Abstract
Motivation: To create, verify and maintain pathway models, curators must discover and assess knowledge distributed over the vast body of biological literature. Methods supporting these tasks must understand both the pathway model representations and the natural language in the literature. These methods should identify and order documents by relevance to any given pathway reaction. No existing system has addressed all aspects of this challenge.
Method: We present novel methods for associating pathway model reactions with relevant publications. Our approach extracts the reactions directly from the models and then turns them into queries for three text mining-based MEDLINE literature search systems. These queries are executed, and the resulting documents are combined and ranked according to their relevance to the reactions of interest. We manually annotate document-reaction pairs with the relevance of the document to the reaction and use this annotation to study several ranking methods, using various heuristic and machine-learning approaches.
Results: Our evaluation shows that the annotated document-reaction pairs can be used to create a rule-based document ranking system, and that machine learning can be used to rank documents by their relevance to pathway reactions. We find that a Support Vector Machine-based system outperforms several baselines and matches the performance of the rule-based system. The success of the query extraction and ranking methods are used to update our existing pathway search system, PathText.
Availability: An online demonstration of PathText 2 and the annotated corpus are available for research purposes at http://www.nactem.ac.uk/pathtext2/.
Contact: makoto.miwa@manchester.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The scale and speed with which biological literature is published introduces multiple challenges for the creation, verification, maintenance and further development of formal, comprehensive and up-to-date models of the physical entities and reactions involved in biological systems (Ananiadou et al., 2006; Kitano, 2002). Efforts to understand a complex biological system in detail need to incorporate knowledge that may be distributed over a large number of scientific publications from among the tens of millions available today. To reduce the demands of discovering and integrating this knowledge, several text mining systems have been proposed (Park et al., 2001; Rajagopalan and Agarwal, 2005; Rzhetsky et al., 2004; Yao et al., 2004; Yuryev et al., 2006; Zhang et al., 2009), which address many of the aspects of pathway curation. However, despite the obvious potential that automatic analysis of the literature holds for assisting large-scale integration of biological knowledge, pathway curation efforts remain largely manual (Herrgård et al., 2008; Swainston et al., 2011; Thiele and Palsson, 2010), carried out with limited or no support from advanced text mining methods. One way to try to remedy this situation is through the use of semantic search systems that make use of these methods. However, there are several technical challenges to overcome, such as interfacing with such systems and integrating information from various systems in a coherent way, as well as the fact that few semantic search systems directly involve either the representations or the semantics (physical entity and reaction definitions) used in pathway model curation efforts.
To address these issues and support pathway curation, we have developed PathText 2, an integrated search system designed to link biological pathways with supporting knowledge in the vast body of literature. Given a pathway model and a reaction, the system is able to find documents that are relevant to the given reaction from MEDLINE. The literature search in PathText 2 is implemented by translating each reaction into a set of queries that are then executed using several semantic search systems. The results of the queries are then combined, ranked and presented to the user in a unified user interface. With these pathway-specific functions, i.e. the reaction-based retrieval of documents and ranking of unified documents by their relevance to pathway reactions, PathText 2 aims to reduce the number of documents curators need to focus on, thus increasing the productivity of pathway curation. The documents retrieved by PathText 2 can help curators to maintain the textual evidence for reactions and to extend pathways by finding related reactions from the documents. PathText 2 is designed to read formal pathway models represented in the Systems Biology Markup Language (SBML) (Hucka et al., 2003) with CellDesigner (Funahashi et al., 2003) extensions. SBML is a major standard format for pathway representation and interchange, and its use assures compatibility with a large ecosystem of existing pathway curation tools and resources. CellDesigner extensions are essential for determining the correct participants in the reaction in our system; SBML itself does not define a fixed set of semantic types for its primitives but instead defines an extension mechanism (e.g. Courtot et al., 2011), allowing such types to be defined (Le Novre et al., 2005).
PathText 2 is a comprehensive enhancement of a previously released system, PathText (Kemper et al., 2010). PathText 2 extends the core functionality of PathText by combining the multiple results received for a single document from the various semantic search systems into a single result, introducing a new document ranking heuristic based on the relevance of the document to the given reaction, offering a new API allowing other systems to easily interface with the system and supporting a novel interface for human users to access the system. Furthermore, we have updated the individual semantic search systems that are queried by PathText 2 and updated the generation of queries applicable to these systems. This is done by implementing a query generation and expansion system based on reaction-event mapping (Ohta et al., 2011). As we are trying to support improved automatic association between pathway models and documents, we have removed previous PathText functionality for the manual annotation and association of documents with pathways. The improved query generation and ranking of the results should make this functionality obsolete, thus reducing the burden on curators.
We evaluate PathText 2 in detail on two corpora annotated by domain experts, both containing judgments on the relevance of various documents to specific pathway model reactions. The evaluation shows that use of PathText 2 substantially improves on PubMed search for discovering relevant documents, that the annotations can support the development of heuristics for document ranking and that the task of determining document-reaction relevance is feasible using machine learning methods.
