) of Staphylococcus aureus nuclease (Boehringer Mannheim) and incubation for 4 h at room temperature followed by the addition of 6 μl of 0.5 M EDTA to inactivate all nucleases. The second aliquot served as an undigested control. No exogenous nuclease was added, and to prevent endogenous nuclease activity, 6 μl of 0.5 M EDTA was added prior to incubation on ice for 4 h. After incubation, samples were either embedded in 1% SeaPlaque agarose for FIGE analysis (described above) or pelleted again (5 min at 800 × g) and fixed for electron microscopy (described below).
Electron microscopy.
Infected cell pellets or cell lysates (described above) were fixed overnight in 5 ml of 2% glutaraldehyde and 0.1 M cacodylic acid buffer at 4°C, embedded in paraffin, sectioned, and micrographed with a Zeiss-Emloca transmission electron microscope and Kodak EM4487 film.
RESULTS
GPCMV is sensitive to BDCRB.
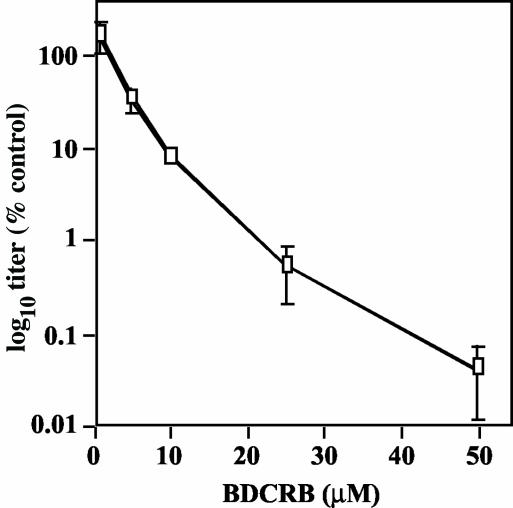
To evaluate the ability of BDCRB to inhibit GPCMV, cells were infected with GPCMV at an MOI of 0.2 in the presence of 0.0, 1.0, 5.0, 25, or 50 μM BDCRB and the titers of the resulting culture supernatants were determined at 7 days postinfection (p.i.). Titer decreases (compared to the no-drug control) were plotted versus drug concentration (Fig. 2). A dose of 1.0 μM BDCRB had no effect on virus titer, but with higher doses, titers deceased in a dose-dependent manner. The highest dose, 50 μM, reduced the virus titer 1,000-fold. A 50% inhibitory concentration (IC50) of approximately 4.7 μM was calculated from the slope of the curve.
FIG. 2.
Sensitivity of GPCMV to BDCRB. Cells were infected with GPCMV at an MOI of 0.2 in the presence of increasing doses of BDCRB. At 7 days p.i., the culture supernatants were titrated with endpoint dilution assays. Titers were converted to a percentage of the titer obtained without drug and plotted on a log10 scale versus drug concentration.
The cytotoxicity of BDCRB was not apparent within this dose range. Uninfected confluent cells maintained in 50 μM BDCRB for up to 2 weeks exhibited no morphological signs of toxicity. Moreover, the time required for subconfluent cells to reach confluence was not altered by 50 μM BDCRB, despite a modest twofold decrease in [3H]thymidine incorporation (data not shown); lower doses of BDCRB (10 or 25 μM) had no effect on incorporation (data not shown). Thus, the inhibitory effects of BDCRB on GPCMV are unlikely to arise from cytotoxicity.
BDCRB does not inhibit the formation of genome-sized (230 kb) GPCMV DNA.
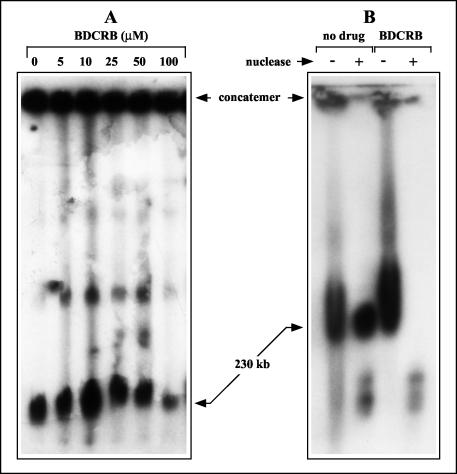
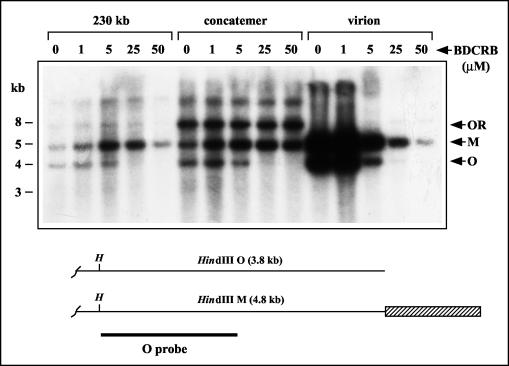
BDCRB and other related members of the halogenated benzimidazole family are known as maturational inhibitors of HCMV because they prevent cleavage of progeny HCMV genomes from concatemeric DNA (37, 66). To determine whether BDCRB has similar effects on GPCMV, cells were infected in the presence of BDCRB for 4 days. Infected cell DNAs were then separated by FIGE and hybridized with the O probe to specifically detect viral DNA species. Previous experiments have shown that GPCMV concatemeric DNA fails to migrate into pulsed-field gels and remains in the sample plug, whereas genomic DNA migrates with an apparent size of 230 kb (47). Doses of BDCRB of up to 100 μM did not alter the quantity of concatemeric DNA detected (Fig. 3A), indicating that the drug does not significantly inhibit concatemer synthesis. No effect of BDCRB on the formation of 230-kb DNA was observed at 50 μM, and a slight decrease was observed at 100 μM (Fig. 3A). Therefore, BDCRB does not significantly block formation of genome-length (230 kb) GPCMV DNA. This is in sharp contrast to its effect on HCMV, where 10 μM is sufficient to eliminate the formation of HCMV genomes (66). Furthermore, partially inhibitory doses of BDCRB have been reported to result in the formation of a monomer-plus HCMV DNA species that migrates with an apparent molecular size of 270 kb (66). Although supergenomic GPCMV species (500 kb and greater) were observed in the experimental results shown in Fig. 3A, their presence was not consistently associated with BDCRB treatment in numerous replicate experiments (data not shown).
FIG. 3.
