This work shows that CRK5, a plasma membrane–associated member of the Arabidopsis Ca2+/calmodulin-dependent kinase-related protein family, phosphorylates the hydrophilic loop of PIN2 and is required for proper polar localization of PIN2 in the transition zones of roots. Inactivation of CRK5 inhibits primary root elongation and delays gravitropic bending of roots and shoots.
Abstract
CRK5 is a member of the Arabidopsis thaliana Ca2+/calmodulin-dependent kinase-related kinase family. Here, we show that inactivation of CRK5 inhibits primary root elongation and delays gravitropic bending of shoots and roots. Reduced activity of the auxin-induced DR5–green fluorescent protein reporter suggests that auxin is depleted from crk5 root tips. However, no tip collapse is observed and the transcription of genes for auxin biosynthesis, AUXIN TRANSPORTER/AUXIN TRANSPORTER-LIKE PROTEIN (AUX/LAX) auxin influx, and PIN-FORMED (PIN) efflux carriers is unaffected by the crk5 mutation. Whereas AUX1, PIN1, PIN3, PIN4, and PIN7 display normal localization, PIN2 is depleted from apical membranes of epidermal cells and shows basal to apical relocalization in the cortex of the crk5 root transition zone. This, together with an increase in the number of crk5 lateral root primordia, suggests facilitated auxin efflux through the cortex toward the elongation zone. CRK5 is a plasma membrane–associated kinase that forms U-shaped patterns facing outer lateral walls of epidermis and cortex cells. Brefeldin inhibition of exocytosis stimulates CRK5 internalization into brefeldin bodies. CRK5 phosphorylates the hydrophilic loop of PIN2 in vitro, and PIN2 shows accelerated accumulation in brefeldin bodies in the crk5 mutant. Delayed gravitropic response of the crk5 mutant thus likely reflects defective phosphorylation of PIN2 and deceleration of its brefeldin-sensitive membrane recycling.
INTRODUCTION
By sensing the Earth’s gravity, plants adjust the growth of their shoots and roots with opposite polarity along the direction of gravity vector. Both positive and negative gravitropic responses, directing downward and upward bending of horizontally placed roots and shoots, respectively, are controlled by asymmetric distribution of the plant hormone auxin (Estelle, 1996). As hypothesized originally by Colodny and Went (Went, 1974), in response to altered gravity stimulus, auxin is transported from upper to lower sections of bending organs stimulating differential cell elongation responses. Cellular transport of auxin is controlled by the AUX/LAX influx and PIN-FORMED (PIN) efflux carriers, and the PGP/ABCB (for P-glycoprotein/ATP binding cassette protein subfamily B) transporters, several of which function in conjunction with PINs (reviewed in Kramer, 2004; Bandyopadhyay et al., 2007; Titapiwatanakun and Murphy, 2009). Whereas regulation of polar localization, activity, and stability of auxin carriers and transporters is being deciphered in detail (Friml, 2010; Ganguly et al., 2012), it is less clear how primary sensing of gravity is linked to specific switches in polar auxin transport.
Gravity is perceived by specific starch-containing statocyte cells in the root columella and stem endodermis (Morita, 2010). Mutations impairing starch biosynthesis, biogenesis, and sedimentation of starch-containing plastids (i.e., statoliths) and their interactions with actin filaments, endoplasmic reticulum, and plasma membrane highlight the importance of mechanosensitive ion channels and components of calcium/calmodulin and inositol-phosphate signaling pathways that connect gravisensing with the regulation of polar localization of PINs and PGPs (Baldwin et al., 2013; Blancaflor, 2013; Kurusu et al., 2013). Emerging data indicate that cortical actin accumulation regulates clathrin-dependent endocytosis (Lin et al., 2012; Nagawa et al., 2012), whereas enhanced inositol triphosphate and Ca2+ levels decelerate exocytosis of PIN1 and PIN2 similarly to mutations of inositol polyphosphate 1-phosphatase and phosphatidylinositol monophosphate 5-kinase genes (Zhang et al., 2011; Mei et al., 2012). Furthermore, signaling through the 3-phosphoinositide-dependent kinase1 and interactions with Ca2+ binding or calmodulin-like proteins appear to regulate the activity of AGC kinases that phosphorylate central hydrophilic loops of PINs, as well as ABCB/PGPs (Benjamins et al., 2003; Zegzouti et al., 2006; Henrichs et al., 2012; Rademacher and Offringa, 2012).