2 PATHTEXT 2 ARCHITECTURE
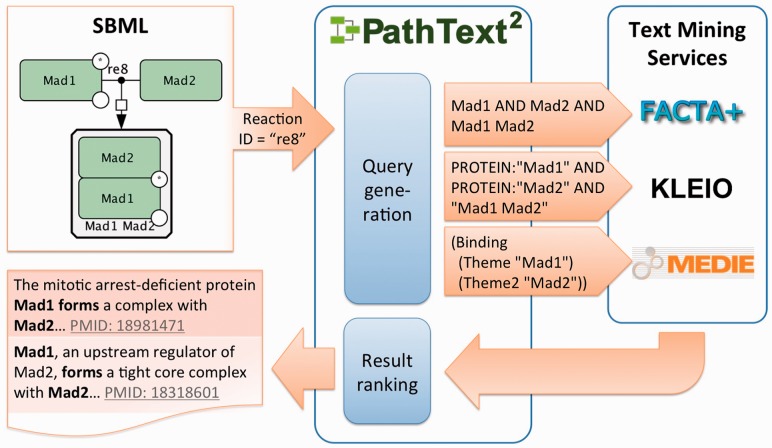
The overall architecture of PathText 2 is illustrated in Figure 1. PathText 2 currently supports SBML, which is the primary format used for major pathway repositories such as BioModels (Novere et al., 2006) and has been characterized as the most successful standard model exchange format for encoding pathway models (Li et al., 2010). PathText 2 specifies reaction semantics using the CellDesigner (Funahashi et al., 2003) types in SBML. CellDesigner is a popular tool for pathway model curation, and the SBML models in major resources such as PANTHER DB (Mi et al., 2007) contain CellDesigner types. We discuss generalization from SBML/CellDesigner to other representations in Section 5.
Fig. 1.
Illustration of PathText 2 architecture
PathText 2 interprets the SMBL models and uses the information to interact with various semantic search systems (see Section 2.1). To interact with these systems, PathText 2 contains query generation modules, which translate a given pathway reaction from the model into a set of queries formatted for use with each of the search systems (Section 2.2). In response to these queries, each system returns a set of documents; these result sets are merged (so that each document only occurs once), ranked and returned back to the user. The user interface is covered in Section 2.3.
2.1 Text mining-based semantic search systems
PathText 2 integrates three state-of-the-art text mining-based semantic search systems: FACTA+ (Tsuruoka et al., 2011), KLEIO (Nobata et al., 2008) and MEDIE (Miyao et al., 2006). Each of these systems is presented briefly later in the text; we refer readers to the publications introducing the systems for detailed descriptions.
FACTA+, an extension of the FACTA system, provides real-time search of direct and indirect associations between biological concepts in MEDLINE abstracts as well as abstracts discussing these concepts. FACTA+ indexes concepts from the abstracts including genes, proteins, diseases, symptoms, drugs, enzymes and simple chemical compounds, identified using biological databases and thesauri such as UniProt, BioThesaurus, Unified Medical Language System (UMLS), KEGG and DrugBank. The user interacts with the system by issuing queries in the form of a word, a concept identifier or any Boolean combination of words and/or identifiers. The web-based user interface shows the results grouped by concepts and ranked according to a choice of co-occurrence statistics (frequency, pointwise mutual information or symmetric conditional probability). FACTA+ additionally includes an indirect search function, which can find an association between a query and a target concept via another concept even if the query and target never co-occur in any publication.
KLEIO is a semantic search system for MEDLINE. It incorporates methods for acronym recognition and disambiguation (Okazaki and Ananiadou, 2006; Okazaki et al., 2010), term normalization (Tsuruoka et al., 2007), gene/protein name recognition (Okanohara et al., 2006) and species recognition and gene/protein species disambiguation (Wang et al., 2010) to improve and expand standard literature querying with semantic categories and faceted search. This allows the user to limit the results by specifying semantic categories to which the query words belong (e.g. ‘PROTEIN:hedgehog’ would find occurrences of the hedgehog protein, as opposed to the animal of the same name). The retrieved MEDLINE abstracts are annotated with biological concepts including genes, proteins, metabolites, bacteria, organs, symptoms, diseases and species. The recognized proteins and genes are additionally augmented by automatically disambiguated species information.
MEDIE is a search engine that can be used to query MEDLINE abstracts based on named entity recognition and normalized syntactic structures produced by deep parsing. The indexing component is supported by the automatic entity taggers NEMine (Sasaki et al., 2008) and NERsuite (http://nersuite.nlplab.org) and the deep parser Enju (Miyao and Tsujii, 2008), which abstracts over syntactic variability by analysing text in terms of predicate-argument structures. MEDIE can search for documents on the basis of subject-verb-object (SVO) triples expressing associations between entities. SVO searches are expressed in terms of three fields: the subject, verb and object. Any of these fields can be left empty, in which case the search engine will ignore that field, thus allowing for queries such as ‘what causes cancer’, by leaving the subject blank, setting the verb to ‘cause’ and the object to ‘cancer’. Analyses from the event extraction system EventMine (Miwa et al., 2012) were also recently incorporated into the MEDIE index. This system produces events, which are typed n-ary associations that involve an event trigger (usually a verb signifying an action, such as ‘induce’) and a set of participants identified as playing specific roles in the event (e.g. Theme, Cause). Event participants may be either named entities, such as proteins and genes, or other events (Ananiadou et al., 2010). Figure 2 provides an illustration of an event structure. To allow for the querying of arbitrary event structures, a new event-based search interface was added to MEDIE, alongside the existing SVO-based interface. This interface also supports queries in which only a portion of an event structure has been specified.
