Abstract
The eukaryotic genome is in a constant state of modification and repair. Faithful transmission of the genomic information from parent to daughter cells depends upon an extensive system of surveillance, signaling, and DNA repair, as well as accurate synthesis of DNA during replication. Often, replicative synthesis occurs over regions of DNA that have not yet been repaired, presenting further challenges to genomic stability. DNA polymerase δ (Pol δ) occupies a central role in all of these processes: catalyzing the accurate replication of a majority of the genome, participating in several DNA repair synthetic pathways, and contributing structurally to the accurate bypass of problematic lesions during translesion synthesis. The concerted actions of pol δ on the lagging strand, pol ε on the leading strand, associated replicative factors, and the mismatch repair (MMR) proteins results in a mutation rate of less than one misincorporation per genome per replication cycle. This low mutation rate provides a high level of protection against genetic defects during development and may prevent the initiation of malignancies in somatic cells. This review explores the role of Pol δ in replication fidelity and genome maintenance.
Keywords: Fidelity, Cancer, DNA synthesis, DNA repair
INTRODUCTION
There are 15 known DNA-dependent polymerases in eukaryotic cells [Hubscher et al. 2002; Sweasy et al. 2006]. Bulk DNA replication is accomplished by a suite of three polymerases: the primase DNA polymerase α (Pol α), and the main replicative polymerases DNA polymerase δ (Pol δ) and DNA polymerase ε (Pol ε), which catalyze DNA synthesis on opposite strands [Nick McElhinny et al. 2008]. Others, known as specialized, bypass, or translesion polymerases, participate in various DNA transactions related to repair, genome stability, and the generation of antibody diversity. The eukaryotic polymerases are well-conserved in terms of overall architecture and sequence, especially within the catalytic domain. In this review, we focus on the main replicative polymerase, Pol δ, and its roles as a highly accurate DNA replicase, repair enzyme, and tumor suppressor.
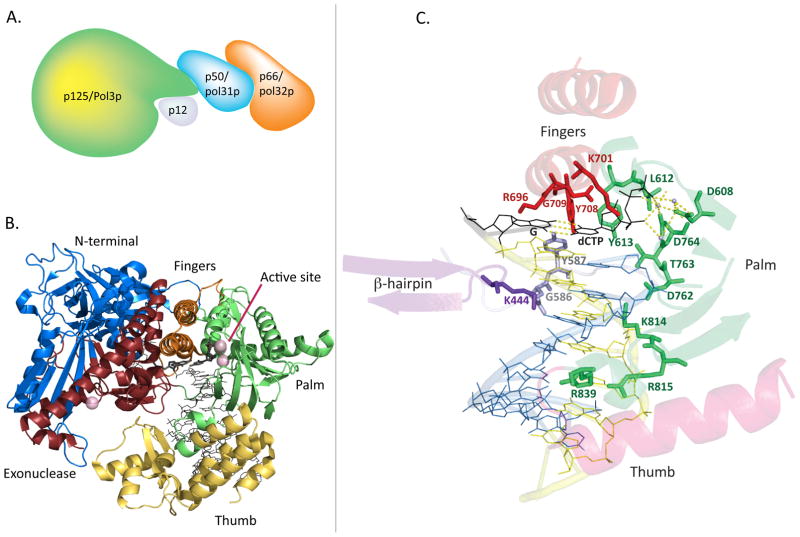
In Saccharomyces cerevisiae, Pol δ is composed of 3 separately encoded subunits—the catalytic subunit Pol3p, and structural subunits Pol31p and Pol32p [Boulet et al. 1989; Gerik et al. 1998; Sitney et al. 1989]. Small angle X-ray scattering studies of yeast Pol δ demonstrate that the holoenzyme is a heterotrimer [Jain et al. 2009]. The C-terminus of the catalytic subunit possesses a cysteine-rich domain at the end of a flexible tether; Pol31p associates with this cysteine-rich domain, and Pol32p is tightly associated with Pol31p. Mammalian Pol δ is composed of 4 subunits: the catalytic subunit p125 (encoded by POLD1, corresponding to yeast POL3), p50 (POLD2, corresponding to yeast POL31), p68 (POLD3 corresponding to yeast POL32), and p12 (POLD4), for which there is no known homologue in S. cerevisiae [Byrnes et al. 1976; Hughes et al. 1999; Lee et al. 1984; Liu et al. 2000] (Fig. 1A).
Fig. 1.
Structure of the Pol δ holoenzyme, catalytic subunit, and DNA binding pocket. A. A conceptual depiction of the four-subunit human Pol δ holoenzyme, based on demonstrated interactions of subunits and a small-angle X-ray scattering study (see text). B. Cartoon representation of the crystal structure of the p125 catalytic subunit in complex with DNA and the incoming dCTP (black) bound at the active site. Ca2+ ions are shown as purple spheres, representing the location of the Mg2+ atoms at the polymerase and exonuclease active sites. C. Pol δ active site and DNA binding channel, highlighting important side chains for polymerase fidelity, as well as purported “sensing” side chains along the minor groove. Palm residues are green, fingers residues are red, N-terminal domain residues are silver, β-hairpin site is purple, DNA template strand is yellow, and DNA primer strand is blue. The incoming dCTP and its template G are shown in black, and active site metals are shown as light blue spheres. Hydrogen bonds (yellow) are shown for the nascent base pair and for the active site metals. (Structure images generated in The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC. from PDB accession code 3IAY).
Pol δ is an essential protein in eukaryotes, and it has a major role in genome maintenance through its involvement in replicative DNA synthesis and multiple synthetic repair processes [Bell and Dutta 2002; Burgers 2009; Loeb and Monnat 2008]. In addition to its 5′→3′ DNA-directed polymerase activity, Polδ possesses 3′→5′ exonuclease activity that imparts the ability to remove newly added non-complementary nucleotides during replication [Byrnes et al. 1976; Morrison et al. 1993; Simon et al. 1991]. The distinguishing characteristics of the replicative polymerases Pol δ and Pol ε are high fidelity and high processivity, but with limited capacity to copy damaged template DNA.
Structure of Pol δ
The Pol δ holoenzyme participates in replicative synthesis in concert with the processivity factor PCNA (proliferating cell nuclear antigen) [Bravo et al. 1987]. A heterodimer composed of p125 and p50 comprises the core mammalian enzyme, which is capable of being stimulated by PCNA [Wang et al. 2011b; Zhou et al. 2012b]. The minimal combination of mammalian subunits for processive DNA synthesis is the core enzyme complexed with either p68 or p12, as demonstrated in the presence of the processivity factor PCNA on M13 gapped plasmid [Zhou et al. 2012a]. The p12 subunit can enhance processivity of the p125-p50-p68 subassembly by up to 15-fold [Podust et al. 2002]. The enhanced rate of DNA synthesis in vitro afforded by the contributions of PCNA and p12 come at the cost of reduced base selection fidelity, possibly by increasing the likelihood of bypass of DNA lesions that would otherwise stall the polymerase [Hashimoto et al. 2003; Meng et al. 2010; Mozzherin et al. 1996; Mozzherin et al. 1997]. This has biological significance as both factors may be targeted by the DNA damage response [Freudenthal et al. 2011; Kirchmaier 2011; Prives and Gottifredi 2008; Ulrich 2009; Zhang et al. 2007]. While most of the factors that influence the fidelity of Pol δ-directed DNA synthesis are intrinsic to the catalytic subunit, there are many potentially important interactions between the non-catalytic subunits and nuclear signaling and repair proteins that may also contribute to fidelity and genome stability (see [Bell and Dutta 2002; Rahmeh et al. 2012; Thommes and Hubscher 1990]).
The p125 catalytic subunit of Pol δ, as with almost all known DNA polymerases, has a distinctive architecture that resembles a right hand [Joyce and Steitz 1995] (Fig 1B). The downstream single stranded DNA is covered by a cleft between the exonuclease and N-terminal domains [Swan et al. 2009]. Nucleotide addition is catalyzed in a pocket formed by the amino acids of the palm, fingers, and thumb domains. The palm makes contact with the nascent duplex DNA, providing the catalytic side chains, and the thumb covers the duplex like a clamp. The fingers and thumb are mobile domains. Based on comparisons of structures of RB69 polymerase, the twin α-helices of the fingers domain rotate away from the catalytic site to create the open conformation, and perform the reverse to create the closed, active conformation [Franklin et al. 2001]. The thumb secures the DNA duplex, and contributes to chaperoning the primer strand to the exonuclease domain for exonucleolytic editing to occur [Franklin et al. 2001; Swan et al. 2009]. The C-terminal domain contains two cysteine-rich sites at the end of an unstructured tether, and contains a subdomain that is critical for interactions with the p50 subunit [Brocas et al. 2010; Cullmann et al. 1993] (see section on Translesion Synthesis for more discussion of this region).
The roles of Polδ in DNA replication
A large body of evidence supports the major role of Pol δ as that of the primary lagging strand replicase, while the somewhat more processive and accurate Pol ε copies the leading strand [Burgers 2009]. A substitution at the corresponding catalytic site residue in yeast Pol δ (L612) and Pol ε (M644) imparts lower fidelity and a distinct error signature to each enzyme [Li et al. 2005; Pursell et al. 2007a; Pursell et al. 2007b; Venkatesan et al. 2006]. Importantly, each of these polymerase mutants makes a specific error with elevated frequency, but rarely makes the reciprocal error. A strand preference of the two polymerases during DNA replication has been demonstrated by analyzing the sequence of DNA that is copied by either of these polymerase mutants for the signature mutations produced by each. Sequence changes between replication origins in yeast cells expressing the mutant polymerases reveal that the Pol δ error signature is predominantly on the lagging strand, and the Pol ε error signature is predominantly on the leading strand [Nick McElhinny et al. 2008]. These data have been determined both in vitro, using constructs containing defined yeast origins, and in vivo, using deep sequencing to examine regions surrounding yeast origins [Larrea et al. 2010]. Considering the essentiality of DNA replication, it is surprising that Pol ε catalytic activity is dispensable in yeast [Kesti et al. 1999]. Presumably, Pol δ is capable of copying the leading strand in the absence of Pol ε. An alternate model of the processing replication fork suggests that portions of the leading strand are copied by Pol δ in normal cells. It is possible that Pol δ can replace Pol ε in situations when the leading strand is unoccupied by a polymerase, such as at replication restart after a stalling event [Kunkel 2011; Pavlov and Shcherbakova 2010](Fig. 2). Each of these possibilities is open to experimental verification by sequencing DNA replicated in Pol ε mutants.