Effects of BDCRB on intracellular GPCMV DNA. (A) Replicate cultures were infected with equal inocula of GPCMV and incubated in the presence of the indicated doses of BDCRB. Intracellular DNA was prepared at 4 days p.i., separated by FIGE, transferred to nylon membranes, and hybridized with the O probe to detect GPCMV replicative DNA forms. (B) Replicate cultures were infected in the presence or absence of 50 μM BDCRB. At 4 days p.i., the cells were subjected to hypotonic lysis and Dounce homogenization. The lysates were then either mock treated (−) or treated with staphylococcal nuclease (+) prior to DNA preparation and analysis as described for panel A. The locations of viral concatemeric and genomic 230-kb DNAs are indicated.
Genome-sized (230 kb) DNA formed in the presence of BDCRB is packaged but is not protected from nuclease.
In numerous studies, resistance to exogenous nuclease has been taken to indicate that DNA is packaged within capsids and thereby protected. It was previously shown that the vast majority of newly formed HCMV genomic DNA within infected cells is nuclease resistant (46). To determine whether the 230-kb GPCMV DNA formed in the presence of BDCRB is protected from nuclease, infected cells were cultured in the presence or absence of BDCRB and hypotonically lysed. The lysates were either mock treated or incubated with staphylococcal nuclease prior to DNA preparation, FIGE separation, and hybridization as described above. As expected, the 230-kb GPCMV DNA from cells that were not treated with BDCRB was fully protected from nuclease digestion. In contrast, 230-kb DNA from BDCRB-treated cells was fully digested by the nuclease (Fig. 3B).
This result suggested that the 230-kb DNA formed in the presence of BDCRB was either not packaged within capsids or was within capsids but that these capsids were somehow permeable to nuclease. To address this issue, cells infected in the presence or absence of BDCRB were examined by transmission electron microscopy.
The nuclei of herpesvirus-infected cells exhibit three types of capsids: C capsids contain DNA (17, 25), B capsids contain proteolytically cleaved scaffold proteins, and A capsids appear to be empty (55). All three originally derive from an unstable procapsid precursor that is built around a core of scaffold proteins that have not been proteolytically cleaved (52). Spontaneous proteolytic cleavage of the scaffolding proteins is thought to give rise to B capsids; the scaffolding is removed during the DNA packaging process that gives rise to C capsids; and A capsids are thought to arise from C capsids that have spontaneously expelled their DNA (for a review, see reference 11).
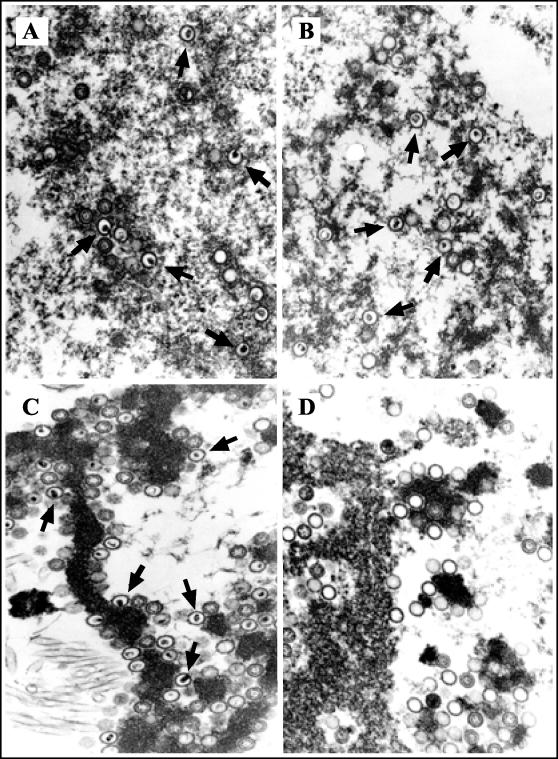
Electron microscopy revealed no differences in the prevalence of A, B, or C capsids within cells that were mock treated or treated with BDCRB (Fig. 4A and B and Table 1). In particular, the presence of numerous C capsids clearly indicated that DNA was packaged within capsids in the presence of BDCRB (Fig. 4B; Table 1). In combination with the nuclease sensitivity results described above, these results suggested that C capsids were formed in the presence of BDCRB but were unable to protect their DNA from exogenous nuclease. To confirm this hypothesis, replicates of the lysates described above were treated with staphylococcal nuclease prior to electron microscopy. As expected, DNA packaged within C capsids in the absence of BDCRB was protected from nuclease (Fig. 4C). In the presence of BDCRB, however, nuclease treatment rendered C capsids very rare while A capsids became significantly more abundant, suggesting that nuclease digestion of DNA within C capsids had given rise to A capsids (Fig. 4D; Table 1). The rare C capsids that were observed were clustered within structures of unknown material (data not shown), suggesting that the nuclease may have failed to adequately penetrate these areas. From these results, we conclude that BDCRB treatment results in packaging of 230-kb GPCMV DNA within capsids but that these capsids are abnormal, in that they allow nuclease to enter and digest the packaged DNA.
FIG. 4.
Electron microscopic analysis of BDCRB-treated GPCMV-infected cells. Replicate cultures were infected at an MOI of 3, incubated for 3 days in the absence or presence of 50 μM BDCRB, pelleted, hypotonically lysed, mock treated or treated with Staphylococcus nuclease (as described for Fig. 3B), fixed, sectioned, and analyzed by electron microscopy. (A) Mock nuclease treatment with no drug; (B) mock nuclease treatment with BDCRB; (C) nuclease treatment with no drug; (D) nuclease treatment with BDCRB. Magnification, ×50,000. Arrows indicate representative C capsids.
TABLE 1.
Effects of BDCRB on A, B, and C capsid frequencies within infected cell nuclei
| Nuclease and drug treatment | No. of capsids (% of total) of typea:
|
||
|---|---|---|---|
| A | B | C | |
| No nuclease | |||
| No drug | 78 (39) | 56 (28) | 66 (33) |
| BDCRB | 93 (24) | 133 (34) | 161 (42) |
| Nuclease | |||
| No drug | 107 (18) | 149 (26) | 329 (56) |
| BDCRB | 285 (72) | 106 (26) | 6 (1.5) |
Intranuclear capsids observed in electron micrographs from the experiment shown in Fig. 4 were classified as A, B, or C capsids based on morphology. Numbers shown are cumulative for five nuclei examined from each group.
BDCRB alters the termini formed on GPCMV genomes.