Fig. 2.
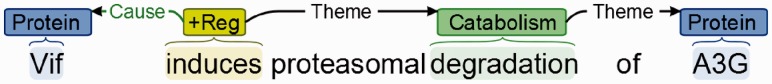
Illustration of event representation
In addition to a web-based user interface, each of the systems described additionally exposes a web service interface that can be used to access the systems programmatically. These web services are used by PathText 2 when executing queries. The use of these web service calls makes it relatively easy to extend the system to use other semantic search systems as they become available. The only requirement would be to write a module to interface between PathText 2 and the new system.
2.2 Query generation
For each semantic search system, PathText 2 provides a query generation module that understands the capabilities of the system and implements a mapping between the semantics of the source pathway model and those applied in the semantic search system. To realize these modules, a set of query generation rules was constructed based on the reaction-event mapping identified in our previous study (Ohta et al., 2011). Table 1 illustrates some of these mappings. As an example, consider that pathway representations in models typically explicitly identify all reaction participants, such as the gene and mRNA entities in Transcription reactions. However, not all such elements need to be stated in text, where expressions such as ‘transcription of p53’ can (implicitly) identify both DNA and mRNA entities. The text-oriented event representation thus only requires a single Theme entity for transcription events. Mapping Transcription reaction reactants to the Theme participants of events permits comparable interpretation.
Table 1.
Top-level correspondences for reaction-event mapping
| Pathway reaction | Event representation | ||
|---|---|---|---|
| Type | Participant | Type | Arguments |
| Truncation | Reactant, Product | Catabolism | Theme:Reactant |
| Transcription | Reactant, Product | Transcription, Gene_expression | Theme:Reactant |
| Translation | Reactant, Product | Translation, Gene_expression | Theme:Reactant |
| Heterodimer association | Reactant:Biomolecule, Product:Complex | Binding | Theme:Reactant |
| Dissociation | Reactant:Complex, Product:Biomolecule | Dissociation | — |
| Transport | Reactant:Biomolecule, from/to:Compartment | Localization | Theme:Reactant, atLoc/toLoc:from/to |
| Degradation/Truncation | Reactant:Biomolecule | Catabolism | Theme:Reactant |
The queries are generated from a given reaction by applying a series of rules; these use information such as the reaction type, reactants, products, modification type and modifiers, as well as the states of the reactants and products. The full set of detailed query generation rules and query specifications are provided in the Supplementary Information.
PathText 2 includes the following modules:
Query generation for FACTA+:
Depending on the reaction type, the FACTA+ query terms are made up of a conjunction of a subset of the reactants, products and modifiers of the reaction. The query may not contain all of these items, as, depending on the type of reaction being described, the text may not be expected to contain all of the details of the reaction. For example, CellDesigner degradation reactions have products of the type degraded, which is not expected to appear in text. When a reaction does not match any of the known types, the FACTA + query consists of the conjunction of all of the reactants and products of the reaction.
Query generation for KLEIO:
As with FACTA+, the KLEIO query terms are made up of a conjunction of a subset of the reactants, products and modifiers for a reaction depending on the reaction type, but with the addition of KLEIO’s semantic types (PROTEIN, DISEASE, METABOLITE, etc.) to identify the correct semantic type for each query term. A mapping is provided between the species types present in the pathway models and these semantic types; for example, a pathway model species with name ‘p53’ and the SBML/CellDesigner type celldesigner:protein is mapped to produce the KLEIO query term ‘PROTEIN:p53’. Types for which no correspondence is defined in the KLEIO type system are mapped to basic keyword queries, similar to those generated for FACTA+. As with FACTA+, where the reaction type does not match the known type, the query will consist of the conjunction of all of the products and reactants of the reaction.
Query generation for MEDIE:
The system generates queries for both SVO and event-structure modules. SVO queries are generated from the mapping shown in Table 1 as follows: the Cause argument in the event representation, when present, is used as the subject, the most frequent verb for each reaction type as annotated in the GENIA corpus (Kim et al., 2008) is used as the verb and the Theme argument is used as the object. In addition, specific rules are applied to a few reaction types. For example, the most frequent verb of the event Localization is ‘localize’, which only takes a subject; therefore, the Theme argument is mapped to the subject, and atLoc and toLoc, specific to this reaction type, are added as keywords. MEDIE event-structure queries are generated directly following the event-reaction mapping provided in Table 1.
Query Expansion for MEDIE Event Query:
MEDIE event queries can optionally be expanded using two complementary approaches: entity expansion and event expansion. Entity expansion performs a rule-based semantic decomposition of the entities participating in the reaction, e.g. ‘Mink1/2’ will become ‘Mink1’ and ‘Mink2’. Event expansion introduces additional queries that are combinatorial variants of the unexpanded form, e.g. a query for a Binding event with n Theme arguments will be augmented with additional queries for Binding events with 1 to 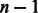 Theme arguments.
Theme arguments.
2.3 User interface and programmatic access
PathText 2 offers a web-based user interface and a web service API for programmatic access. Both offer the same functionality for searching the literature for a given reaction in a provided pathway model. They take as input an SMBL/CellDesigner model in XML format and a reaction ID that identifies the reaction of interest in the model and return a list of ranked references. Each returned reference consists of a PubMed ID, a snippet of text that provides evidence supporting the reaction and a confidence level with which the reference matches the query (Fig. 1). Additionally, two Boolean parameters control whether to perform entity expansion and/or event expansion (Section 2.2). A screenshot of the PathText 2 web interface is shown in Figure 3.