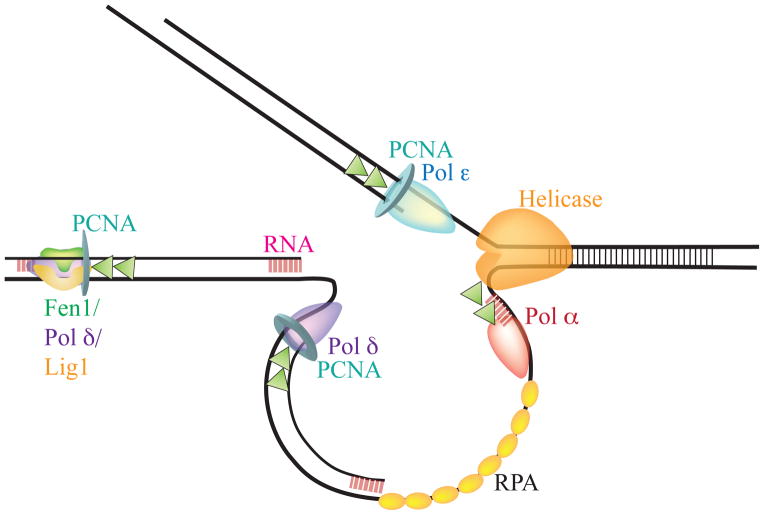
Fig. 2.
Model of the Eukaryotic Replication Fork. The current model, showing Okazaki fragments at three stages of formation. Next to the MCM helicase, the primase Pol α synthesizes an RNA primer (pink box with lines) and a small amount of DNA, beginning lagging strand synthesis. Replication protein A (RPA) coats the single stranded DNA between the Pol α-catalyzed primers. The next primer has been extended by the lagging strand replisome, represented by Pol δ and PCNA. The third Okazaki fragment has been completely extended, and Okazaki fragment maturation, directed by Pol δ, Fen1, and Lig1, is underway. The leading strand is shown as being copied by Pol ε and PCNA, although Pol δ may be responsible for some leading strand synthesis as well. Several important cofactors are not shown for simplicity. Figure inspired by and adapted from [Burgers 2009].
Genetic and biochemical studies in yeast have identified the enzymes involved in the coordinate processes of lagging strand synthesis and Okazaki fragment maturation [Zheng and Shen 2011]. The lagging strand is first primed by Pol α, which catalyzes the incorporation of a short RNA fragment, followed by 20–30 deoxynucleotides. Pol δ is loaded at the 3′-OH of this primer, bound to PCNA, and it replicates 200–300 bases until it encounters the next primer. There, it works in concert with the flap endonuclease, Fen1, to undertake two distinct activities: strand displacement synthesis and nick translation. In strand displacement synthesis, pol δ displaces the entire RNA primer while synthesizing DNA complementary to the template strand. Nick translation refers to the coordinate activities of pol δ strand displacement and Fen1 endonuclease activity regularly resecting a 1–5 nucleotide flap of RNA to move a single nucleotide nick along the length of the RNA primer, until the entire RNA/DNA duplex has been replaced and the nick can be ligated [Ayyagari et al. 2003; Jin et al. 2003].
The 3′→5′ exonuclease activity of Pol δ may direct an alternative pathway for flap processing [Jin et al. 2001]. Pol δ is capable of restricting the extent of strand displacement synthesis by idling on the DNA, carrying out repetitive extension and removal of one nucleotide, until the flap of displaced DNA or RNA can be resected by Fen1, or by a combination of Fen1 and Dna2 [Garg et al. 2004]. This inhibition of strand displacement synthesis protects against the generation of single strand segments of DNA, the cleavage of which could result in double strand breaks. In the absence of Fen1, idling may also provide a means of presenting a ligatable nick to achieve fragment maturation and prevent genomic instability
POL δ IN DNA REPLICATION
Base selection and proofreading
The rate of nucleotide misincorporation by budding yeast Pol δ has been reported in the range of 1.8 × 10−8 to 4.4 × 10−7 errors per generation, depending on the assay used [Daee et al. 2010; Fortune et al. 2005; Li et al. 2005; Morrison and Sugino 1994; Nick McElhinny et al. 2007; Pavlov et al. 2001; Venkatesan et al. 2006]. This low base substitution rate is a result of the serial processes of base selection, proofreading, and mismatch repair. The loss-of-function mutants for any combination of these processes often exhibit a synergistic increase in mutation rate over that of the single mutants [Morrison et al. 1993; Pavlov et al. 2001]. In vitro mutation rate measurements from purified wild type yeast Pol δ and proofreading-deficient Pol δ (Pol3-01) on M13mp2-lacZ gapped plasmid indicate that base discrimination alone catalyzes approximately 0.3 to 1 base substitutions per 100,000 nucleotides, and that proofreading contributes 10-fold or more to the overall fidelity of Pol δ [Fortune et al. 2005; Nick McElhinny et al. 2007]. Purified human Pol δ holoenzyme exhibits slightly higher fidelity in the same assay, with a similar enhancement by proofreading [Schmitt et al. 2009].
A few mechanisms that govern replicative fidelity appear to be universal among DNA polymerases, as demonstrated in kinetic and structural studies using enzymes from a wide range of phyla. These basic steps and constraints to the synthetic reaction, elaborated below in brief, provide a basis for understanding the Pol δ mutants described in this review. With respect to base selection fidelity, the initial step is discrimination of the incoming nucleotide at the active site (reviewed in [Kunkel and Bebenek 2000]. The exact details of the next step are not fully elaborated, but evidence from kinetics studies suggest that there is a brief opportunity for the nucleotide to adopt a favorable conformation with the template base. After the nucleotide binds in the active site, the fingers are triggered to close. Closure depends on correct nucleotide binding, as demonstrated by tracking variances in fluorescence from labeled primer-template constructs in Pol δ-related T7 DNA polymerase [Luo et al. 2007]. Based on kinetic measurements of a fluorescence-labeled E514C substitution in the T7 DNA polymerase, binding of the incorrect nucleotide results in a mismatch-specific conformational change in the fingers domain, and a sharp reduction in the rate of nucleotide addition, which favors nucleotide disassociation [Tsai and Johnson 2006]. Binding of the correct nucleotide induces a different conformational change, and increases the rate of nucleotide addition. The nucleotidyl transferase reaction then occurs at a pair of magnesium ions that are coordinated by amino acids of the palm domain [Joyce and Steitz 1995; Swan et al. 2009]. Studies in T7 polymerase indicate that pyrophosphate release triggers another conformational change prior to translocation to the next template position to begin the cycle over again [Patel et al. 1991].
Early studies of polymerases identified an inverse relationship between the extent of base misincorporation and the exonuclease activity of the enzyme [Muzyczka et al. 1972] (reviewed in [Reha-Krantz 2010]). Partitioning of activity between the polymerase and exonuclease domains is usually described in the context of kinetics. The rate of the nucleotide addition reaction dictates the likelihood of whether the enzyme will remain in polymerase mode or switch to an editing mode [Beard et al. 2002; Johnson 1993]. Mispaired nucleotides usually adopt a distorted structure in relation to the templating base, due to non-canonical hydrogen bonding geometries, leading to a momentary delay in DNA synthesis [Goodman 1997; Johnson and Beese 2004; Kool 2002]. A delay might also indicate a mismatch in one of the upstream base pairs, which could change the presentation of the primer or template strand, making it difficult for any incoming nucleotide to adopt a favorable orientation. Alternatively, the amino acid side chains that line the catalytic pocket and make extensive contacts with the minor groove of the nascent DNA duplex might induce a delay if a distortion is detected [Carver et al. 1994; Doublie et al. 1998; Ng et al. 1989; Thompson et al. 2002]. Purified polymerases often stall or are delayed at positions where a mismatch is forced, as in primer extension assays in which deoxynucleotide pool choice is restricted. [Echols and Goodman 1991]. Conversely, increasing the rate of polymerization appears to result in (or from) a reduced level of proofreading and, subsequently, a higher rate of errors [Mozzherin et al. 1996]. A reduced level of proofreading can result from a relaxed base selectivity, from reduced exonuclease partitioning, or from higher concentrations of free deoxynucleotides (reviewed in [Mathews 2006] and [Reha-Krantz 2010]).
Frameshift mutagenesis
Microsatellite instability (MSI) describes the tendency of short repeated tracts of sequence in DNA to undergo shrinkage and expansion, usually due to errors in DNA synthesis that result in a loss or gain of one or more repeat units [Sia et al. 1997; Streisinger et al. 1966]. It is a measurable phenotype that is used as a marker in human disease, such as colorectal cancer, and has been linked to defects in mismatch repair [Ionov et al. 1993; Leach et al. 1993; Umar et al. 2004]. Polymerases have much lower fidelity with respect to insertions and deletions when copying repetitive sequence than when copying non-repetitive sequences. The effect of iterative DNA sequence on insertion/deletion (indel) fidelity depends on the length of the repeat unit (inverse), the number of repeat units (inverse), and, in the case of homopolymeric runs, on whether the template is purinic or pyrimidinic [Harfe and Jinks-Robertson 2000; Kroutil et al. 1996; Sia et al. 1997].
Forward mutation assays and reversion assays with either wild type or exonuclease-deficient Pol δ in yeast indicate that, generally, Pol δ is very good at proofreading base selection errors and single base additions or deletions in non-repetitive sequence, but has limited capacity to proofread single base deletions in homopolymeric runs. For example, in a run of five thymidines in the LacZ sequence in M13mp2 gapped plasmid, wild type yeast Pol δ has a reversion frequency of 6.6 × 10−4, and this increases to 1.7 × 10−3 when the length of the run increases to seven thymidines [Fortune et al. 2005]. In a reporter assay featuring a 10-unit run of dinucleotide repeats, Pol δ exhibited an 8–30-fold increase in frameshift mutagenesis in the repeat region compared to a control region [Abdulovic et al. 2011]. Interestingly, this rate only increased slightly when an exonuclease-deficient Pol δ was used in the same assay, highlighting the limited capacity of Pol δ for detecting these types of errors, and the dependence of eukaryotic cells on mismatch repair for suppression of frameshift mutagenesis.