To determine whether BDCRB affects the relative prevalence of type I versus type II GPCMV genomes, cells were infected in the presence of increasing doses of BDCRB for 5 days. Extracellular virion DNA was then prepared from culture supernatants, and concatemeric and intracellular 230-kb DNAs were isolated from cells by FIGE. The DNAs were digested with HindIII, and the resulting fragments were separated electrophoretically, transferred to nylon membranes, and hybridized with the O probe to detect the two terminal fragments from the right end of the genome: HindIII O, resulting from type I genomes, and HindIII M, resulting from type II genomes (Fig. 5). These terminal fragments are normally present in nearly equal amounts. One micromolar BDCRB did not alter the ratio of O to M termini; however, 5.0 μM BDCRB markedly reduced O termini with no reduction of M termini (Fig. 5). At 25 and 50 μM, BDCRB virtually eliminated O termini while M termini were still present. A similar effect was observed on extracellular virion DNA; however, the amount of extracellular virion DNA was dramatically reduced in a dose-dependent fashion, consistent with the decrease in virus titer. These findings indicate that BDCRB dramatically alters the type of termini found at the right end of GPCMV genomes and concatemers.
FIG. 5.
Effects of BDCRB on the right end of the GPCMV genome. Replicate cultures were infected for 4 days in the presence of the indicated doses of BDCRB (micromolar). Intracellular concatemeric and 230-kb DNAs were separated by FIGE and excised from the gel in agarose blocks. Virion DNA was prepared from culture supernatants by centrifugation. All DNAs were then digested with HindIII, separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with the O probe to detect terminal M and O fragments. Arrows indicate the HindIII M and O terminal fragments as well as the O-R junction fragment present in concatemeric DNA. A schematic below illustrates the right ends of type I and type II genomes and shows the HindIII sites (H) that define the HindIII O and M terminal fragments. The terminal repeat is indicated by a hatched box. A thick line indicates the sequences contained within the O probe.
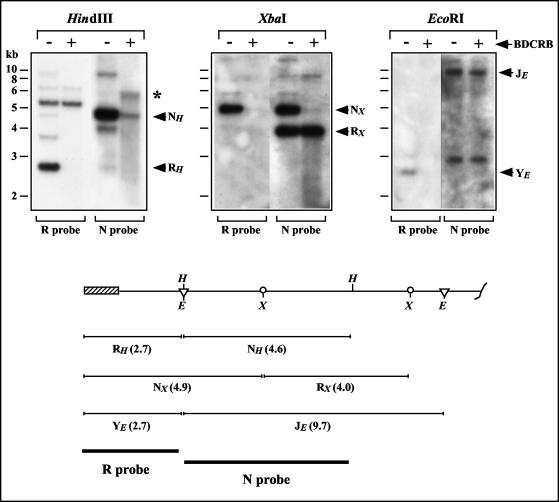
Similar analyses were conducted with probes from the left end of the GPCMV genome. The 230-kb DNA produced in the presence of 50 μM BDCRB was isolated by FIGE and digested with HindIII, EcoRI, or XbaI. The resulting fragments were separated by electrophoresis, transferred to nylon membranes, and hybridized with the R or N probes, consisting of the terminal HindIII R or penultimate HindIII N regions, respectively. BDCRB treatment abolished any signal for the terminal HindIII R fragment and greatly diminished the signal for the penultimate HindIII N fragment (Fig. 6). Hybridization with N probe detected an anomalous fragment specific to BDCRB-treated DNA that migrated with an apparent molecular size (∼5.5 kb) larger than that of HindIII N (4.6 kb). Smearing was evident below HindIII N and this anomalous fragment, suggestive of heterogeneous fragment sizes (Fig. 6).
FIG. 6.
Effects of BDCRB on the left end of the GPCMV genome. Replicate cultures were infected for 4 days in the presence or absence of 50 μM BDCRB. Intracellular 230-kb DNAs were separated by FIGE and excised from the gel in agarose blocks. The DNAs were then digested with HindIII, XbaI, or EcoRI, separated by agarose gel electrophoresis, transferred to nylon membranes, and hybridized with the R probe. The membranes were then stripped and rehybridized with the N probe. An anomalously migrating BDCRB-specific HindIII fragment in panel A is indicated with an asterisk. The schematic below illustrates the restriction sites at the left end of the genome. The terminal repeat is shown as a hatched box. Vertical lines indicate HindIII (H) recognition sites, circles represent XbaI (X) recognition sites, and triangles indicate EcoRI (E) recognition sites. The sequences contained within the R and N hybridization probes are indicated with thick bars, and the positions and sizes of HindIII, XbaI, and EcoRI fragments to which these probes are predicted to hybridize are indicated.
The XbaI and EcoRI digestions yielded similar results. BDCRB induced a complete loss of the left-end-terminal XbaI N fragment but did not significantly affect levels of the penultimate XbaI R fragment; again, smearing of the penultimate fragment was detected (Fig. 6). Similarly, BDCRB induced complete loss of the left-end-terminal EcoRI Y fragment but did not noticeably affect the penultimate EcoRI J fragment (Fig. 6). Taken together, these data demonstrate that BDCRB treatment results in the formation of abnormal genomes that lack sequences from the left end of the genome. Our failure to detect novel BDCRB-associated terminal fragments with three different restriction enzymes indicates that the novel termini induced by BDCRB are likely to be heterogeneous in location. These data further suggest that the BDCRB-induced termini range between approximately 2.7 and 4.9 kb from the normal left end of the genome. While this interpretation would seem the simplest, the retarded migration of the anomalously migrating fragment (relative to HindIII N) may suggest an unusual DNA structure. Thus, we cannot rule out the possibility of a specific BDCRB-induced terminus that has abnormal migration properties in agarose gels due to an atypical structure (e.g., hairpin, branch, or lariat, etc.).