Fig. 3.
Screenshot of PathText 2 web interface
3 DOCUMENT-REACTION RELEVANCE RANKING
User feedback for the PathText system indicated that it would be useful to combine the results of the various semantic search systems, ensuring that no document appeared more than once. It was also requested that this set should be ranked according to the relevance of the document to the reaction. To address these desiderata, PathText 2 performs document combination and also includes various functions for document ranking. In this section, we present the details of the document-reaction relevance ranking.
We formulate the task as follows. Firstly, we assume that the ranking of documents by relevance to a given reaction is independent of other reactions in the pathway, and we consider a single reaction at a time. The ranking method is provided with a reaction, the set of queries generated for each semantic search system for the reaction (Section 2.2) and the response of the systems to these queries. The response to each query consists of an ordered set of documents, each of which may additionally be associated with system-specific information, such as the entities recognized in the document. The ranking method must produce a document list containing the union of document sets provided. The aim, then, is to order this list so that documents most relevant to the given reaction occur first.
To address this task, we manually annotated reaction-document pairs with the relevance of the documents to each reaction (Section 3.1). These annotations were then used to develop novel methods for combining the results of the semantic search systems and ranking documents (Section 3.2), which were further validated through an independent second round of annotation. The experimental setup is described in Section 3.3.
3.1 Corpus annotation
We initially created a corpus of pathway reaction-document pairs manually annotated using four levels of relevance: Not relevant, Partly relevant, Relevant and Highly relevant (for detailed definitions, see Supplementary Material). These data were used to train the document-reaction ranking in PathText 2 and for the primary evaluation.
To create the corpus, we initially selected from the PANTHER DB a set of prominent pathway models of interest that were familiar to our domain experts: the p38 MAPK, p53, p53 feedback-loops and Wnt signalling pathways. We then selected reactions that activated the specific reaction-query mappings of PathText 2 (Section 2.2). Results obtained from the semantic search systems were then combined and ranked using a simple Average hit ratio ranking heuristic (described in Section 3.2). To ensure that we had a sufficient number of candidate documents to enable the measurement of differences between ranking methods, we selected a set of reactions from each pathway for which at least 10 candidate documents were identified by PathText 2. We then took a random sample of these reactions for manual annotation. This corpus was used only to study the document ranking methods. We did not use this corpus to assess the recall, as PathText 2 itself was used to obtain the set of candidate documents and reactions.
Table 2 shows the annotation statistics for the 45 randomly selected reactions and the 450 evaluated documents (exactly 10 per reaction). Relatively few of the documents state the specified reaction explicitly; this is because in pathway models, combined biological phenomena (e.g. Phosphorylation and Activation) can be represented in a single reaction. Where a document was found to state only part of the reaction, it was judged as Partly relevant.
Table 2.
Summary of the annotated corpus statistics
| Pathway | p38 MAPK | p53 | p53 feedback | Wnt signalling | Total |
|---|---|---|---|---|---|
| Number of reactions | 16 | 12 | 6 | 11 | 45 |
| Number of documents | 160 | 120 | 60 | 110 | 450 |
| Highly relevant | 6 | 13 | 15 | 14 | 48 |
| Relevant | 0 | 17 | 16 | 0 | 33 |
| Partly relevant | 101 | 42 | 8 | 33 | 184 |
| Not relevant | 53 | 48 | 21 | 63 | 185 |
Not relevant is the most frequent single judgment over the four pathways in the corpus. One reason identified for these false positives was a failure in the underlying entity recognition common to each of the search systems; this can be attributed to ambiguous biomolecule names or incorrect synonym expansion. Another possible reason is a failure in the event extraction used to build the MEDIE event search system; this can be attributed to a mismatch between the event type and the reaction, or an incorrect assignment of participant roles (e.g. Theme and Cause might be swapped). In addition, our annotation guidelines are strict; for example, documents retrieved for a Heterodimer/Multimer association reaction were required to explicitly refer to binding involving all the reactants to be judged as Highly relevant. The detailed annotation guidelines are provided as Supplementary Material.
3.2 Ranking methods
We implemented and evaluated the following methods for ranking documents according to their relevance to a given pathway reaction. For all methods, the newer documents (according to the publication date) are ranked first in cases where the scores are equal.
Random:
This method simply takes the union of the documents returned by the semantic search systems and creates a randomly sorted list of the documents. This naive method establishes a lower bound on performance.
BM25:
This method ranks the documents using the Okapi BM25 (Robertson et al., 1999) ranking function, which is often used as a baseline for ranking in keyword-matching-based search systems. To calculate BM25 scores, bag-of-words queries (OR queries) are generated by decomposing FACTA + queries, document frequencies of the queries and the average document length are calculated by using the entire PubMed abstracts, and the parameters 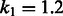 and
and 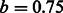 are used. This method is used as a baseline for the performance of document ranking.
are used. This method is used as a baseline for the performance of document ranking.
Average hit ratio:
For a given reaction and semantic search system, we define the hit ratio for each document as the fraction of queries generated by PathText 2 that retrieve the document. This score has the range [0,1] and so can be compared, regardless of the number of queries generated. This method orders documents by the average hit ratio across the semantic search systems for each document.