The prevailing models for frameshift mutagenesis all invoke a strand misalignment mechanism in which the primer terminus realigns to form the correct base pairing with either the +1 or −1 base on the template strand [Kunkel and Soni 1988] and reviewed in [Bebenek and Kunkel 2000]. The unpaired base would be flipped out of the duplex—a wrinkle in the strand—evading detection by intrinsic proofreading, but detectable by mismatch repair. Evidence for this mechanism in polymerases comes from a set of crystal structures from the error-prone human X-family polymerase Pol λ, which capture a slippage-mediated deletion event [Garcia-Diaz et al. 2006]. This highly deletion-prone polymerase is capable of realigning on the template to permit the next template base (originally at the −1 position) to pair with the bound nucleotide by flipping the intervening base into an extrahelical position. This configuration does not alter the conformation of the enzyme, and does not distort the position of the primer-template at the active site, as it is stabilized by a correct base pair at the T1P1 position. While it is unknown if the same structure can be accommodated by the tighter binding pocket of pol δ, and hence would represent a common mechanism for −1 frameshift mutagenesis, it fulfills the criteria of a structure that could evade detection by intrinsic proofreading.
DETERMINANTS OF FIDELITY IN THE POL δ CATALYTIC SUBUNIT
Multiple amino acids that line the catalytic pocket, as well as others that are located outside the catalytic site, contribute to the base selection fidelity of the enzyme (Fig 1C). Although the sequences corresponding to the conserved motifs A and B in DNA polymerases have been extensively modified through targeted or random mutagenesis in a variety of prokaryotic organisms, only a limited number of conserved side chains in the catalytic site have received close scrutiny in eukaryotic replicative polymerases. These are summarized in Table I, and an extensive list of antimutator pol δ variants can be found in [Herr et al. 2012]. Small, but important differences exist among the various polymerase families, so the following survey of informative fidelity mutants in the various regions of the catalytic site is restricted to Pol δ wherever possible, and to other B-family polymerases when necessary. Unless otherwise stated, all amino acid positions will refer to the S. cerevisiae sequence.
Table I.
Mutation Rates of Wild type and Mutant Pol δ Enzymes.
| Source | Substitution | Domain | B.s.rate in vivo (×10−7) | Assay | B.s.rate in vitro (×10−6) | Insertion (×10−6) | Deletion (×10−6) | Assay | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Budding yeast | WT | 0.2--1.2 | URA3 | 1 | |||||
| D641N | Palm | 630 | 1 | ||||||
| Pol3-01 | Exo | 24 | 2 | ||||||
|
|
|||||||||
| WT | 1.9-4.4 | CANr | 3 | ||||||
| D396N | Exo | 14 | 3 | ||||||
| S268F | N-terminal | 10 | 3 | ||||||
| P942L | Thumb | 9.7 | 3 | ||||||
| L612M | Motif A | 11 | 4 | ||||||
| L612F | Motif A | 7 | 5 | ||||||
| L612M | Motif A | 15 | 5 | ||||||
| L612G | Motif A | 33 | 5 | ||||||
| L612N | Motif A | 73 | 5 | ||||||
| L612K | Motif A | 26 | 5 | ||||||
| L612I | Motif A | 3.4 | 5 | ||||||
| L612T | Motif A | 3.7 | 5 | ||||||
| L612V | Motif A | 3 | 5 | ||||||
| Pol3-01 | Exo | 56 | 5 | ||||||
| Y708A | Motif B | 28 | 6 | ||||||
| Y613A | Motif A | lethal | 6 | ||||||
| D641N | Palm | 15.93 | 7 | ||||||
| S268F | N-terminal | 7.83 | 7 | ||||||
| R542C | Exo | 3.78 | 7 | ||||||
| D250K | N-terminal | 40.5 | 7 | ||||||
| T655I | Palm | 5.94 | 7 | ||||||
| C1074S | Thumb | 12.96 | 7 | ||||||
| Pol3-01 | Exo | 297 | 7 | ||||||
| D321V | Exo | 232.2 | 7 | ||||||
| D407A | Exo | 148.5 | 7 | ||||||
| D407V | Exo | 205.2 | 7 | ||||||
| D520E | Exo | 213.3 | 7 | ||||||
| D520V | Exo | 54 | 7 | ||||||
| R696W | Motif B | 67 | 8 | ||||||
|
|
|||||||||
| WT | 1.9 | CANr | 26 | 0.2 | 3.2 | M13--lacZ | 9 | ||
| L612M | Motif A | 17.4 | 130 | 0.63 | 51 | 9 | |||
|
|
|||||||||
| D250V | Exo | 170 | 0.68 | 31 | 9 | ||||
| WT | 13 | 10 | 13 | 10 | |||||
| Pol3-01 | Exo | 130 | 12 | 57 | 10 | ||||
|
| |||||||||
| Mouse | WT | 5 | Ouabain | 11 | |||||
| D400A | Exo | 72 | 11 | ||||||
|
|
|||||||||
| WT | 0.4 | HPRT | 12 | ||||||
| L604G | Motif A | 2 | 12 | ||||||
| L604K | Motif A | 1.7 | 12 | ||||||
|
| |||||||||
| Human | WT | 4.5 | 0.77 | 1.1 | M13--lacZ | 13 | |||
| D402A | Exo | 44 | 12 | 20 | 13 | ||||
| L606M | Motif A | 72 | 98 | 160 | 13 | ||||
| L606G | Motif A | 520 | 90 | 77 | 14 | ||||
| L606K | Motif A | 3.2 | 0.52 | 1.3 | 14 | ||||
B.s.—Base substitution; Pol3-01 — D421A/E423A; Insertions and deletions are ±1 frameshifts.
References:
[Gordenin et al. 1993],
The palm: motifs A and C
The structure and sequence of the palm domain is the most conserved feature of DNA polymerases, and overlaying structures of polymerases from different families reveals remarkable agreement with respect to the spatial organization of key sequences and the catalytic center [Venkatesan et al. 2006; Wang et al. 1997]. Two Mg2+ ions contribute to the polymerization reaction by activating the 3′ primer hydroxyl for nucleophilic attack on the α-phosphate of the incoming nucleotide, and a third Mg2+ ion binds the pyrophosphate leaving group [Swan et al. 2009]. The two active site aspartate residues (D608 and D764), in conjunction with E802, coordinate three Mg2+ ions in yeast Pol δ [Swan et al. 2009]. Substitution at cognate metal-coordinating sites in other polymerases, such as the Klenow fragment of Pol I, results in a dramatic decrease in polymerase activity and diminished fidelity in vitro [Joyce and Steitz 1995; Patel and Loeb 2000; Polesky et al. 1992].
Discrimination between dNTPs and ribonucleotides in DNA polymerases has been postulated to depend on a simple steric mechanism involving a single active site position and the 2′ OH of the ribonucleotide [Joyce 1997]. A surprisingly high level of ribonucleotide incorporation has been demonstrated to occur in wild type yeast, but an increase in this level can lead to genomic instability [Nick McElhinny et al. 2010a; Nick McElhinny et al. 2010b]. In Pol δ, a palm-associated residue, Y613, appears to interact directly with the sugar of the incoming deoxynucleotide [Swan et al. 2009]. Modification of this position in a variety of prokaryotic and viral polymerases results in a sharp reduction in ribonucleotide discrimination, leading to an increase in the rate of rNTP incorporation into replicating or damaged DNA [Astatke et al. 1998; DeLucia et al. 2006; Joyce 1997]. Modification of the identical position in the B-family polymerase RB69 (Y415A) drastically reduces discrimination against ribonucleotide incorporation [Yang et al. 2002a]. A Y613A substitution in haploid yeast, however, is lethal, and the same substitution in yeast pol ε is severely growth limited. The neighboring residue in pol ε, M644 (L612 in Pol δ) exhibits an 11-fold reduction in ribonucleotide selectivity when changed to a glycine, but a 4-fold higher level of discrimination when changed to a leucine, suggesting that the site of sugar discrimination is unclear in eukaryotic B-family polymerases [Nick McElhinny et al. 2010b]. The lethal and growth-restricted phenotypes seen for Y613A substitutions for Pol δ and Pol ε, respectively, suggest that Y613 may be required for catalysis in eukaryotes.
Yeast Pol δ L612 has been extensively studied as a site that is important for fidelity in T4, Taq, and E. coli Pol1, and has been modified in the mouse (L604) and human (L606) proteins [Niimi et al. 2004; Patel et al. 2001; Reha-Krantz and Nonay 1994; Schmitt et al. 2010; Shinkai et al. 2001; Venkatesan et al. 2007; Zhong et al. 2008]. Early work on an analogous position in T4 DNA polymerase revealed a sensitivity to the pyrophosphate analogue phosphonacetic acid (PAA) [Reha-Krantz and Nonay 1994]. This L612M mutation, when modeled in yeast cells, exhibits a phenotype that is consistent with reduced proofreading [Li et al. 2005]. The crystal structure of yeast Pol δ in ternary complex with DNA and dCTP indicates that L612 forms a hydrogen bond with the incoming nucleotide [Swan et al. 2009]. This intimate association with the primer end could indicate a mechanism for the reduced proofreading observed in the L612M variant that results in a 60–80 fold increase in base substitutions and 1000-fold more frameshifts in a mismatch repair deficient background, compared with mismatch repair-deficiency alone [Li et al. 2005]. Additionally, the L612M mutant exhibited a 75-fold increase in frameshift mutations on an Exo1-deficient background, suggesting that the role of Pol δ exonuclease activity in mismatch repair may be affected by this mutation.
A screen of all possible amino acid substitutions at L612 in yeast revealed eight viable mutants [Venkatesan et al. 2006]. The mutants had a variety of cell cycle phenotypes, from near-normal to a few (L612G and L612N) undergoing G2/M arrest. Most of the variants exhibited a characteristic error spectrum from CAN1 forward mutation experiments, showing a strong bias towards G->A and T->C transitions, but not C->T or A->G. The wild type enzyme has a similar bias, though its frequency of substitutions is much lower. The range of phenotypes that can be generated by manipulating this position is astonishing, and reflects the sensitivity of the active site to perturbation.