Cellular activities of ABCB/PGPs, PINs, and AUX1 determine the polarity and threshold of auxin transport. Thus, in combination with auxin-sensing fluorescent reporters, cellular localization of PINs provides correlative information on directional transport and distribution of auxin in different tissues and cell types (reviewed in Friml, 2010; Grunewald and Friml, 2010). In the roots, auxin moves through the stele reaching a maximum in the meristem and columella and then is transported upwards to the elongation zone through the epidermis and flows backward to the root tip in the cortex (Blilou et al., 2005). PIN1, 3, and 7 are localized toward the root tip in basal membranes of stele cells, whereas PIN4 shows basal localization in stem cells (reviewed in Kleine-Vehn and Friml, 2008). In the columella, apolar localization of PIN3 and 7 facilitate auxin flow toward the epidermis in synergism with AUX1, which is located in the basal membranes of epidermal cells and directs shootward auxin transport from the columella and lateral root cap (Swarup et al., 2001). PIN2 displays apical (shootward) and basal (rootward) localizations in epidermal and cortex cells, respectively, consistent with its key role in upward epidermal transport and cortex-mediated downward recycling of auxin (Kleine-Vehn et al., 2008a). In the upper section of horizontally placed gravistimulated roots, which show bending toward the gravity vector, epidermal PIN2 localization in the apical membrane is temporarily reduced due to enhanced PIN2 endocytosis and degradation. Along with a redirection of auxin transport toward the lower root section where elongation is inhibited, PIN2 levels are increased in the lower epidermis and cortex (Abas et al., 2006). At the same time, PIN3 is reported to show transient relocalization (i.e., transcytosis) to the basal membrane of columella cells facing the gravity vector (Kleine-Vehn et al., 2010). Although PIN3, together with overlapping functions of PIN4 and 7, is thought to play an important role in statocyte sedimentation-induced switch in the polarity of columella-derived auxin flow (Friml et al., 2002), the pin3 mutant shows only weak gravitropic defects compared with the pin2 and aux1 mutants (Müller et al., 1998; Swarup et al., 2005). This indicates that PIN2 and AUX1 play pivotal roles in the regulation of root gravitropic response, whereas in comparison, PIN3 is a key regulator of shoot gravitropic and phototropic responses in the stem endodermis (Ding et al., 2011; Rakusová et al., 2011).
PINs are probably secreted to membranes in an apolar fashion and then their basal localization is established through membrane-recycling controlled by the Rab5-GTPase ARA7, VPS9A (vacuolar protein sorting), and brefeldin A (BFA)–sensitive ADP-ribosylation-GDP/GTP-exchange factor GNOM proteins (Dhonukshe et al., 2008). Brefeldin inhibition of GNOM-dependent basal targeting stimulates PIN accumulation in so-called brefeldin compartments and leads ultimately to their relocalization to apical membranes (Kleine-Vehn et al., 2010). Without changing apical localization of PIN2 in the epidermis, low-level BFA treatment stimulates basal to apical PIN2 transcytosis in the cortex, which abolishes root gravitropic response, demonstrating the importance of cortical auxin recycling to the root tip (Rahman et al., 2010). Basal-to-apical switching of PINs is stimulated by the PINOID (PID), WAG1, and WAG2 AGC kinases, which phosphorylate TPRxS motifs in the hydrophilic loops of PINs (Michniewicz et al., 2007; Dhonukshe et al., 2010). Phosphorylated PINs are recycled from the apical membrane by clathrin-dependent endocytosis controlled by the ARA7 GTPase and brefeldin-insensitive GNOM-LIKE1 proteins and their regulatory partners (reviewed in Löfke et al., 2013). Remarkably, endocytosis of PIN2 from the apical membrane is inhibited by brefeldin in gln1 mutant roots, suggesting that the apical endocytosis pathway is either dependent on or coregulated with the BFA-sensitive GNOM-dependent basal recycling pathway (Teh and Moore, 2007). Recently, Zhang et al. (2010) identified two PID-independent phosphorylation sites in the hydrophilic loop of PIN1, which indicates that in addition to already known classes of AGCs (Ganguly et al., 2012), other protein kinases might also be involved in cell type–specific localization of some of the PINs.
Here, we show that CRK5, a member of the Arabidopsis thaliana Ca2+/calmodulin-dependent kinase-related (CRK) protein family (Harper et al., 2004), is required for proper polar localization of PIN2 in the transition zones of roots. Inactivation of CRK5 causes a root gravitropic defect and stimulates lateral root formation. PIN2 is depleted from apical membranes of epidermal cells and shows mixed apolar and apical localization in the cortex of root transient zone of crk5 mutant. CRK5 is localized to the plasma membrane (PM). BFA stimulates CRK5 internalization and accelerates the accumulation of PIN2 in brefeldin bodies in the crk5 mutant compared with the wild type. CRK5 undergoes autoactivation and phosphorylates the hydrophilic T-loop of PIN2 in vitro, suggesting that it plays a role in the regulation of PIN2 membrane recycling.