Priority ranking:
Scoring by average hit ratio implicitly assumes that the effectiveness of the various semantic search systems at recognizing relevant documents is equal, which may not hold in practice. We therefore define a heuristic that gives priority to systems whose evaluation on training data indicates to be more effective (Section 4). Specifically, the hit ratio for MEDIE event is considered first, followed by MEDIE SVO, then KLEIO and finally FACTA+. When choosing the order between any two documents, the hit ratios of the documents are compared in this order in turn; the document that comes first is the one with the highest value for the first ratio that is not equal. Thus, if a pair of documents has the same hit ratio for the MEDIE event system, MEDIE SVO scores will be used, unless these are equal, in which case KLEIO scores are used and so on.
Machine learning with annotated corpus:
Several machine-learning methods have been used in merging results from several search engines (Shokouhi and Si, 2011). This task has been treated as classification (Si and Callan, 2003), regression (He et al., 2011; Shokouhi and Zobel, 2009) and ranking (Joachims, 2002) problems. We also treated the task according to these three problem types and solved each problem individually, using the primary annotated corpus data (Section 3.1) for training. We implemented three machine learning-based ranking components using the following learning methods: support vector machines (SVM) (Vapnik, 1998), support vector regression (SVR) (Drucker et al., 1996) and ranking SVM (RankSVM) (Joachims, 2002). We used documents annotated with the relevance labels as input to the machine-learning methods, and we trained the SVM classifier to predict the relevance label for each document and used these predictions for ranking. The SVR method is similarly trained to predict the document relevance label, but it uses a regression model. For SVR training, we mapped the relevance labels to scores, from 0 (Not relevant) to 3 (Highly relevant). In contrast to these point-wise methods, the RankSVM method constructs a binary SVM classifier that predicts which of a given pair of inputs should be ranked higher. RankSVM was trained on document pairs, with the order of the pair derived from the relevance labels; those with the same relevance label are ignored.
All of the machine-learning methods were applied using the following set of features: (i) hit ratio of each system without query expansion; (ii) hit ratio of MEDIE event with query expansion; (iii) number of hits for each system without query expansion and (iv) number of hits for MEDIE event with query expansion. We chose these features, as they match the elements that make up the heuristic rules; the comparison between the heuristic rules and machine-learning methods can then show if machine-learning methods perform better with the same information.
Machine learning with curated pathways:
To explore the potential for previously released pathway annotations to support the ranking task, we extracted reaction-document pairs from PubMed IDs found in comments attached to pathway model reactions. In all, 3586 reaction-document pairs were annotated in five manually curated pathway models (Caron2010 mTOR SignalingNetwork, TLR ICSB, Kaizu2010 BuddingYeastCellCycle, Rb pathway3, EGFR signaling for RTKC) in the BioModels database. PathText 2 was used to retrieve up to 1000 documents for each reaction, giving 530 reaction-document pairs. We note that the relevance of these documents is based on the full texts, not abstracts, that this approach does not provide a way to distinguish between different levels of relevance and that the relevance is not determined according to our criteria. Nevertheless, these reaction-document pairs may provide useful supervision for ranking. We used two different approaches to make use of the pairs, using the same features mentioned above, focusing on the SVM-learning methods based on the results of preliminary evaluation. The first approach (Pathway) is to train an SVM classifier on the pairs with pseudo-negative pairs randomly selected from the pairs retrieved by PathText 2. The second approach (SVM + Pathway) is to train an SVM classifier on the annotated corpus with an additional feature, which denotes the prediction by the SVM classifier in the first approach (stacking).
3.3 Experimental setup
We divided the annotated corpus into training and test sets by placing a randomly selected 80% of the reactions into the training set and 20% into the test set. Ensuring that the reactions are kept separate eliminates the risk of over-fitting of the algorithms to the corpus. Table 3 presents some statistics regarding the split data. The test set was not used during the development of the methods.
Table 3.
Statistics of the train/test data split
| Category | Train | Test | All |
|---|---|---|---|
| Number of reactions | 36 | 9 | 45 |
| Number of documents | 360 | 90 | 450 |
| Highly relevant | 38 | 10 | 48 |
| Relevant | 26 | 7 | 33 |
| Partly relevant | 148 | 36 | 184 |
| Not relevant | 148 | 37 | 185 |
We evaluate ranking performance using the normalized discounted cumulative gain (nDCG) metric (Järvelin and Kekäläinen, 2002). This metric is commonly used for measuring performance on graded relevance judgments where the gold standard does not provide a strict total ordering of the data. It also has the benefit that it can compare the ranking performance between queries, owing to it producing a fixed range metric (with values between 0.0 and 1.0), with larger values indicating better performance.
Feature vectors were (L2-) normalized before applying the machine-learning methods. The regularization parameter (C) values of the machine-learning methods were left at their default value of 1.0.
4 RESULTS
We initially evaluated the individual contribution of each semantic search system on the training data by measuring the average hit ratio of each system separately (Table 4). This evaluates the ranking performance of each semantic search system on fixed document sets (i.e. system recall is not evaluated); documents that were not retrieved by a semantic search system were assigned the lowest ranking. The results show that the semantic search systems vary in their effectiveness, with MEDIE event best matching the gold standard. These results were used to define the ordering for the priority ranking heuristic (Section 3.2). We note that although some systems show similar overall ranking performance, detailed analysis indicates substantial differences in the document sets retrieved by the systems (see Supplementary Material).