Motif C is a β-turn-β-loop located in the palm domain that contains the second aspartate (D764) responsible for coordinating Mg2+ ions at the active site. Additionally, D762 and T763 are positioned to have a role in sensing distortions in the minor groove at the first and second positions of the duplex (T1P1 and T2P2) [Swan et al. 2009]. While no studies of Pol δ have concentrated on motif C specifically, a mutagenesis screen in motif C of Taq polymerase revealed that different combinations of substitutions at 782–784 (Pol δ 759–761) yielded a range of phenotypes with respect to mismatched primer extension, highlighting the importance of this region for enzyme-duplex interactions that affect fidelity [Strerath et al. 2007].
There are residues outside of the three conserved motifs that also appear to make functional contacts with the nascent duplex, particularly in the minor groove. These include K444 in the exonuclease domain, Q586 and Y587 in an unstructured loop near the N-terminal domain, and K814, R815, and R839 in the palm [Swan et al. 2009]. With the exception of Q586 and R839, these positions are absolutely conserved in B-family polymerases [Copeland et al. 1993; Jacewicz et al. 2007; Yang et al. 2005]. It is possible that these sites are indeed necessary for sensing mismatches to engage the active site switch to exonucleolytic proofreading. Alternatively, they may be simply required to stabilize the duplex DNA, and mismatch-induced distortions in the DNA molecule itself are transmitted to the active site via the template strand, as suggested by Johnson and Beese [Johnson and Beese 2004].
The fingers: motif B
The fingers domain is a highly mobile part of the catalytic subunit, and its structure varies greatly among the various polymerase families and in different organisms [Sawaya et al. 1994; Swan et al. 2009; Wang et al. 1997]. However, a more highly conserved sequence, motif B, is usually found on one of the α-helices of the fingers domain. The extensive work that has been done with mutagenesis of motif B side chains of RB69, of family A polymerases (Pol I from E. coli and T. aquaticus), as well as the family B polymerase Pol α from S. cerevisiae, has revealed the major determinants of fidelity in the fingers domain [Bell et al. 1997; Ogawa et al. 2003; Suzuki et al. 1996; Suzuki et al. 2000; Yang et al. 2002b].
The basic structure of motif B is remarkably conserved, in that certain key residues, following the periodicity of the helix, occur from virus to eukaryotes at the same positions along the helical edge [Ogawa et al. 2003]. These side chains (R696, K701, N705, Y708) bind to and orient the incoming nucleotide at the active site. Structural analysis suggests that Y708 interacts with the primer end and may contribute to creating a favorable electrostatic environment for correct base pairing [Swan et al. 2009; Wang et al. 2011a]. Pavlov, et al. introduced an alanine substitution at this position, and found that the mutant strain exhibited a mild mutator phenotype with genomic instability that was greatly exacerbated in the context of reduced mismatch repair [Pavlov et al. 2001]. Attempts at randomly mutagenizing the entire region in DNA polymerase α suggested that two residues, K944 and G952 (pol δ K701 and G709 ), are essential [Ogawa et al. 2003]. Ogawa, et al. made and purified variants at G952, and found that they had at least 80-fold less activity than wild type Pol α.
The N-terminal amino acid of motif B in human Pol δ, R689, was found to be substituted by tryptophan in human DLD-1 colon cancer cell lines [Flohr et al. 1999; Popanda et al. 1999]. Modeling this mutant in yeast (R696W) revealed it to be a lethal substitution in haploid cells [Daee et al. 2010]. Titrating the relative expression of R696W with wild type Pol δ in diploid yeast demonstrated a strong mutator phenotype, as shown by scoring mutations and sequencing the CAN1 locus in Canr mutant cells. The error spectrum was strikingly similar to that of the hprt locus in DLD-1 cells, suggesting that this mutant polymerase exerts a detectable influence on the mutation profile of these cells. Purified R696W showed a strong bias toward incorporation of thymidine opposite guanine, and had much lower catalytic activity than wild type Pol δ. Structural analysis of R696W by modeling the substitution on the published yeast Pol δ ternary structure suggests that its location between the polymerase active site and the exonuclease site may affect active site switching [Swan et al. 2009]. Alternatively, a slight change in its interactions with residues on the N-terminal domain may alter the position of amino acids at the end of the rigid P-helix that are more proximal to the catalytic site.
The thumb: DNA binding and active site switching
The primary function of the thumb domain appears to be to stabilize the nascent duplex, as mutations in the thumb of T7 RNA polymerase and E. coli Pol I cause a loss of processivity and catalytic activity in copying an undamaged template [Beese et al. 1993; Bonner et al. 1994; Minnick et al. 1996]. However, the thumb also has a major function in enhancing fidelity by contributing to primer translocation during proofreading, and possibly by aiding in the detection of mismatches in the duplex. The thumb domain in B-family polymerases has two subdomains connected by a flexible hinge. One subdomain is proximal to the palm, and encloses the DNA double helix, and the other (the “tip” of the thumb) is packed against the exonuclease domain, which is opposite the palm domain and polymerase active site [Swan et al. 2009]. In Pol δ, a long β-hairpin from the exonuclease domain extends into the DNA major groove, while a shorter loop appears in crystal structure of RB69 polymerases [Shamoo and Steitz 1999; Wang et al. 1997] (Fig 1B).
A comparison of crystal structures of RB69 in the polymerizing and proofreading states reveals a change in thumb position suggesting that the thumb tip may be involved in translocation of the primer end [Franklin et al. 2001]. Mutational analysis of T4 DNA polymerase showed that active site switching mutants were primarily located in the two active sites, in the thumb and in the β-hairpin [Stocki et al. 1995]. β-hairpin mutants in the B-family polymerase RB69 exhibit a strong mutator phenotype that is based on reduced exonuclease activity [Trzemecka et al. 2009], and a genetic study of yeast Pol δ with a base substitution at L523 in the exonuclease domain that exhibited a strong defect in active site switching revealed mutants that were similar to Pol δ mutants that lack exonuclease activity [Jin et al. 2005]. The current model of B-family polymerase proofreading holds that the thumb tip associates with the primer strand, uses the β-hairpin placement along the major groove as a guide to melt the nascent duplex, and relocates the primer end to the exonuclease active site [Hadjimarcou et al. 2001; Reha-Krantz 1998; Stocki et al. 1995; Subuddhi et al. 2008]. The amount of primer end translocation appears to be limited to just a few base pairs, and the partitioning involves coordinated movements of the thumb tip, the fingers domain, and the exonuclease domain, as shown in crystal structures of RB69 complexed with DNA containing an abasic site analogue in the active site [Hogg et al. 2004].
Exonuclease domain
Pol δ exonuclease activity is a major factor affecting the fidelity of DNA synthesis, allowing for the removal of terminally mismatched primer strand bases, and is likely required for extrinsic editing of sequence catalyzed by Pol α [Morrison et al. 1991; Pavlov et al. 2006; Perrino and Loeb 1990; Simon et al. 1991]. The exonuclease domain of Pol δ is adjacent to the major groove of the duplex DNA, and it harbors the metal-coordinating site that catalyzes the removal of the terminal nucleoside from the primer strand [Beese and Steitz 1991; Swan et al. 2009]. Amino acid substitutions for either the coordinating aspartate or the glutamate residues abolishes exonucleolytic activity, effectively rendering the enzyme proofreading-deficient [Morrison et al. 1991; Morrison et al. 1993; Simon et al. 1991]. The pol3-01 mutant (D321A/E323A), lacking exonuclease activity, consistently exhibits a 10–100-fold higher rate of mutation than wild type Pol δ [Fortune et al. 2005; Jin et al. 2001; Morrison et al. 1993; Venkatesan et al. 2006]. Careful analysis of the types of mutations that arise from in vitro synthesis of a lacZ reporter in a gapped plasmid shows that proofreading by Pol δ exonuclease activity reduces base substitution errors that are introduced by the polymerase active site by approximately 60-fold [Fortune et al. 2005]. Abolishing exonuclease activity has a much less dramatic effect on single base and larger deletions between repeat sequences, at least in vitro, indicating that these errors are more likely to be detected by mismatch repair surveillance (see section on frameshift mutagenesis).
Across the catalytic subunit: insights on fidelity from antimutator variants
While the majority of variants discussed in this review contain substitutions that lead to an increased mutation rate, there is an extensive literature devoted to polymerase variants that exhibit decreased mutation rates [Drake and Allen 1968; Herr et al. 2011; Reha-Krantz 1998]. These antimutator variants can illuminate intrinsic determinants of fidelity because they seem to often arise in response to a specific mutation or environmental condition that leads to reduced fidelity [Drake 1993]. A notable exception is the β-hairpin residue G447S, modeled onto yeast Pol δ from an analogous site on T4 polymerase. On T4, this substitution caused an increase in mutations and a defect in partitioning to the exonuclease domain, while in Pol δ, it produced an antimutator phenotype, especially toward frameshift mutations [Hadjimarcou et al. 2001]. The L612M substitution in Motif A that results in PAA sensitivity can be partially rescued by another substitution, L758M, which attenuates its increase in mutation rate, its PAA sensitivity, and synthetic lethality with mismatch repair deficiency [Li et al. 2005]. Herr, et al, examined pol3-01/Msh6Δ yeast, which are synthetic lethal in the haploid state, to look for variants that escape lethality [Herr et al. 2012]. Most of the variants they identified on the Pol3 subunit were in or near the catalytic pocket or DNA “sensing” regions, implying that these substitutions probably affect base discrimination and/or the ability to extend a terminal mismatch.
POL δ AS A REPAIR PROTEIN
The contribution of Pol δ to genome stability and prevention of mutagenesis goes beyond its role in replicative fidelity. Most of the DNA repair processes in eukaryotic cells appear to involve Pol δ in mechanisms that still are not adequately defined.
Mismatch repair
Base substitution errors and frameshifts that escape polymerase proofreading are usually detected and corrected by post-replicative mismatch repair (MMR) [Kunkel and Erie 2005; Li 2008; Modrich 2006]. In canonical MMR, DNA distortions that indicate mispaired bases, or loops that are formed by frameshift mutations, are detected by a complex of either Msh2/Msh6 or Msh2/Msh3. On the lagging strand, the exonuclease Exo1 is responsible for the removal of tens to thousands of nucleotides, followed by polymerase-directed repair synthesis and ligation of the 3′ nick by the DNA ligase Lig1. Genetic analysis of MMR deficient yeast and biochemical analysis of human MMR proteins suggest that Msh2/Msh6 or Msh2/Msh3 use nicks at the 5′ end of Okazaki fragments, and/or interactions with PCNA, to identify the nascent strand [Iams et al. 2002; Pavlov et al. 2003; Umar et al. 1996]. These nicks may serve as an entry point for exonuclease digestion of regions harboring mismatches by Exo1 on the lagging strand [Modrich 1997].