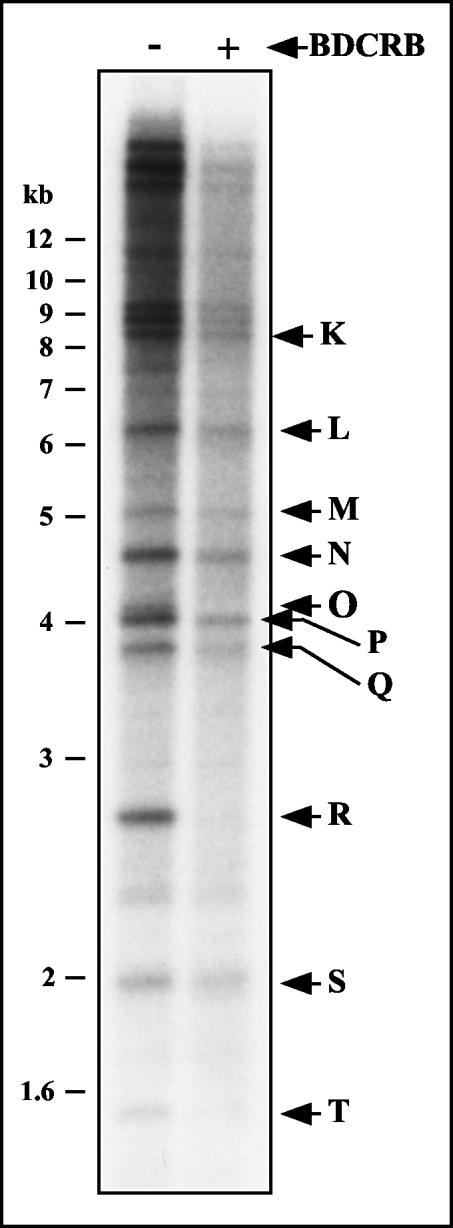
To determine whether other regions of the GPCMV genome are altered by BDCRB, infected cell cultures were mock treated or treated with 50 μM BDCRB between 72 and 96 h p.i. and viral DNA was labeled by the addition of [32P]orthophosphate to the culture media between 80 and 96 h p.i. Intracellular 230-kb DNA was then isolated by FIGE and digested with HindIII. The resulting fragments were separated electrophoretically and autoradiographed. It was not possible to confirm the absence of HindIII O because it was occluded by HindIII P, which has almost the same molecular weight as HindIII O (32). However, consistent with the hybridization studies (Fig. 6), HindIII R fragments were absent from BDCRB-treated DNA (Fig. 7). With this exception, the restriction fragment patterns were qualitatively identical between BDCRB-treated and untreated DNAs, indicating that BDCRB does not grossly alter the restriction pattern of GPCMV DNA but rather its effects appear to be limited to specific alterations at the genomic termini. Quantitatively, however, HindIII S appeared to be overrepresented relative to other fragments in the BDCRB-treated lane, being nearly equal in intensity to its counterpart in the untreated lane while most of the other fragments were weaker (Fig. 7). While this may suggest an anomaly of fragment molar ratios within the viral DNA, we cannot rule out the possibility that some of the signal that appears to be derived from HindIII S may be nonviral in origin (e.g., mitochondrial DNA). Hence, quantitative comparisons of fragment molar ratios from this experiment may be unreliable.
FIG. 7.
Restriction fragment pattern analysis of GPCMV genomes formed in the presence of BDCRB. Duplicate cultures were infected with GPCMV at an MOI of 3. The culture media was replaced at 72 h p.i. with media alone or media containing 50 μM BDCRB. Viral DNA was labeled between 80 and 96 h p.i. by the addition of [32P]orthophosphate to the media of both cultures. Intracellular DNA was prepared at 96 h p.i. and separated by FIGE. Intracellular 230-kb GPCMV DNA was excised in agarose blocks, extracted from the agarose, and digested with HindIII. The resulting restriction fragments were separated by agarose electrophoresis, transferred to a nylon membrane, and autoradiographed. Arrows indicate the locations of GPCMV HindIII restriction fragments surmised from their published molecular weights (32).
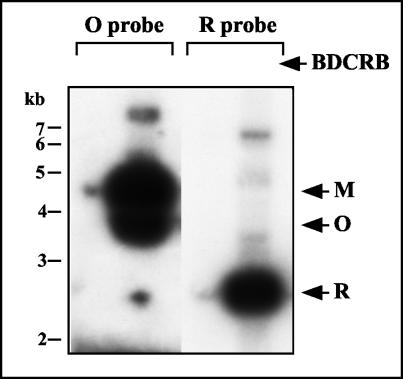
To determine whether the abnormal genomes formed in the presence of BDCRB are able to mature into extracellular virions, terminal analysis, as described above, was performed on DNA derived from extracellular virions. Consistent with previous experiments (Fig. 5), the amount of extracellular virion DNA was greatly reduced in the presence of 50 μM BDCRB; however, extensive exposure of the autoradiographs revealed that the small amount of virion DNA present contains exclusively M termini and no O termini at the right end of the genome. R termini were detected, and their signals approximately equaled those of the M termini (Fig. 8). That these weak signals are not derived from spillover from the adjacent lanes during loading can be deduced from the facts that HindIII O was not present in the BDCRB-treated lane (Fig. 8), whereas, upon shorter exposure (data not shown), nearly equimolar amounts of both HindIII O and M were present in the adjacent untreated lane. This conclusion extends to the R fragment, as the same membrane was rehybridized with the R probe. Therefore, the small amount of DNA that is able to mature into extracellular virions in the presence of BDCRB is comprised of normal type II genomes and lacks the left end BDCRB-associated truncations described for intracellular 230-kb DNA described above.
FIG. 8.
Terminal structure of extracellular virion DNA produced in the presence of BDCRB. Replicate cultures were infected and incubated for 4 days in the presence (+) or absence (−) of 50 μM BDCRB. Virions were isolated from the culture supernatants by centrifugation. Virion-associated DNAs were extracted, digested with HindIII, separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with the O probe (shown in Fig. 5) to detect terminal HindIII M and O fragments. After autoradiography, the membrane was stripped and rehybridized with the R probe (shown in Fig. 6) to detect terminal HindIII R fragments. Arrows indicate the positions of M, O, and R fragments. Exposure times were adjusted to match signal intensities from M and R fragments.
C capsids formed in the presence of BDCRB are retained in the nucleus.
The results of experiments shown in Fig. 5 and 8 demonstrated that inhibitory doses of BDCRB dramatically reduced the amount of extracellular virion DNA. This reduction approximately paralleled the decreases in infectious titers (Fig. 2 and 5). However, within the nucleus, large amounts of C capsids and 230-kb DNA were present. This suggested that a block existed to the egress of C capsids to form extracellular virion particles. To determine the stage at which egress was inhibited, electron micrographs of nine untreated and nine BDCRB-treated infected cells were examined, and the total numbers of cytoplasmic and nuclear C capsids were counted for each group. Although the numbers of intranuclear C capsids were similar for each group, BDCRB treatment reduced the frequency of cytoplasmic C capsids by 100-fold: 203 C capsids were identified in cytoplasmic regions of untreated cells, whereas only two cytoplasmic C capsids were identified in BDCRB-treated cells. This suggests that the block lies at the level of the nuclear envelope. Hence, while complete genomes (having normal M and R termini), although rare, successfully reached the extracellular compartment, incomplete genomes (lacking R termini) remained within the nucleus. These observations suggest that a highly sensitive quality control mechanism may exist to prevent transit from the nucleus of capsids containing even slightly truncated genomes.