Table 4.
Contribution of each semantic search system on training data in nDCG
| FACTA | KLEIO | MEDIE SVO | MEDIE EVENT |
|---|---|---|---|
| 0.829 | 0.847 | 0.850 | 0.859 |
Table 5 shows the nDCG score of each ranking system. For the machine-learning methods, we found that the SVM method achieved the best results, notably outperforming the average hit ratio heuristic and BM25. RankSVM slightly outperformed the average hit ratio method, but SVR did not. The results for SVR are to be expected, as we treated relevance judgments as linearly increasing absolute scores, which imposes considerable constraints on SVR. In contrast, SVM and RankSVM do not have these constraints. The priority ranking heuristic shows competitive performance on the test data, outperforming two of the machine-learning methods and showing a notable performance improvement compared with the average hit ratio heuristic. For machine-learning methods, using the pairs curated in pathway models, an SVM trained on the pairs (Pathway) performed as well as RankSVM, and the pairs slightly improved performance when used for creating an additional feature for training with the data specifically annotated for ranking (SVM + Pathway). These results demonstrate that existing pathway annotations can be beneficially combined with data annotated for the ranking task.
Table 5.
Evaluation of test data and rule-based scoring method on training data in nDCG
| Data | SVM + Pathway | Pathway | SVM | RankSVM | SVR | Priority ranking | Average hit ratio | BM25 | Random |
|---|---|---|---|---|---|---|---|---|---|
| Test | 0.788 | 0.719 | 0.777 | 0.719 | 0.672 | 0.775 | 0.696 | 0.747 | 0.542 |
| Train | – | – | – | – | – | 0.865 | 0.846 | 0.842 | 0.641 |
We note that the performance of the methods on the test data appears somewhat lower than the results obtained for the individual systems on the training data. However, comparison on training and test data results confirms that this is owing to differences between the two datasets; for example, the priority ranking heuristic achieves a higher level of performance (0.865 nDCG) than any single semantic search system on the training data (Table 4). The results confirm that it is effective to aggregate the various semantic search systems and indicate that the systems provide complementary information regarding document relevance.
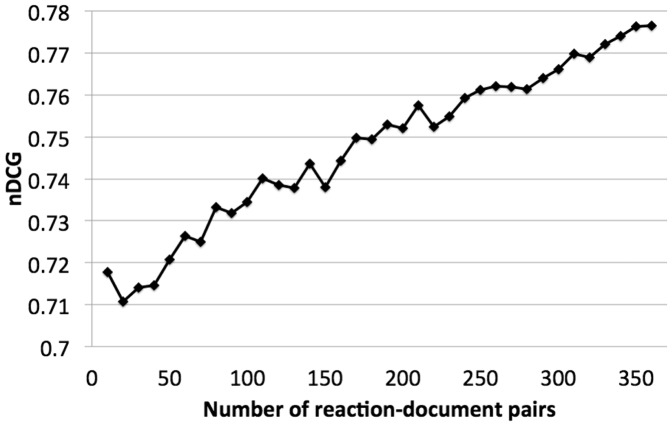
The priority ranking heuristic produced comparable results to the SVM-based ranking on the test dataset. This indicates that heuristic ranking informed by an analysis of the performance of the semantic search methods remains competitive with machine learning for the current training data size. To assess the degree to which this result depends on the specific size of the training dataset, we calculated a learning curve on the test dataset for the SVM method (Fig. 4). We find that the performance of the SVM increases with the size of the training data up to the point of using all of the available 360 training documents, and the curve does not appear to have flattened out up to this point. This suggests that if given more annotated data, the SVM has the potential to outperform the heuristics, which do not require training and therefore do not benefit from additional data. We note also that we used a somewhat limited set of features, and that there is potential for improved performance by incorporating more detailed query-result pair-specific features, such as detailed information on how a query matches, or the rarity of the query and matched reactions.
Fig. 4.
Learning curve on SVM-based ranking
Table 6 gives the results of manual evaluation of PathText 2 with three settings contrasted with PubMed search, showing the top 10 precision and nDCG of the manual evaluations on five reactions. Each reaction had three manual evaluations, and the score shows the means of scores calculated for the three evaluations. PubMed provides keyword-based search and returns documents in a reverse chronological ordering (Lu, 2011). We refer readers to the Supplementary Material for the details of the evaluation setting. These results indicate that (i) documents retrieved by PathText 2 are more relevant than those retrieved by querying PubMed; (ii) query expansion is effective in retrieving relevant documents and (iii) priority ranking outperforms the average hit ratio heuristic.
Table 6.
Manual evaluation of PathText 2 and PubMed in Top 10 precision and nDCG
| Evaluation Metric | Priority ranking + Query expansion | Average hit ratio + Query expansion | Priority ranking | PubMed |
|---|---|---|---|---|
| Top 10 precision | 0.493 | 0.347 | 0.393 | 0.280 |
| nDCG | 0.419 | 0.373 | 0.376 | 0.215 |
In summary, our results show that the annotated corpus can benefit both the development of ranking based on heuristic rules and the training of machine-learning methods. In addition, either of these approaches could result in a substantial improvement in the document ranking functionality in PathText 2, compared with the average hit ratio heuristic.