Recent evidence using fluorescently-tagged MMR components suggests that replication and MMR may be temporally and spatially coupled [Hombauer et al. 2011]. Modrich showed in 1997 that HeLa cell nuclear extracts that lacked mismatch repair activity could be complemented either by a subfraction of repair proficient cell-extract that co-purified with Pol δ, or by purified calf thymus Pol δ alone, demonstrating the role of Pol δ as the repair polymerase in MMR [Longley et al. 1997]. Further, an Exo1-independent mechanism for MMR depends on the ability of Pol δ to undertake strand displacement synthesis, whereby a nicked strand is melted away from its complementary strand by the advance of a replicating Pol δ, creating a flap of DNA that is cleaved by Flap Endonuclease 1 (Fen1) [Kadyrov et al. 2009]. This strand displacement repair also requires the p68 subunit. Thus, the few errors that are generated by Pol δ during replication and which escape proofreading are usually addressed by Pol δ again as part of mismatch repair.
Translesion synthesis
Although cells have adapted highly efficient repair systems to limit the amount of spontaneous and endogenously-generated DNA damage, the replication machinery will nonetheless encounter these lesions [Branzei and Foiani 2010; Friedberg 2005]. Many of these lesions are capable of blocking progression of Pol δ in vitro, though sequencing of genomic sites after treatment with oxidative or methylating agents demonstrates that error-prone synthesis by Pol δ can still occur past such lesions in vivo [Pavlov et al. 2002]. Blocking lesions, replication stress, and alternate DNA structures can stall the replication fork, which can lead to fork collapse, strand breaks, and genomic rearrangements [Tourriere and Pasero 2007]. Commonly encountered base modifications such as 8-oxo-deoxyguanine or 6-methyl-deoxyguanine can be used as templates by a replicating Pol δ with reduced efficiency, but the altered base pairing properties of these lesions usually results in a mispair that pol δ is not able to extend [Fazlieva et al. 2009; McCulloch et al. 2009]. When Pol δ encounters an abasic site, it has a tendency to incorporate an adenine, possibly due to a highly conserved tyrosine (Y708) which acts as a mock template, and which has a more favorable electrostatic interaction with adenine [Obeid et al. 2010; Schaaper et al. 1983]]. Thus, Pol δ can occasionally bypass lesions encountered during replication, but it does so with reduced fidelity.
Translesion, or specialized polymerases can copy correctly across from specific lesions, but are generally not processive or accurate (see [Sale et al. 2012] for a review). However, for specific lesions, they can be more accurate than the replicative polymerases [Zahn et al. 2011]. For translesion polymerases to function on the lagging strand, Pol δ must be displaced from the replisome or template. Following bypass of the lesion, Pol δ would need to be reloaded onto the template. A model was recently proposed in which a cysteine-rich region in the C-terminal domain of the catalytic (p125) subunit serves as a point of regulation for the rest of the holoenzyme by controlling the interaction of the p125 subunit with the non-catalytic subunits (p50 and p66) [Netz et al. 2011]. Decades of work on various yeast mutants have supported a role for the p125 cysteine-rich domains in holoenzyme stability and, subsequently, a role for Pol δ in protecting genome stability [Chanet and Heude 2003; Cullmann et al. 1993; Giot et al. 1997; Hanna et al. 2007; Huang et al. 2002; Sanchez Garcia et al. 2004]. Netz, et al demonstrated that the cysteine-rich region, Cys B, formed an iron-sulfur cluster, while Cys A, the other cysteine-rich sequence, is likely a Zn-binding region [Netz et al. 2011].
The catalytic subunit of Pol ζ has a C-terminal region that is very similar to that of Pol δ, and was shown to be able to bind to p50-p66 with an affinity similar to that of the Pol δ catalytic subunit, leading to the proposal of a model where the catalytic subunits of Pol δ and Pol ζ exchange at the site of a blocking lesion [Baranovskiy et al. 2012]. Pol ζ was not capable of binding to subunits of Pol α or Pol ε. Johnson, et al. demonstrated that a four-subunit complex of Rev1p, Rev7p, Pol31p and Pol32p could be purified from yeast in a 1:1:1:1 stoichiometry, and put forth a competing model in which the Pol ζ holoenzyme normally exists as a four-subunit complex containing Pol31 and Pol32 [Johnson et al. 2012]. There is an entire exchange of Pol δ and Pol ζ holoenzymes in this model, which is supported by their observation that Rev1 and Rev7 aggregate in the absence of Pol31/Pol32. Further experimentation is required to refine the model of polymerase exchange at sites of damage, both with Pol ζ and with other translesion polymerases and the replicative polymerases.
Base excision repair
Base excision repair (BER) involves the removal of specific damaged bases, and generally requires a stepwise application of glycosylase and phosphodiesterase activity to excise the base and introduce a nick in the phosphodiester backbone (for review, see [Parikh et al. 1999; Wilson et al. 2000]. Depending on the type of lesion and the proteins involved, BER could involve the non-processive replacement of a single nucleotide, or strand displacement synthesis to replace a stretch of nucleotides [Dianov et al. 1992; Matsumoto and Bogenhagen 1991]. Left unrepaired, such damaged bases can cause Pol δ to mispair or stall, which could lead to genomic instability. While it is clearly established that Pol β is the major polymerase responsible for base excision repair (BER), there is strong evidence that Pol δ can play a situational role in repair of methylation damage.
The first evidence for a potential role for Pol δ in BER came from yeast cells harboring a temperature sensitive mutant of Pol δ. These were shown to be sensitive to methyl methanesulfonate, but not to UV light, at the elevated temperature. This sensitivity was found to be suppressed by introduction of a plasmid bearing mammalian Pol β [Blank et al. 1994]. Parsons, et al., demonstrated the requirement for Pol δ in the repair of single base adducts that are 3′ proximal to a single strand break [Parsons et al. 2007]. The BER glycosylases NTH1 and OGG1 cannot excise such complex lesions in Pol δ immunodepleted HeLa cell extracts. Repair activity was restored by addition of purified Pol δ, and was shown to depend on Pol δ exonuclease activity. This was further confirmed by using a damaged luciferase reporter in mouse embryonic fibroblasts (MEFs) lacking Pol δ exonuclease activity. Another group also used Pol δ immunodepleted cell extracts from Pol β-deficient MEFs and complemented the extracts with recombinant Pol δ, uncovering a possible role for the p12 subunit in modulating the activity of Pol δ in BER with respect to short or long-patch repair [Zhou et al. 2012a]. Also, yeast Pol δ mutants lacking the pol32 subunit were synthetically lethal in combination with mutant repair glycosylases, revealing a possible requirement for this subunit in abasic site synthesis by Pol δ, or for subsequent extension by Pol ζ [Auerbach and Demple 2010].
Nucleotide excision repair
Some DNA adducts, such as those produced by UV light, are refractory to BER, and can cause replication fork stalling if left unrepaired. Nucleotide excision repair (NER) addresses these lesions by removing a 25–30 bp segment of the strand that contains the lesion, followed by repair synthesis [Lehmann 2011]. The first evidence that Pol δ was a potential polymerase in NER came from quantitation of nucleotide incorporation in UV irradiated human fibroblasts using an inhibitor, butylphenyl-2′dGTP that discriminates between Pol δ and Pol α, and aphidicolin, which restricts Pol δ activity [Dresler et al. 1988; Hunting et al. 1991; Wright et al. 1994]. Subsequent reconstitution of NER repair from purified components confirmed that both Pol δ and Pol ε, as well as Pol κ, could contribute to repair synthesis, though they are recruited via different mechanisms, and possibly to different types of lesions [Araujo et al. 2000; Ogi et al. 2010].
Double strand break repair
In the event of double strand breaks and some alternate DNA structures, cells undergo non-homologous end joining (NHEJ) or homologous recombination (HR) to repair the break, depending on the type of lesion and the timing of the cell cycle [Daley et al. 2005; Maher et al. 2011]. Failure to promptly and effectively repair these breaks can lead to genomic instability or apoptosis. Much of the early evidence that Pol δ is involved in (HR) came from genetics studies demonstrating synthetic lethality of a variety of conditional Pol δ mutants in yeast with loss of recombination factors like rad51, rad52, and rad53 [Giot et al. 1997]. However, these data came from studies of pol3-13, which is multiply substituted in the region of the Fe-S cluster involved in maintaining holoenzyme integrity. It is more likely that the lethal phenotype in these cells is due to an increase in stalled or collapsed forks due to instability of the holoenzyme [Brocas et al. 2010; Chanet and Heude 2003].
Studies of the pol3-ct mutant (deletion of C-terminal LSKW), which also perturbs holoenzyme stability, but to a lesser extent than pol3-13, reveal the following phenotypes: a mild phenotype in meiotic crossing over resulting in shorter crossover tracts; an increase in spontaneous inter- and intra-chromosomal rearrangements; and a failure to complete break-induced recombination, resulting in damage-prone half-crossover structures [Galli et al. 2003; Maloisel et al. 2004; Smith et al. 2009]. Deletion of the pol32 subunit in yeast, which is thought to coordinate recruitment of Pol δ to PCNA on DNA, results in a reduced base substitution rate, but increased deletions and breaks at the replication fork that may reflect collapsed fork structures [Hanna et al. 2007; Huang et al. 2002]. These results suggest that pol3-ct and pol3-13 are holoenzyme destabilizing mutations that reduce processivity, resulting in a failure to complete repair synthesis, particularly in recombinatorial processes [Brocas et al. 2010].
More direct evidence for participation of Pol δ in recombination comes from a reconstituted D-loop extension system, whereby PCNA binds to a hybrid DNA loop and recruits Pol δ to achieve loop extension [Li et al. 2009]. Pol δ was also shown by 2-D electrophoresis of MMS-treated samples to be required for the formation of an X-structure that could correspond to an early step in template-switch recombination [Vanoli et al. 2010]. Break-induced recombination is a form of HR that occurs when an advancing replication fork encounters a double strand break. Pol δ can perform the synthesis portion of the repair, and the pol32 subunit, but not exonuclease activity, is essential, as demonstrated by use of a mating type switching-based assay in budding yeast [Holmes and Haber 1999; Lydeard et al. 2007]. This work also demonstrated the effect of PCNA on mating type switching, suggesting that the mode of recruitment of Pol δ to recombination intermediates that result from different types of lesions may be similar.