DISCUSSION
The halogenated benzimidazoles, including BDCRB, were the first compounds shown to inhibit herpesvirus DNA cleavage and packaging (66). They provide valuable tools for investigating these processes. To take advantage of the simple genome structure of GPCMV in such investigations, we evaluated the sensitivity of GPCMV to BDCRB. Titer reduction assays determined an IC50 against GPCMV of 4.7 μM, indicating that GPCMV is sensitive to BDCRB but is about eightfold less sensitive than HCMV (IC50 of ∼0.6 μM) (66). Detailed analyses revealed dramatic differences between the molecular effects of BDCRB on HCMV and GPCMV. When HCMV-infected cells are treated with BDCRB, concatemeric DNA accumulates but is not cleaved to form intracellular 230-kb genomes (66). In stark contrast, we found that BDCRB does not significantly reduce the quantity of intracellular GPCMV 230-kb DNA. This DNA, however, exclusively has type II termini at the right end, is truncated heterogeneously near the left end, and is packaged into capsids that fail to protect it from nuclease and fail to transit to the cytoplasm.
The reasons that underlie these differences are unclear. In HCMV, resistance to BDCRB is conferred by two amino acid substitutions (D344E and A355T) in pUL89 (66), and resistance to the closely related compound TCRB is further conferred by a Q204R mutation within pUL56 (37). In GPCMV, the predicted amino acid sequences for the pUL56 and pUL89 homologs indicate that the pUL56 Q204 residue is conserved and within pUL89 both the D344 and A355 residues are not conserved (Mark Schleiss, personal communication). Hence, the GPCMV pUL89 homolog may have some inherent resistance to BDCRB. Indeed, the 8-fold resistance that we observed (relative to HCMV) is consistent with the 10-fold level of resistance reported for the D344E mutation within HCMV pUL89 (37). Thus, one hypothesis to explain the differences in the effects that BDCRB has on the two viruses is that the primary drug targets may be different. While in HCMV, BDCRB inhibits both pUL89 and pUL56, in GPCMV its effects may be manifested primarily through inhibition of pUL56 (GPCMV pUL89 being naturally resistant).
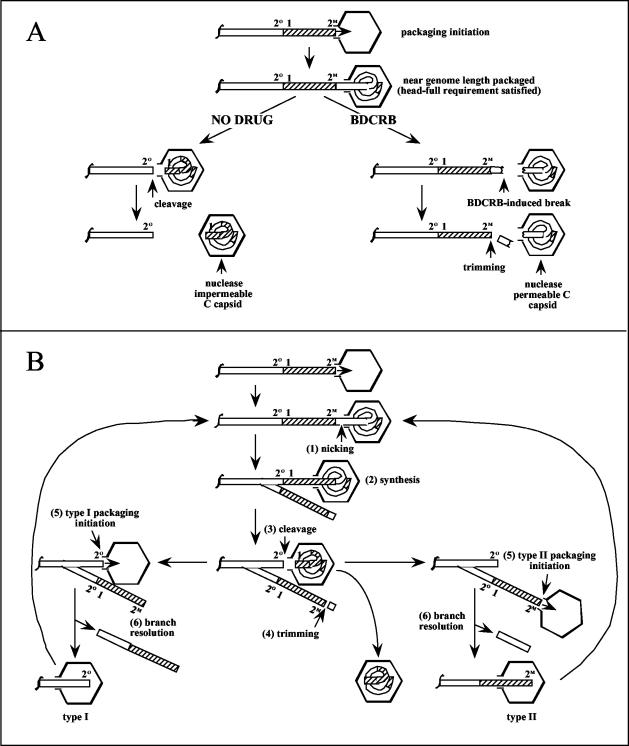
Figure 9A shows a simple model of GPCMV DNA packaging and a hypothesis for how BDCRB could induce both a predominance of type II termini at the right end and heterogeneous loss of sequences from the left end. The initial steps are based on a general model of herpesvirus DNA packaging in which procapsids associate with pac2-containing concatemer ends to initiate packaging and packaging proceeds in a right-to-left (relative to the GPCMV genome) direction. DNA cleavage is suppressed by a head-full mechanism until close to a genome-length has entered. In the absence of drug, cleavage occurs at the next cleavage site encountered and the newly formed genome is sealed within the capsid. In the presence of BDCRB, however, premature cleavage or breaks occur just prior to entry of the repeat, resulting in left-end-terminally truncated genomes within capsids. These capsids fail to be sealed and remain nuclease permeable. The premature cleavages also result in extra sequences on the newly formed concatemer termini. As abundant M termini were observed both on concatemeric and cleaved DNA, a trimming step must be invoked to generate concatemer ends with normal M termini. These M termini then serve as the substrates for initiation of additional packaging events, resulting in M termini exclusively on packaged genomes.
FIG. 9.
(A) Proposed model for the BDCRB mechanism of action. DNA packaging initiates on concatemers at pac2-containing right-end termini. Packaging commences until almost a full genome has been packaged, satisfying the head-full requirement. In the absence of drug (left), cleavage is then free to commence at the next cleavage site encountered and the resulting C capsid is sealed to become nuclease impermeable. In the presence of BDCRB (right), heterogeneous breaks occur once the head-full requirement is satisfied just prior to packaging of the left-terminal repeat. The resulting C capsid remains nuclease permeable. Trimming of the concatemer end reconstitutes a right-end terminus suitable for initiation of packaging. The locations of GPCMV pac1 elements (1) and the proposed pac2 elements (47, 49) found at O (2O) and M (2M) termini are indicated. (B) Proposed model for terminal repeat duplication. Packaging initiates and proceeds as shown in panel A. Although for simplicity a type II concatemer end is shown, either type I or type II ends can serve to initiate packaging. As the capsid nears what will become the left-terminal repeat, a nick occurs prior to the repeat (1). DNA synthesis primes at the nick, and the replication fork migrates leftward, copying through the repeat (2). Packaging continues and cleavage occurs at the left end of the repeat (3). Trimming as shown in panel A restores a type II right-end terminus (4). The next round of packaging initiates on either the type I or the type II terminus on the concatemer (5). Resolution of the branched concatemer allows continued packaging while generating small DNA fragments (6). The locations of GPCMV pac1 elements (1) and the proposed pac2 elements (47, 49) found at O (2O) and M (2M) termini are indicated.