5 DISCUSSION AND FUTURE WORK
Our evaluation of the performance of the machine-learning system with respect to the training data size suggests that the machine-learning methods could benefit from additional data. One idea is to gain feedback from users on the perceived quality of the results while simultaneously increasing the amount of data available for training the ranking component, as for our SVM + Pathway model. This feedback might be gained by allowing users to annotate documents with relevance annotations. Additionally, there remain many opportunities for further improvements to the feature representation of the SVM method, which has the potential to allow for substantial increases in ranking performance.
PathText 2 relies on pathways being specified in SBML with CellDesigner semantics. Although this representation is commonly used, it is not the only possible representation. One possibility here is to generalize the support to include SBML with alternative physical entity and reaction semantics, such as Systems Biology Ontology (SBO) (Courtot et al., 2011), as well as alternative pathway representations such as BioPAX (Demir et al., 2010). This could be done using established mappings between different pathway model semantics (Strömbäck and Lambrix, 2005) as well as automatic conversions such as that between SBML and BioPAX (Mi et al., 2011). Such support could further allow integration of PathText 2 with numerous domain tools such as Cytoscape (Smoot et al., 2011).
We finally note that there remain opportunities for improvement of the semantic search systems themselves, in particular in the text-mining algorithms to recognize entities and events.
6 CONCLUSION
We have presented PathText 2, an integrated search system designed to link biological pathways with supporting knowledge in the vast body of literature. This system allows direct access to the most relevant documents from the literature, thus supporting various tasks in the creation, verification, maintenance and extension of pathway models. The system implements SBML parsing, the conversion of reactions into system-specific queries, query result combination and ranking by relevance to the given pathway reaction using heuristic or machine learning-based methods and an API supporting programmatic access to the search functionality. A major focus of our efforts has been the development of ranking functionality that combines the candidate document information retrieved by the various text mining-based search systems into a list ordered in a way where documents most likely to be relevant to the query reaction are presented first. We created a corpus of 450 judgments that identify on a four-point scale the relevance of documents to reactions randomly selected from a set of four PANTHER DB pathways and used it to evaluate simple ranking heuristics, advanced heuristics informed by evaluation of the training set and three machine learning-based ranking methods. Our results show that an SVM-based ranking with annotations in pathway models can notably outperform the simple ranking heuristics, achieving a 0.788 nDCG score. An online demonstration of the PathText 2 system is accessible, and the annotated corpus is available for research purposes from the project homepage (http://www.nactem.ac.uk/pathtext2/). Access to the PathText 2 API is available on request.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Yonghwa Jo and Hyeyeon Choi for their contributions to the development of the relevance judgment annotation criteria. This work is a part of joint research of KISTI and NaCTeM. They also thank Georgios V. Gkoutos (Department of Computer Science, University of Aberystwyth), Paul N. Schofield (Department of Physiology, Development and Neuroscience, University of Cambridge), Loukia Tsaprouni (Wellcome Trust, Sanger Institute), Yukiko Matsuoka (The Systems Biology Institute and JST ERATO Kawaoka infection-induced host response network project) and Manami Katoh (JST ERATO Kawaoka infection-induced host response network project) for the evaluation and Paul Thompson for his helpful comments on the manuscript.
Funding: Part of this work is supported by the Biotechnology and Biological Sciences Research Council (BBSRC) [BB/G013160/1].
Conflict of Interest: none declared.
REFERENCES
- Ananiadou S, et al. Text mining and its potential applications in systems biology. Trends Biotechnol. 2006;24:571–579. doi: 10.1016/j.tibtech.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Ananiadou S, et al. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 2010;28:381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Courtot M, et al. Controlled vocabularies and semantics in systems biology. Mol. Syst. Biol. 2011;7:543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker H, et al. 1996. Support vector regression machines. In: NIPS’96. MIT Press, Cambridge, MA, USA, pp. 155–161. [Google Scholar]
- Funahashi A, et al. Celldesigner: a process diagram editor for gene-regulatory and biochemical networks. Biosilico. 2003;1:159–162. [Google Scholar]
- He C, et al. 2011. A weighted curve fitting method for result merging in federated search. In: Proceedings of SIGIR’11. ACM, New York, NY, USA, pp. 1177–1178. [Google Scholar]
- Herrgård MJ, et al. A consensus yeast metabolic network obtained from a community approach to systems biology. Nat. Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Järvelin K, Kekäläinen J. Cumulated gain-based evaluation of ir techniques. ACM Trans. Inf. Syst. 2002;20:422–446. [Google Scholar]
- Joachims T. 2002. Optimizing search engines using clickthrough data. In: Proceedings of the 8th ACM SIGKDD. ACM, New York, NY, USA, pp. 133–142. [Google Scholar]