POL δ IN CANCER
The role of mutations in Pol δ in human cancer remains to be defined. In the case of DNA Pol β, a variety of mutant enzymes possessing a wide range of activity have been found in tumors arising in diverse tissue types [Donigan et al. 2012]. One might expect that mutations in Pol δ would be a common finding in cancers that express a mutator phenotype and contain large numbers of mutations. However, mutations in the catalytic domain of Pol δ almost invariably reduce catalytic activity, and cells harboring these mutant genes may replicate more slowly and be outcompeted during the evolution of a tumor. Thus, the advantage that is conferred by the expression of a mutator would be greatest early in tumorigenesis, and the presence of Pol δ variants would only be detected in subclones using deep DNA sequencing methods [Beckman and Loeb 2006]. Fortunately, accurate methods for next-generation DNA sequencing are being developed and can be applied to identifying rare mutations within human cancers. A recent publication from the Cancer Genome Atlas Network reported that 25% of colorectal cancer biopsies had coding substitutions in Pol ε or mismatch repair genes, correlating with the more hypermutated, MSI-positive samples. Of the eighteen Pol ε substitutions, three were found in the exonuclease domain, one had been previously reported in dbSNP (www.ncbi.nlm.nih.gov/SNP), and one was in the conserved Pol VI region. Two Pol δ substitutions were reported, one of which (human R808, yeast R815) is predicted to make extensive contacts with the minor groove of the duplex (Fig. 1C).
Mouse models of Pol δ
While there have been no clear links established between Pol δ and human carcinogenesis, several mouse models of Pol δ mutants demonstrate that Pol δ is a tumor suppressor [Preston et al. 2010]. Mice homozygous null for Pol δ exhibited peri-implantation lethality, with poor blastocyst outgrowth in vitro, confirming the essentiality of the gene that was noted in yeast [Uchimura et al. 2009]. A point mutation that inactivates the exonuclease domain of Pol δ was exploited by gene targeting to generate Pold1D400A/D400A mice [Goldsby et al. 2002; Goldsby et al. 2001]. Mice lacking Pol δ proofreading develop normally to adulthood, but then succumb to a variety of cancers, notably thymic lymphomas and squamous cell carcinomas. A similar phenotype was seen in Pold1D400A/− mice [Uchimura et al. 2009]. It is notable that the tumor types that were observed in the absence of Pol δ proofreading were distinct from those seen in mouse models of MMR deficiency and those seen in proofreading-deficient Pol ε mice, indicating that the mutations introduced by the mutant Pol δ contribute to tissue-specific tumorigenesis [Albertson et al. 2009].
The motif A yeast Pol δ mutants L612G and L612K were modeled in mouse (L604), and resulted in remarkably different phenotypes [Venkatesan et al. 2007]. In both cases, the homozygous mutants died in utero, but the heterozygotes were viable. Mouse embryonic fibroblasts demonstrated 4 to 5-fold elevated spontaneous mutation rates via hprt forward mutation analysis, but the lysine substitution exhibited nearly twice the incidence of chromosome instability in metaphase spreads (38-fold over wild type), and a much shorter lifespan brought on, in part, by accelerated tumor formation. Further analysis of these mutant polymerases with purified human Pol δ preparations revealed that the lysine substitution imparted an antimutator phenotype with regard to base selection, but was a hypermutator with regard to deletions, while the glycine substitution induced a general mutator phenotype with slightly enhanced lesion bypass capability [Schmitt et al. 2010]. The lower lesion bypass capability and impaired replication fork progression demonstrated by the lysine substitution strongly suggests that its higher base substitution fidelity comes at the cost of an increase in stalled replication forks, which can lead to DNA breaks. Thus, the more aggressive tumor presentation and lower lifespan of the L604K mice is likely due to high genomic instability.
Pol δ in tumors and tumor cell lines
Studies of small cell lung cancer biopsies revealed a consistent reduction in the mRNA and protein levels of the p12 subunit [Huang et al. 2010]. This subunit is targeted by the DNA damage response to disassociate from the replicative complex, resulting in slower synthesis with reduced bypass capability. Much like the effect of the lysine substitution at L604 in mice, reducing the level of the p12 subunit in HCT116 colon cancer cells by siRNA treatment results in a marked increase in broken chromosomes. Reducing the amount of the p125 subunit is sufficient to induce genomic instability, as reduced expression of POL3 in yeast results in a striking mutator phenotype in which over half of the errors found by sequencing the CAN1 locus are deletion events between direct repeats [Kokoska et al. 2000]. Another study linked lower expression of POL3 to fragile site instability in yeast, presumably by the induction of double strand breaks at stalled replication forks. [Lemoine et al. 2008].
Mutations in Pol δ have been found in a handful of cancer cell lines but few with changes in the polymerase active site or exonuclease domain [da Costa et al. 1995; Flohr et al. 1999]. In addition to the R689W variant mentioned earlier, the R506H (exonuclease domain) variant was reported in the DLD-1 colon cancer cell line. The purified yeast version of this mutant was found to have a mild (2.5-fold) increase in mutation rate in an MMR-deficient background [Daee et al. 2010]. The R648Q variant, purified from rat hepatoma cells, is located adjacent to the fingers domain, and exhibits an increased sensitivity to a commonly used polymerase inhibitor, butylphenyl-dGTP, and to antineoplastic drugs, and may have an increased tendency to mispair opposite O6-methyl-dG [Popanda et al. 1999].
Inhibitors to Pol δ in the treatment of cancer
Given the central role for Pol δ in replication and DNA repair, and its many interactions with other key proteins in cell proliferation and survival, it would seem to be an excellent target for small molecule inhibitors of replication. Since cancer cells have been demonstrated, in many cases, to accumulate mutations at a greater rate than healthy cells, use of mutagenic polymerase substrates could increase the rate of lethal mutations in these error-prone, rapidly dividing cells [Prindle et al. 2010]. Nucleoside analogs have been in use as antineoplastic agents for decades, and they can often act as mutagens, in addition to their more established role as inhibitors of deoxynucleotide metabolism [Longley et al. 2003; Shao et al. 2006]. Alternatively, inhibitors could be developed that block replication, either by binding to the active site or targeting the interaction domains between p125 and p68. Alternatively, molecules that compete with p12 for binding to the catalytic subunit, or that block the PCNA-holoenzyme interaction could be valuable, particularly in cancers in which homologous recombination is already compromised, and which can be sensitized by ionizing radiation [Punchihewa et al. 2012]. Inhibitors already exist for BRCA2, PARP, ERCC1, and other DNA repair proteins in the treatment of cancer due to their roles in recognition and repair of strand breaks [Janssen et al. 2009; Ljungman 2009]. Inhibiting Pol δ may have a broader reach, as it could be synthetically lethal with a variety of DNA repair defects.
Perspective
The field of DNA polymerase research began over 50 years ago, and Pol δ has been studied for over 35 years, but new and fascinating properties continue to emerge about this protein. It has been a story of continual development and application of new biochemical and genetic tools to address old questions--leading to the discovery of new tools, and new questions. We are just beginning to get a clearer picture of the role of the non-catalytic subunits of Pol δ, and of the role of Fe-S clusters in polymerase biology. Genomic technologies hold some promise to address the role of Pol δ in repair processes, as well as to define the role of replicative polymerases in cancer and other diseases. Studies of the kinetics of Pol δ, and computer modeling of structural and kinetic data have shed light and raised questions about the rate limiting steps and manner of base selection and translocation, and these methods could be used to address a model of active site switching with various DNA mismatches. We also look forward to more structures of yeast and human Pol δ, with mismatches in the active site, and a ternary structure in the open conformation. Eventually, a three-dimensional, dynamic understanding of Pol δ in replication and error correction at the atomic level will emerge.
Acknowledgments
The authors would like to thank our colleagues Alan Herr, Jiang-Cheng Shen, Scott Kennedy, and Eddie Fox for their careful reading of the manuscript, and for informative discussions. This work is supported by NIH grants CA15802, CA102029, and POLCA77852. The manuscript was prepared by MJP with significant intellectual input from LAL.