How BDCRB might induce premature cleavages is not known; however, one attractive hypothesis is that it relaxes the cleavage site sequence specificity of the terminase without relieving the head-full requirement. Consequently, packaging would proceed without cleavage until the head-full requirement was satisfied; thereafter, the cleavage machinery would be free to cleave the DNA without sequence specificity, producing heterogeneous ends just short of the normal left end of the genome. From our data, this hypothesis predicts that the head-full requirement in GPCMV is satisfied when 2.7 to 4.9 kb of DNA remains to be packaged. This amount (1.1 to 2.1% of the GPCMV genome length) is consistent with studies of EBV amplicon packaging, which found that efficient packaging only occurred with genomes less than 2% undersized or 10% oversized relative to the normal EBV genome length (9).
The model also fits well with the hypothesis discussed above that BDCRB inhibits GPCMV via pUL56 and not through pUL89. HCMV pUL56 and its homolog in HSV-1, pUL28, have been reported to bind to pac1 sequences in vitro (2, 10), whereas nucleolytic activity is primarily associated with HCMV pUL89 (58). Thus, inhibition of GPCMV pUL56 could relax cleavage site specificity while permitting pUL89-mediated non-sequence-specific cleavages, precisely as predicted by our model. Even so, these hypotheses remain speculative. Identification of GPCMV genes containing mutations that confer resistance to BDCRB is in progress and should help to elucidate the proteins that are targeted. What is clear from our data is that BDCRB does not inhibit translocation of GPCMV DNA into capsids. By analogy to bacteriophage, GPCMV DNA translocation is presumed to be mediated by the terminase complex, comprised of both pUL56 and pUL89. Thus, BDCRB does not profoundly inhibit the GPCMV terminase but rather is highly specific for certain terminase activities.
Another striking effect observed in HCMV with partially inhibitory doses of BDCRB and TCRB is the formation of a 270-kb monomer-plus DNA species (37, 66). Little is known about the structure of monomer-plus DNA. One hypothesis suggests that it results from cleavage site skipping, manifested perhaps by the combined effects of relaxation of cleavage site specificity via inhibition of pUL56 and impaired cleavage via inhibition of pUL89. In HCMV, skipping one cleavage site would result in a 270-kb DNA species consisting of one long genome segment flanked by two short genome segments. Because the GPCMV genome lacks an internal cleavage site, cleavage site skipping would result in formation of a 460-kb dimer, which may exceed the packaging capacity of the capsid and hence not be formed. While our failure to observe any analogous supergenomic DNA species in BDCRB-treated GPCMV-infected cells is consistent with this hypothesis, this mechanism for monomer-plus formation remains unproven and other explanations may be equally probable.
In a previous paper (53), we reported evidence indicating that cleavage of GPCMV concatemeric DNA to form type II genomes is likely to involve a duplication of the terminal repeat, as concatemers contain almost exclusively single copies of the terminal repeat between genomes, whereas two copies (one at each end) are needed to form type II genomes. Analogous duplicative events have been proposed for the HSV-1 a sequence (21, 67) as well as for bacteriophage T3 and T7 (22, 23). In phage T7, detection of abundant short hairpin fragments derived from a potential stem-loop adjacent to the terminal repeat inspired a model in which a nick at the base of the stem-loop primes DNA synthesis back through the repeat. A trimming step analogous to that shown in Fig. 9A then removes the hairpin fragment, leaving a duplicated repeat and a new genomic terminus on the concatemer (16). Figure 9B shows an adaptation of this model to GPCMV. As we have no evidence for a stem-loop, a simple nick is proposed to occur just prior to packaging of the terminal repeat (step 1). A replication fork primes at the nick and copies through the terminal repeat (step 2). Cleavage at the left side of the repeat releases the packaged genome (step 3) and leaves an O terminus on one branch of the concatemer. On the other branch, a trimming reaction identical to that proposed in Fig. 9A removes the extra sequences to form an M terminus (step 4). Subsequent rounds of packaging can then initiate on either the O or the M end of the branched concatemer (step 5). Of note, the equimolar nature of these two ends predicts equimolar amounts of the two genome types, which is experimentally observed (47). Branch resolution (step 6), possibly mediated by the viral alkaline nuclease (43), may be required for continuation of DNA packaging.
This model explains a number of seemingly conflicting observations reported for different herpesviruses. First, data from several herpesviruses that have terminal repeats indicate that their genomic termini have complementary single-base overhangs (42, 47, 50, 51, 64), a feature that was difficult to reconcile with previous repeat duplication models that involved strand separation and fill-in reactions (67). In our model, all termini are formed by internal cleavages that could easily produce single-base staggered ends.
Second, our model is able to accommodate the differences in arrangements of pac1 and pac2 that exist for viruses with different genome structures. In herpesviruses that lack terminal repeats, pac1 and pac2 lie at opposite ends of the genome such that cleavage only requires a simple staggered cut at a site between pac1 and pac2 (15, 18, 26, 27). This is analogous to the cleavage shown in step 3 and would not involve branch formation or duplication. In other viruses, such as HSV-1, HCMV, and GPCMV, pac1 and pac2 lie at opposite ends of a terminal repeat. In HSV-1 and HCMV, these repeats are called a sequences. Where two such repeats are adjacent, simple cleavage would again suffice, as it could occur between the pac1 in one repeat and the pac2 in the adjacent repeat. But in concatemers of HSV-1 and GPCMV, single copies of the terminal repeat predominate (19, 40, 53, 69) and evidence suggests that single copies can be duplicated during cleavage (21, 53, 67). While the model shown in Fig. 9B illustrates GPCMV, it applies equally to other viruses having a sequence-like repeats. MCMV, however, is unique in that its genome has unusually small terminal repeats (30 bp) and pac1 and pac2 are located within unique sequences adjacent to but not within the repeats, resulting in either pac2-30-bp repeat-pac1 or pac2-30-bp repeat-30-bp repeat-pac1 structures at concatemer junctions (41, 48). Even so, the model in Fig. 9A needs only minor modification to work with the MCMV junction structure: the cleavage in step 3 would still occur to the left of pac1, giving rise to a pac2 end with no repeat (assuming a single repeat-containing junction), while the trimming in step 4 would occur to the right of pac2, giving rise to a pac2 end with a repeat. Because both concatemer ends retain a copy of pac2 (one with and one without a repeat), both ends can presumably serve to initiate the next round of packaging. Consistent with this, type I MCMV genomes that lack a 30-bp repeat at the pac2 end have been reported (42).