- Kemper B, et al. PathText: a text mining integrator for biological pathway visualizations. Bioinformatics. 2010;26:i374–i381. doi: 10.1093/bioinformatics/btq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, et al. Corpus annotation for mining biomedical events from literature. BMC Bioinformatics. 2008;9:10. doi: 10.1186/1471-2105-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Le Novre N, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat. Biotechnol. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- Li C, et al. Biomodels database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst. Biol. 2010;4:92. doi: 10.1186/1752-0509-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. Pubmed and beyond: a survey of web tools for searching biomedical literature. Database. 2011;2011:baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, et al. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35(Suppl. 1):D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, et al. BioPAX support in CellDesigner. Bioinformatics. 2011;27:3437–3438. doi: 10.1093/bioinformatics/btr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M, et al. Boosting automatic event extraction from the literature using domain adaptation and coreference resolution. Bioinformatics. 2012;28:1759–1765. doi: 10.1093/bioinformatics/bts237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao Y, et al. 2006. Semantic retrieval for the accurate identification of relational concepts in massive textbases. In: Proceedings of ACL’06. ACL, Sydney, Australia, pp. 1017–1024. [Google Scholar]
- Miyao Y, Tsujii J. Feature forest models for probabilistic HPSG parsing. Comput. Linguist. 2008;34:35–80. [Google Scholar]
- Nobata C, et al. 2008. Kleio: a knowledge-enriched information retrieval system for biology. In: Proceedings of SIGIR’08. ACM, New York, NY, USA, pp. 787–788. [Google Scholar]
- Novere L, et al. Biomodels database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34(Suppl. 1):D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, et al. 2011. From pathways to biomolecular events: opportunities and challenges. In: Proceedings of BioNLP’11. ACL, Portland, OR, USA, pp. 105–113. [Google Scholar]
- Okanohara D, et al. 2006. Improving the scalability of semi-markov conditional random fields for named entity recognition. In: Proceedings of ACL’06. ACL, Sydney, Australia, pp. 465–472. [Google Scholar]
- Okazaki N, Ananiadou S. Building an abbreviation dictionary using a term recognition approach. Bioinformatics. 2006;22:3089–3095. doi: 10.1093/bioinformatics/btl534. [DOI] [PubMed] [Google Scholar]
- Okazaki N, et al. Building a high-quality sense inventory for improved abbreviation disambiguation. Bioinformatics. 2010;26:1246–1253. doi: 10.1093/bioinformatics/btq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, et al. Bidirectional incremental parsing for automatic pathway identification with combinatory categorial grammar. Pac. Symp. Biocomput. 2001;6:396–407. [PubMed] [Google Scholar]
- Rajagopalan D, Agarwal P. Inferring pathways from gene lists using a literature-derived network of biological relationships. Bioinformatics. 2005;21:788–793. doi: 10.1093/bioinformatics/bti069. [DOI] [PubMed] [Google Scholar]
- Robertson SE, et al. 1999. Okapi at trec-7: automatic ad hoc, filtering, vlc and interactive track. In: Proceedings of The Seventh Text REtrieval Conference (TREC-7). NIST Special Publication 500-242, Gaithersburg, MD, pp. 253–264. [Google Scholar]
- Rzhetsky A, et al. Geneways: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J. Biomed. Inform. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, et al. How to make the most of ne dictionaries in statistical NER. BMC Bioinformatics. 2008;9(Suppl. 11):S5. doi: 10.1186/1471-2105-9-S11-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokouhi M, Si L. Federated search. Found. Trends Inf. Retr. 2011;5:1–102. [Google Scholar]
- Shokouhi M, Zobel J. Robust result merging using sample-based score estimates. ACM Trans. Inf. Syst. 2009;27:14:1–14:29. [Google Scholar]
- Si L, Callan J. A semisupervised learning method to merge search engine results. ACM Trans. Inf. Syst. 2003;21:457–491. [Google Scholar]
- Smoot ME, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strömbäck L, Lambrix P. Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. Bioinformatics. 2005;21:4401–4407. doi: 10.1093/bioinformatics/bti718. [DOI] [PubMed] [Google Scholar]
- Swainston N, et al. The subliminal toolbox: automating steps in the reconstruction of metabolic networks. Integr. Bioinformatics. 2011;8:186. doi: 10.2390/biecoll-jib-2011-186. [DOI] [PubMed] [Google Scholar]
- Thiele I, Palsson BO. Reconstruction annotation jamborees: a community approach to systems biology. Mol. Syst. Biol. 2010;6:361. doi: 10.1038/msb.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka Y, et al. Learning string similarity measures for gene/protein name dictionary look-up using logistic regression. Bioinformatics. 2007;23:2768–2774. doi: 10.1093/bioinformatics/btm393. [DOI] [PubMed] [Google Scholar]
- Tsuruoka Y, et al. Discovering and visualizing indirect associations between biomedical concepts. Bioinformatics. 2011;27:i111–i119. doi: 10.1093/bioinformatics/btr214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN. Statistical Learning Theory. New York: Wiley-Interscience; 1998. [Google Scholar]
- Wang X, et al. Disambiguating the species of biomedical named entities using natural language parsers. Bioinformatics. 2010;26:661–667. doi: 10.1093/bioinformatics/btq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, et al. 2004. Pathwayfinder: paving the way towards automatic pathway extraction. In: Proceedings of APBC’04. Australian Computer Society, Inc., Darlinghurst, Australia, pp. 53–62. [Google Scholar]
- Yuryev A, et al. Automatic pathway building in biological association networks. BMC Bioinformatics. 2006;7:171. doi: 10.1186/1471-2105-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. Pathbinder–text empirics and automatic extraction of biomolecular interactions. BMC Bioinformatics. 2009;10(Suppl. 11):S18. doi: 10.1186/1471-2105-10-S11-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.