References
- Abdulovic AL, Hile SE, Kunkel TA, Eckert KA. The in vitro fidelity of yeast DNA polymerase delta and polymerase epsilon holoenzymes during dinucleotide microsatellite DNA synthesis. DNA Repair (Amst) 2011;10(5):497–505. doi: 10.1016/j.dnarep.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106(40):17101–4. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14(3):349–59. [PMC free article] [PubMed] [Google Scholar]
- Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci U S A. 1998;95(7):3402–7. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach PA, Demple B. Roles of Rev1, Pol zeta, Pol32 and Pol eta in the bypass of chromosomal abasic sites in Saccharomyces cerevisiae. Mutagenesis. 2010;25(1):63–9. doi: 10.1093/mutage/gep045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278(3):1618–25. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA Polymerase delta and zeta Switch by Sharing Accessory Subunits of DNA Polymerase delta. J Biol Chem. 2012;287(21):17281–7. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Shock DD, Vande Berg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J Biol Chem. 2002;277(49):47393–8. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- Beckman RA, Loeb LA. Efficiency of carcinogenesis with and without a mutator mutation. Proc Natl Acad Sci U S A. 2006;103(38):14140–5. doi: 10.1073/pnas.0606271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260(5106):352–5. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JB, Eckert KA, Joyce CM, Kunkel TA. Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J Biol Chem. 1997;272(11):7345–51. doi: 10.1074/jbc.272.11.7345. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blank A, Kim B, Loeb LA. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994;91(19):9047–51. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner G, Lafer EM, Sousa R. The thumb subdomain of T7 RNA polymerase functions to stabilize the ternary complex during processive transcription. J Biol Chem. 1994;269(40):25129–36. [PubMed] [Google Scholar]
- Boulet A, Simon M, Faye G, Bauer GA, Burgers PM. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989;8(6):1849–54. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–19. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326(6112):515–7. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- Brocas C, Charbonnier JB, Dherin C, Gangloff S, Maloisel L. Stable interactions between DNA polymerase delta catalytic and structural subunits are essential for efficient DNA repair. DNA Repair (Amst) 2010;9(10):1098–111. doi: 10.1016/j.dnarep.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284(7):4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes JJ, Downey KM, Black VL, So AG. A new mammalian DNA polymerase with 3′ to 5′ exonuclease activity: DNA polymerase delta. Biochemistry. 1976;15(13):2817–23. doi: 10.1021/bi00658a018. [DOI] [PubMed] [Google Scholar]
- Carver TE, Jr, Hochstrasser RA, Millar DP. Proofreading DNA: recognition of aberrant DNA termini by the Klenow fragment of DNA polymerase I. Proc Natl Acad Sci U S A. 1994;91(22):10670–4. doi: 10.1073/pnas.91.22.10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet R, Heude M. Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae. Curr Genet. 2003;43(5):337–50. doi: 10.1007/s00294-003-0407-2. [DOI] [PubMed] [Google Scholar]
- Copeland WC, Lam NK, Wang TS. Fidelity studies of the human DNA polymerase alpha. The most conserved region among alpha-like DNA polymerases is responsible for metal-induced infidelity in DNA synthesis. J Biol Chem. 1993;268(15):11041–9. [PubMed] [Google Scholar]
- Cullmann G, Hindges R, Berchtold MW, Hubscher U. Cloning of a mouse cDNA encoding DNA polymerase delta: refinement of the homology boxes. Gene. 1993;134(2):191–200. doi: 10.1016/0378-1119(93)90093-i. [DOI] [PubMed] [Google Scholar]
- da Costa LT, Liu B, el-Deiry W, Hamilton SR, Kinzler KW, Vogelstein B, Markowitz S, Willson JK, de la Chapelle A, Downey KM, et al. Polymerase delta variants in RER colorectal tumours. Nat Genet. 1995;9(1):10–1. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase delta variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci U S A. 2010;107(1):157–62. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–51. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- DeLucia AM, Chaudhuri S, Potapova O, Grindley ND, Joyce CM. The properties of steric gate mutants reveal different constraints within the active sites of Y-family and A-family DNA polymerases. J Biol Chem. 2006;281(37):27286–91. doi: 10.1074/jbc.M604393200. [DOI] [PubMed] [Google Scholar]
- Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992;12(4):1605–12. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan KA, Sun KW, Nemec AA, Murphy DL, Cong X, Northrup V, Zelterman D, Sweasy JB. Human POLB Gene Is Mutated in High Percentage of Colorectal Tumors. J Biol Chem. 2012;287(28):23830–9. doi: 10.1074/jbc.M111.324947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391(6664):251–8. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Drake JW. General antimutators are improbable. J Mol Biol. 1993;229(1):8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- Drake JW, Allen EF. Antimutagenic DNA polymerases of bacteriophage T4. Cold Spring Harb Symp Quant Biol. 1968;33:339–44. doi: 10.1101/sqb.1968.033.01.039. [DOI] [PubMed] [Google Scholar]
- Dresler SL, Gowans BJ, Robinson-Hill RM, Hunting DJ. Involvement of DNA polymerase delta in DNA repair synthesis in human fibroblasts at late times after ultraviolet irradiation. Biochemistry. 1988;27(17):6379–83. doi: 10.1021/bi00417a028. [DOI] [PubMed] [Google Scholar]
- Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Fazlieva R, Spittle CS, Morrissey D, Hayashi H, Yan H, Matsumoto Y. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37(9):2854–66. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohr T, Dai JC, Buttner J, Popanda O, Hagmuller E, Thielmann HW. Detection of mutations in the DNA polymerase delta gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer. 1999;80(6):919–29. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280(33):29980–7. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105(5):657–67. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Freudenthal BD, Brogie JE, Gakhar L, Kondratick CM, Washington MT. Crystal structure of SUMO-modified proliferating cell nuclear antigen. J Mol Biol. 2011;406(1):9–17. doi: 10.1016/j.jmb.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6(12):943–53. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- Galli A, Cervelli T, Schiestl RH. Characterization of the hyperrecombination phenotype of the pol3-t mutation of Saccharomyces cerevisiae. Genetics. 2003;164(1):65–79. doi: 10.1093/genetics/164.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124(2):331–42. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18(22):2764–73. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273(31):19747–55. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase delta in DNA repair in vivo. Genetics. 1997;146(4):1239–51. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc Natl Acad Sci U S A. 2002;99(24):15560–5. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7(6):638–9. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- Goodman MF. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci U S A. 1997;94(20):10493–5. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjimarcou MI, Kokoska RJ, Petes TD, Reha-Krantz LJ. Identification of a mutant DNA polymerase delta in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics. 2001;158(1):177–86. doi: 10.1093/genetics/158.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M, Ball LG, Tong AH, Boone C, Xiao W. Pol32 is required for Pol zeta-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat Res. 2007;625(1–2):164–76. doi: 10.1016/j.mrfmmm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. Sequence composition and context effects on the generation and repair of frameshift intermediates in mononucleotide runs in Saccharomyces cerevisiae. Genetics. 2000;156(2):571–8. doi: 10.1093/genetics/156.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Shimizu K, Nakashima N, Sugino A. Fidelity of DNA polymerase delta holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42(48):14207–13. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Ogawa M, Lawrence NA, Williams LN, Eggington JM, Singh M, Smith RA, Preston BD. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2012;7(10):e1002282. doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Williams LN, Preston BD. Antimutator variants of DNA polymerases. Crit Rev Biochem Mol Biol. 2011;46(6):548–70. doi: 10.3109/10409238.2011.620941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23(7):1483–93. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96(3):415–24. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147(5):1040–53. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ME, Rio AG, Galibert MD, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160(4):1409–22. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QM, Tomida S, Masuda Y, Arima C, Cao K, Kasahara TA, Osada H, Yatabe Y, Akashi T, Kamiya K, et al. Regulation of DNA polymerase POLD4 influences genomic instability in lung cancer. Cancer Res. 2010;70(21):8407–16. doi: 10.1158/0008-5472.CAN-10-0784. [DOI] [PubMed] [Google Scholar]
- Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–63. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- Hughes P, Tratner I, Ducoux M, Piard K, Baldacci G. Isolation and identification of the third subunit of mammalian DNA polymerase delta by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res. 1999;27(10):2108–14. doi: 10.1093/nar/27.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunting DJ, Gowans BJ, Dresler SL. DNA polymerase delta mediates excision repair in growing cells damaged with ultraviolet radiation. Biochem Cell Biol. 1991;69(4):303–8. doi: 10.1139/o91-046. [DOI] [PubMed] [Google Scholar]
- Iams K, Larson ED, Drummond JT. DNA template requirements for human mismatch repair in vitro. J Biol Chem. 2002;277(34):30805–14. doi: 10.1074/jbc.M200846200. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363(6429):558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Jacewicz A, Makiela K, Kierzek A, Drake JW, Bebenek A. The roles of Tyr391 and Tyr619 in RB69 DNA polymerase replication fidelity. J Mol Biol. 2007;368(1):18–29. doi: 10.1016/j.jmb.2007.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Hammel M, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural insights into yeast DNA polymerase delta by small angle X-ray scattering. J Mol Biol. 2009;394(3):377–82. doi: 10.1016/j.jmb.2009.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci U S A. 2009;106(45):19108–13. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J Biol Chem. 2003;278(3):1626–33. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- Jin YH, Garg P, Stith CM, Al-Refai H, Sterling JF, Murray LJ, Kunkel TA, Resnick MA, Burgers PM, Gordenin DA. The multiple biological roles of the 3′-->5′ exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol Cell Biol. 2005;25(1):461–71. doi: 10.1128/MCB.25.1.461-471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′-->5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci U S A. 2001;98(9):5122–7. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc Natl Acad Sci U S A. 2012;109(31):12455–60. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116(6):803–16. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A. 1997;94(5):1619–22. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Steitz TA. Polymerase structures and function: variations on a theme? J Bacteriol. 1995;177(22):6321–9. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci U S A. 2009;106(21):8495–500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3(5):679–85. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- Kirchmaier AL. Ub-family modifications at the replication fork: Regulating PCNA-interacting components. FEBS Lett. 2011;585(18):2920–8. doi: 10.1016/j.febslet.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Kokoska RJ, Stefanovic L, DeMai J, Petes TD. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase delta or by decreases in the cellular levels of DNA polymerase delta. Mol Cell Biol. 2000;20(20):7490–504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- Kroutil LC, Register K, Bebenek K, Kunkel TA. Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry. 1996;35(3):1046–53. doi: 10.1021/bi952178h. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Balancing eukaryotic replication asymmetry with replication fidelity. Curr Opin Chem Biol. 2011;15(5):620–6. doi: 10.1016/j.cbpa.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263(29):14784–9. [PubMed] [Google Scholar]
- Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107(41):17674–9. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75(6):1215–25. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Lee MY, Tan CK, Downey KM, So AG. Further studies on calf thymus DNA polymerase delta purified to homogeneity by a new procedure. Biochemistry. 1984;23(9):1906–13. doi: 10.1021/bi00304a003. [DOI] [PubMed] [Google Scholar]
- Lehmann AR. DNA polymerases and repair synthesis in NER in human cells. DNA Repair (Amst) 2011;10(7):730–3. doi: 10.1016/j.dnarep.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Kokoska RJ, Petes TD. Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Mol Cell Biol. 2008;28(17):5359–68. doi: 10.1128/MCB.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase delta in Saccharomyces cerevisiae. Genetics. 2005;170(2):569–80. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol Cell. 2009;36(4):704–13. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY. Identification of a fourth subunit of mammalian DNA polymerase delta. J Biol Chem. 2000;275(25):18739–44. doi: 10.1074/jbc.M001217200. [DOI] [PubMed] [Google Scholar]