Third, while several reports have shown that both pac1 and pac2 are required for cleavage and packaging, other results indicate that pac1 and pac2 can mediate cleavage events independently: (i) in HSV-1, cleavages at an ectopically inserted a sequence occur a metered distance from either pac1 or pac2 (67); (ii) in MCMV, cleavages occur exactly 27 bp from pac1 but either 1 or 31 bp from pac2 (42, 48); (iii) in HSV-1 amplicon studies, genomes containing pac1 but lacking pac2 are cleaved but not packaged (28); (iv) in equine herpes virus type 1, ends resulting from cleavage at the internal L-IR junction, which has a pac1 but lacks a pac2-like sequence, are abundantly observed on concatemeric DNA (61); and (v) in GPCMV, which we have postulated to have two functional pac2 elements, one at the M terminus (pac2M) and one at the O terminus (pac2O) (47, 49), a mutant (RG5001) in which pac2O is displaced from pac1 by a 1.3-kb insertion at the O-R junction forms normal M and R termini but lacks O termini (47). These last two observations suggest that for GPCMV, the cleavage in step 3 is mediated exclusively by pac1 and that pac2O is only needed in step 5 for packaging initiation to from type I genomes, whereas pac2M mediates both the trimming reaction in step 4 and the packaging initiation in step 5. Thus, the model predicts independent functions for pac1 and pac2 consistent with observations from several different viruses.
Fourth, Umene has reported mysterious a sequence-containing fragments in HSV-1-infected cells (65). The majority of these fragments have the unique feature of being authentically cleaved at the pac2 end while containing various amounts of b sequences or adjacent a sequences at the other end. Assuming that the branch resolution in step 6 is not sequence specific but occurs variable distances from the repeat (as determined by migration of the replication fork), fragments with precisely this structure would result. Thus, the model provides a possible mechanism for the formation of fragments that we propose be termed Umene fragments.
The packaging of nearly full-length viral genomes into nuclease-permeable capsids appears to be unprecedented in the herpesvirus literature. During bacteriophage packaging, the DNA is stabilized within the capsid by the addition of a capping protein (13, 39, 62), and the recent finding that the DNA packaged by an HSV-1 UL25-null mutant is frequently expunged from capsids suggests that the pUL25 protein may serve as such a cap (45). Thus, an attractive possibility is that BDCRB may block capsid association of the GPCMV pUL25 homolog or perhaps another protein functions to seal the capsid after the DNA has entered. As HSV-1 pUL25 has also been reported to have DNA binding activity (54), interaction with the packaged DNA may be necessary for its association with C capsids. Thus, in the presence of BDCRB, the reduced length or the absence of specific R-terminal sequences could indirectly prevent the capping protein from associating with capsids. Experiments to further characterize the protein composition of GPCMV C capsids produced in the presence of BDCRB should prove enlightening.
Finally, our observations suggest that capsids containing truncated genomes are unable to egress from the nucleus. In HSV-1, A and B capsids as well as capsids containing nuclease-protected subgenomic amplicon DNA fragments are retained in the nucleus (56, 68). These observations suggest that the process of capsid envelopment by the inner nuclear membrane somehow excludes capsids that lack sufficient amounts of DNA. In the presence of BDCRB, we observed large quantities of R-terminally truncated genomes in the nucleus. In contrast, genomes in the culture supernatants exhibited normal R termini but were extremely rare. Electron microscopy indicated that the block to capsid egress was at the nuclear membrane. These results suggest that GPCMV nuclear envelopment is highly discriminatory. Given that the truncated genomes are only 1.1 to 2.1% smaller than normal genomes, discrimination on the basis of total DNA content alone would seem unlikely. A more probable explanation is that C capsids produced in the presence of BDCRB lack certain capsid proteins necessary for egress, such as the capping protein discussed above. Curiously, HCMV appears devoid of this quality control measure, as extracellular enveloped capsids lacking DNA are abundantly produced (30, 31).
It is also possible that BDCRB could have a direct impact on nuclear egress separate from its effects on packaging. A recent report revealed that a stereoisomeric derivative of BDCRB called 1263W94 does not block formation of nuclease-impermeable HCMV C capsids (i.e., cleavage and packaging) but does inhibit egress of C capsids from the nucleus (36). Resistance to 1263W94 maps to the HCMV UL97 kinase (7), and similarly, C capsids made by a UL97 deletion mutant fail to egress to the cytoplasm (36, 70). Such an effect may extend to BDCRB; however, this would seem unlikely given that GPCMV is not sensitive to 1263W94 (C. Murphy, D. E. Nixon, and M. A. McVoy, unpublished data). How the machinery that controls egress (nuclear envelopment and/or tegumentation) can discriminate normal capsids from those that contain truncated genomes, and how these compounds interfere with these processes, should prove to be a fascinating area for future investigations.
Acknowledgments
We thank Mark Schleiss for providing plasmid pKTS398 and for sharing unpublished GPCMV sequence data, Kyun Jae Hur for valued technical assistance, and John Drach and Leroy Townsend for kindly providing BDCRB.
This research was supported by grant funding from Virginia's Commonwealth Health Research Board, Public Health Services grants R21AI43527, RO1AI46668, and KO8AI01435 (to D.E.N.) from the National Institutes of Health, American Cancer Society grant IN-105V, and funds from the A. D. Williams Fund of the Medical College of Virginia, Virginia Commonwealth University.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler, S. P. 1986. Molecular epidemiology of cytomegalovirus: evidence for viral transmission to parents from children infected at a day care center. Pediatr. Infect. Dis. 5:315-318. [PubMed] [Google Scholar]
- 4.Bataille, D., and A. Epstein. 1994. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology 203:384-388. [DOI] [PubMed] [Google Scholar]
- 5.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porat, T. 1983. Replication of herpesvirus DNA, p. 81-86. In B. Roizman (ed.), The herpesviruses. Plenum Press, New York, N.Y.
- 7.Biron, K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 9.Bloss, T. A., and B. Sugden. 1994. Optimal lengths for DNAs encapsidated by Epstein-Barr virus. J. Virol. 68:8217-8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, J. C., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids. In E. Bogner and A. Holzenburg (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 12.Buerger, I., J. Reefschlaeger, W. Bender, P. Eckenberg, A. Popp, O. Weber, S. Graeper, H. D. Klenk, H. Ruebsamen-Waigmann, and S. Hallenberger. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens, S., T. Horn, and A. D. Kaiser. 1972. Head assembly steps controlled by genes F and W in bacteriophage lambda. J. Mol. Biol. 64:551-563. [DOI] [PubMed] [Google Scholar]
- 14.Catalano, C. E., D. Cue, and M. Feiss. 1995. Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol. 16:1075-1086. [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury, S. I., H. J. Buhk, H. Ludwig, and W. Hammerschmidt. 1990. Genomic termini of equine herpesvirus 1. J. Virol. 64:873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung, Y. B., C. Nardone, and D. C. Hinkle. 1990. Bacteriophage T7 DNA packaging. III. A “hairpin” end formed on T7 concatemers may be an intermediate in the processing reaction. J. Mol. Biol. 216:939-948. [DOI] [PubMed] [Google Scholar]
- 17.Dargan, D. J. 1986. The structure and assembly of herpes-viruses, p. 359-437. In J. R. Harris and R. W. Horne (ed.), Electron microscopy of proteins. Academic Press, London, England.