- Ljungman M. Targeting the DNA damage response in cancer. Chem Rev. 2009;109(7):2929–50. doi: 10.1021/cr900047g. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9(8):594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3(5):330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272(16):10917–21. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc Natl Acad Sci U S A. 2007;104(31):12610–5. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448(7155):820–3. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Maher RL, Branagan AM, Morrical SW. Coordination of DNA replication and recombination activities in the maintenance of genome stability. J Cell Biochem. 2011;112(10):2672–82. doi: 10.1002/jcb.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L, Bhargava J, Roeder GS. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics. 2004;167(3):1133–42. doi: 10.1534/genetics.104.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20(9):1300–14. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Bogenhagen DF. Repair of a synthetic abasic site involves concerted reactions of DNA synthesis followed by excision and ligation. Mol Cell Biol. 1991;11(9):4441–7. doi: 10.1128/mcb.11.9.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic Acids Res. 2009;37(9):2830–40. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou Y, Lee EY, Lee MY, Frick DN. The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry. 2010;49(17):3545–54. doi: 10.1021/bi100042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnick DT, Astatke M, Joyce CM, Kunkel TA. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J Biol Chem. 1996;271(40):24954–61. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
- Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272(40):24727–30. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281(41):30305–9. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′----5′ exonuclease activity. Proc Natl Acad Sci U S A. 1991;88(21):9473–7. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12(4):1467–73. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Sugino A. The 3′-->5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242(3):289–96. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, McConnell M, Jasko MV, Krayevsky AA, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes misincorporation catalyzed by calf thymus DNA polymerase delta. J Biol Chem. 1996;271(49):31711–7. doi: 10.1074/jbc.271.49.31711. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc Natl Acad Sci U S A. 1997;94(12):6126–31. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzyczka N, Poland RL, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972;247(22):7116–22. [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2011;8(1):125–32. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Weiss SJ, Fisher PA. Recognition and binding of template-primers containing defined abasic sites by Drosophila DNA polymerase alpha holoenzyme. J Biol Chem. 1989;264(22):13018–23. [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010a;6(10):774–81. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 2007;282(4):2324–32. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010b;107(11):4949–54. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24(7):2734–46. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, Marx A. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010;29(10):1738–47. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Limsirichaikul S, Niimi A, Iwai S, Yoshida S, Suzuki M. Distinct function of conserved amino acids in the fingers of Saccharomyces cerevisiae DNA polymerase alpha. J Biol Chem. 2003;278(21):19071–8. doi: 10.1074/jbc.M208605200. [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37(5):714–27. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9(1):37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- Parsons JL, Preston BD, O’Connor TR, Dianov GL. DNA polymerase delta-dependent repair of DNA single strand breaks containing 3′-end proximal lesions. Nucleic Acids Res. 2007;35(4):1054–63. doi: 10.1093/nar/gkl1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Kawate H, Adman E, Ashbach M, Loeb LA. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J Biol Chem. 2001;276(7):5044–51. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
- Patel PH, Loeb LA. Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J Biol Chem. 2000;275(51):40266–72. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30(2):511–25. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16(2):202–7. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13(9):744–8. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10(1):207–13. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork-20 years later. Mutat Res. 2010;685(1–2):45–53. doi: 10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001;159(1):47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino FW, Loeb LA. Hydrolysis of 3′-terminal mispairs in vitro by the 3′----5′ exonuclease of DNA polymerase delta permits subsequent extension by DNA polymerase alpha. Biochemistry. 1990;29(22):5226–31. doi: 10.1021/bi00474a002. [DOI] [PubMed] [Google Scholar]
- Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277(6):3894–901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- Polesky AH, Dahlberg ME, Benkovic SJ, Grindley ND, Joyce CM. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem. 1992;267(12):8417–28. [PubMed] [Google Scholar]
- Popanda O, Flohr T, Fox G, Thielmann HW. A mutation detected in DNA polymerase delta cDNA from Novikoff hepatoma cells correlates with abnormal catalytic properties of the enzyme. J Cancer Res Clin Oncol. 1999;125(11):598–608. doi: 10.1007/s004320050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20(5):281–93. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindle MJ, Fox EJ, Loeb LA. The mutator phenotype in cancer: molecular mechanisms and targeting strategies. Curr Drug Targets. 2010;11(10):1296–303. doi: 10.2174/1389450111007011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C, Gottifredi V. The p21 and PCNA partnership: a new twist for an old plot. Cell Cycle. 2008;7(24):3840–6. doi: 10.4161/cc.7.24.7243. [DOI] [PubMed] [Google Scholar]
- Punchihewa C, Inoue A, Hishiki A, Fujikawa Y, Connelly M, Evison B, Shao Y, Heath R, Kuraoka I, Rodrigues P, et al. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J Biol Chem. 2012;287(17):14289–300. doi: 10.1074/jbc.M112.353201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase epsilon. Nucleic Acids Res. 2007a;35(9):3076–86. doi: 10.1093/nar/gkm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007b;317(5834):127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Zhou Y, Xie B, Li H, Lee EY, Lee MY. Phosphorylation of the p68 subunit of Pol delta acts as a molecular switch to regulate its interaction with PCNA. Biochemistry. 2012;51(1):416–24. doi: 10.1021/bi201638e. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ. Regulation of DNA polymerase exonucleolytic proofreading activity: studies of bacteriophage T4 “antimutator” DNA polymerases. Genetics. 1998;148(4):1551–7. doi: 10.1093/genetics/148.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804(5):1049–63. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ, Nonay RL. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. J Biol Chem. 1994;269(8):5635–43. [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13(3):141–52. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA. The C-terminal zinc finger of the catalytic subunit of DNA polymerase delta is responsible for direct interaction with the B-subunit. Nucleic Acids Res. 2004;32(10):3005–16. doi: 10.1093/nar/gkh623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264(5167):1930–5. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Kunkel TA, Loeb LA. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983;80(2):487–91. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91(9):1163–72. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MW, Venkatesan RN, Pillaire MJ, Hoffmann JS, Sidorova JM, Loeb LA. Active site mutations in mammalian DNA polymerase delta alter accuracy and replication fork progression. J Biol Chem. 2010;285(42):32264–72. doi: 10.1074/jbc.M110.147017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99(2):155–66. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6(5):409–31. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- Shinkai A, Patel PH, Loeb LA. The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J Biol Chem. 2001;276(22):18836–42. doi: 10.1074/jbc.M011472200. [DOI] [PubMed] [Google Scholar]
- Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17(5):2851–8. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10(8):2165–70. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitney KC, Budd ME, Campbell JL. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol. 2009;29(6):1432–41. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocki SA, Nonay RL, Reha-Krantz LJ. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3′--> 5′ exonuclease activities. J Mol Biol. 1995;254(1):15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Strerath M, Gloeckner C, Liu D, Schnur A, Marx A. Directed DNA polymerase evolution: effects of mutations in motif C on the mismatch-extension selectivity of thermus aquaticus DNA polymerase. Chembiochem. 2007;8(4):395–401. doi: 10.1002/cbic.200600337. [DOI] [PubMed] [Google Scholar]
- Subuddhi U, Hogg M, Reha-Krantz LJ. Use of 2-aminopurine fluorescence to study the role of the beta hairpin in the proofreading pathway catalyzed by the phage T4 and RB69 DNA polymerases. Biochemistry. 2008;47(23):6130–7. doi: 10.1021/bi800211f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Baskin D, Hood L, Loeb LA. Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc Natl Acad Sci U S A. 1996;93(18):9670–5. doi: 10.1073/pnas.93.18.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Yoshida S, Adman ET, Blank A, Loeb LA. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J Biol Chem. 2000;275(42):32728–35. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16(9):979–86. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166(5):693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- Thommes P, Hubscher U. Eukaryotic DNA replication. Enzymes and proteins acting at the fork. Eur J Biochem. 1990;194(3):699–712. doi: 10.1111/j.1432-1033.1990.tb19460.x. [DOI] [PubMed] [Google Scholar]
- Thompson EH, Bailey MF, van der Schans EJ, Joyce CM, Millar DP. Determinants of DNA mismatch recognition within the polymerase domain of the Klenow fragment. Biochemistry. 2002;41(3):713–22. doi: 10.1021/bi0114271. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007;6(7):900–13. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Trzemecka A, Plochocka D, Bebenek A. Different behaviors in vivo of mutations in the beta hairpin loop of the DNA polymerases of the closely related phages T4 and RB69. J Mol Biol. 2009;389(5):797–807. doi: 10.1016/j.jmb.2009.04.055. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45(32):9675–87. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura A, Hidaka Y, Hirabayashi T, Hirabayashi M, Yagi T. DNA polymerase delta is required for early mammalian embryogenesis. PLoS One. 2009;4(1):e4184. doi: 10.1371/journal.pone.0004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8(4):461–9. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87(1):65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 2010;6(11):e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J Biol Chem. 2006;281(7):4486–94. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, Ladiges WC, Preston BD, Loeb LA. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27(21):7669–82. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89(7):1087–99. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- Wang M, Xia S, Blaha G, Steitz TA, Konigsberg WH, Wang J. Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2011a;50(4):581–90. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Q, Chen H, Li X, Mai W, Chen K, Zhang S, Lee EY, Lee MY, Zhou Y. P50, the small subunit of DNA polymerase delta, is required for mediation of the interaction of polymerase delta subassemblies with PCNA. PLoS One. 2011b;6(11):e27092. doi: 10.1371/journal.pone.0027092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SH, Sobol RW, Beard WA, Horton JK, Prasad R, Vande Berg BJ. DNA polymerase beta and mammalian base excision repair. Cold Spring Harb Symp Quant Biol. 2000;65:143–55. doi: 10.1101/sqb.2000.65.143. [DOI] [PubMed] [Google Scholar]
- Wright GE, Hubscher U, Khan NN, Focher F, Verri A. Inhibitor analysis of calf thymus DNA polymerases alpha, delta and epsilon. FEBS Lett. 1994;341(1):128–30. doi: 10.1016/0014-5793(94)80254-8. [DOI] [PubMed] [Google Scholar]
- Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002a;41(32):10256–61. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- Yang G, Franklin M, Li J, Lin TC, Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002b;41(8):2526–34. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- Yang G, Wang J, Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44(9):3338–46. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- Zahn KE, Wallace SS, Doublie S. DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions. Curr Opin Struct Biol. 2011;21(3):358–69. doi: 10.1016/j.sbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of dna polymerase delta. J Biol Chem. 2007;282(21):15330–40. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3(1):23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Pedersen LC, Kunkel TA. Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity. Nucleic Acids Res. 2008;36(12):3892–904. doi: 10.1093/nar/gkn312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Chen H, Li X, Wang Y, Chen K, Zhang S, Meng X, Lee EY, Lee MY. Production of recombinant human DNA polymerase delta in a Bombyx mori bioreactor. PLoS One. 2012a;6(7):e22224. doi: 10.1371/journal.pone.0022224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Meng X, Zhang S, Lee EY, Lee MY. Characterization of human DNA polymerase delta and its subassemblies reconstituted by expression in the multibac system. PLoS One. 2012b;7(6):e39156. doi: 10.1371/journal.pone.0039156. [DOI] [PMC free article] [PubMed] [Google Scholar]