- 18.Davison, A. J. 1984. Structure of the genome termini of varicella-zoster virus. J. Gen. Virol. 65:1969-1977. [DOI] [PubMed] [Google Scholar]
- 19.Davison, A. J., and N. M. Wilkie. 1981. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J. Gen. Virol. 55:315-331. [DOI] [PubMed] [Google Scholar]
- 20.Deiss, L. P., J. Chou, and N. Frenkel. 1986. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol. 59:605-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deiss, L. P., and N. Frenkel. 1986. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol. 57:933-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 23.Fujisawa, H., and K. Sugimoto. 1983. On the terminally redundant sequences of bacteriophage T3 DNA. Virology 124:251-258. [DOI] [PubMed] [Google Scholar]
- 24.Garber, D. A., S. M. Beverley, and D. M. Coen. 1993. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459-462. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammerschmidt, W., H. Ludwig, and H. J. Buhk. 1988. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J. Virol. 62:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper, L., J. Demarchi, and T. Ben-Porat. 1986. Sequence of the genome ends and of the junction between the ends in concatemeric DNA of pseudorabies virus. J. Virol. 60:1183-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodge, P. D., and N. D. Stow. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 75:8977-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang, J. S., and E. Bogner. 2002. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem. 277:6943-6948. [DOI] [PubMed] [Google Scholar]
- 30.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 31.Irmiere, A., and W. Gibson. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J. Virol. 56:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isom, H. C., M. Gao, and B. Wigdahl. 1984. Characterization of guinea pig cytomegalovirus DNA. J. Virol. 49:426-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob, R. J., L. S. Morse, and B. Roizman. 1979. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 29:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koslowski, K. M., P. R. Shaver, X. Y. Wang, D. J. Tenney, and N. E. Pederson. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai, A. C., and Y. Chu. 1991. A rapid method for screening vaccinia virus recombinants. BioTechniques 10:564-565. [PubMed] [Google Scholar]
- 39.Lenk, E., S. Casjens, J. Weeks, and J. King. 1975. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology 68:182-199. [DOI] [PubMed] [Google Scholar]
- 40.Locker, H., and N. Frenkel. 1979. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J. Virol. 32:429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marks, J. R., and D. H. Spector. 1984. Fusion of the termini of the murine cytomegalovirus genome after infection. J. Virol. 52:24-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks, J. R., and D. H. Spector. 1988. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology 162:98-107. [DOI] [PubMed] [Google Scholar]
- 43.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGregor, A., and M. R. Schleiss. 2001. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol. Genet. Metab. 72:15-26. [DOI] [PubMed] [Google Scholar]
- 45.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McVoy, M. A., D. E. Nixon, and S. P. Adler. 1997. Circularization and cleavage of guinea pig cytomegalovirus genomes. J. Virol. 71:4209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McVoy, M. A., D. E. Nixon, S. P. Adler, and E. S. Mocarski. 1998. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J. Virol. 72:48-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McVoy, M. A., D. E. Nixon, J. K. Hur, and S. P. Adler. 2000. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J. Virol. 74:1587-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mocarski, E. S., A. C. Liu, and R. R. Spaete. 1987. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain). J. Gen. Virol. 68:2223-2230. [DOI] [PubMed] [Google Scholar]
- 51.Mocarski, E. S., and B. Roizman. 1982. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell 31:89-97. [DOI] [PubMed] [Google Scholar]
- 52.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 53.Nixon, D. E., and M. A. McVoy. 2002. Terminally repeated sequences on a herpesvirus genome are deleted following circularization but are reconstituted by duplication during cleavage and packaging of concatemeric DNA. J. Virol. 76:2009-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perdue, M. L., J. C. Cohen, C. C. Randall, and D. J. O'Callaghan. 1976. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology 74:194-208. [PubMed]
- 56.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-300. In H. Frainkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 3. Plenum, New York, N.Y.
- 57.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 1048-1066. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fundamental virology. Raven Press, New York, N.Y.
- 58.Scheffczik, H., C. G. Savva, A. Holzenburg, L. Kolesnikova, and E. Bogner. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scholz, B., S. Rechter, J. C. Drach, L. B. Townsend, and E. Bogner. 2003. Identification of the ATP-binding site in the terminase subunit pUL56 of human cytomegalovirus. Nucleic Acids Res. 31:1426-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Severini, A., A. R. Morgan, D. R. Tovell, and D. L. Tyrrell. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200:428-435. [DOI] [PubMed] [Google Scholar]
- 61.Slobedman, B., and A. Simmons. 1997. Concatemeric intermediates of equine herpesvirus type 1 DNA replication contain frequent inversions of adjacent long segments of the viral genome. Virology 229:415-420. [DOI] [PubMed] [Google Scholar]
- 62.Sternberg, N., and R. Weisberg. 1977. Packaging of coliphage lambda DNA. I. The role of the cohesive end site and the gene A protein. J. Mol. Biol. 117:717-731. [DOI] [PubMed] [Google Scholar]
- 63.Tamashiro, J. C., D. Filpula, T. Friedmann, and D. H. Spector. 1984. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J. Virol. 52:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamashiro, J. C., and D. H. Spector. 1986. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J. Virol. 59:591-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Umene, K. 1994. Excision of DNA fragments corresponding to the unit-length a sequence of herpes simplex virus type 1 and terminus variation predominate on one side of the excised fragment. J. Virol. 68:4377-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varmuza, S. L., and J. R. Smiley. 1985. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41:793-802. [DOI] [PubMed] [Google Scholar]
- 68.Vlazny, D. A., A. Kwong, and N. Frenkel. 1982. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc. Natl. Acad. Sci. USA 79:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner, M. J., and W. C. Summers. 1978. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J. Virol. 27:374-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 98:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, X., S. Efstathiou, and A. Simmons. 1994. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology 202:530-539. [DOI] [PubMed] [Google Scholar]