RESULTS
Inactivation of CRK5 Results in Delayed Gravitropic Responses
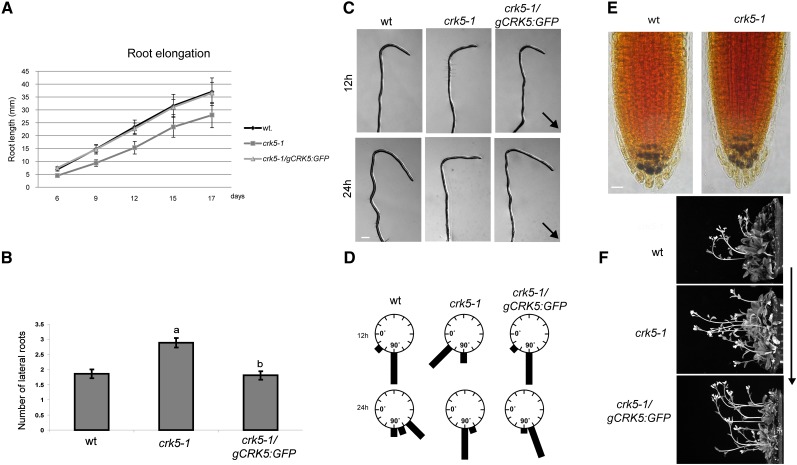
CRK5 (called alternatively PIP-D) was originally identified in yeast two-hybrid and in vitro protein binding assays as a potential interaction partner of the C-terminal WD40-repeat domain of nuclear PRL1 protein, a subunit of the spliceosome-activating complex (Németh et al., 1998; Koncz et al., 2012). Although subsequent studies did not confirm in vivo formation of a CRK5-PRL1 complex under normal growth conditions, we found that inactivation of the CRK5 gene by a T-DNA insertion in the MPIZ mutant line 38225 (crk5-1; Ríos et al., 2002; see Supplemental Figure 1A online) resulted in reduced primary root elongation (Figure 1A), analogously to the prl1 mutation (Németh et al., 1998). By contrast, a second T-DNA insertion in the CRK5 gene (Salk_003774, crk5-2) obtained from the SIGnAL collection (Alonso et al., 2003) failed to confer a root elongation detect. Characterization of the mutant alleles indicated that in crk5-1 a T-DNA insertion was located 54 bp downstream of the ATG codon in the first exon, whereas in crk5-2, a single T-DNA insert was identified 167 bp 5′-upstream of the ATG codon (see Supplemental Figure 1A online). Quantitative RT-PCR (qRT-PCR) analysis of homozygous T-DNA–tagged lines detected no transcript from the crk5-1 allele, indicating that it is a null mutation. On the other hand, insertion of the pROK2 T-DNA (Alonso et al., 2003) into the untranslated leader region fused the coding region of the CRK5 gene with an upstream T-DNA left border carrying a cauliflower mosaic virus 35S promoter. Consequently, the full-length CRK5 coding region was transcribed by the 35S promoter in the crk5-2 line, correlating with its wild-type phenotype (see Supplemental Figure 1B online). To verify that the root elongation defect, reducing the root length by ∼30% in the crk5-1 mutant compared with the wild type, was indeed caused by inactivation of CRK5, we constructed modified versions of CRK5 by replacing its stop codon with coding regions of green fluorescent protein (GFP) and β-glucuronidase (uidA/GUS) reporters (see Supplemental Figure 1C online). Introduction of the gCRK5:GFP construct into the crk5-1 mutant restored root elongation to the wild type, indicating genetic complementation of the mutation (Figure 1A).
Figure 1.
Root Developmental and Gravitropic Defects Caused by the crk5-1 Mutation.
(A) The crk5-1 mutant shows reduced elongation of primary roots. Root elongation rates of the wild type (wt), crk5-1 mutant, and complemented crk5-1/gCRK5:GFP seedlings were compared by measuring root lengths at different time points.
(B) Compared with the wild type, the crk5-1 mutant develops ∼30% more lateral roots. This defect is restored by the gCRK5:GFP construct conferring genetic complementation of the crk5-1 mutation. Bars in (A) and (B) indicate se of measurements performed with at least 50 plants in three biological replicates. Different letters in (B) indicate significant differences at P < 0.05.
(C) Comparison of root gravitropic responses of 7-d-old wild-type, crk5-1, and genetically complemented crk5-1/gCRK5:GFP seedlings. Root bending is shown at 12 and 24 h after rotation of vertically grown seedlings by 135°. Arrows indicate the direction of gravity. Bars = 500 μm.
(D) Quantitative representation of root gravitropic responses of wild-type, crk5-1, and crk5-1/gCRK5:GFP seedlings 7 and 24 h after reorientation by 135°. Length of black bars indicates the fractions of seedlings with various degrees of root bending at different time points. In each case, at least 70 seedlings were examined in three separate experiments.
(E) Comparison of starch accumulation in the columella tiers of wild-type and crk5-1 roots by Lugol staining (blue). Bar = 50 μm.
(F) Gravitropic bending of inflorescence stems of 4-week-old wild-type, crk5-1 mutant, and crk5-1/gCRK5:GFP plants 3 h after rotation by 90° in the dark. From each line at least 50 plants were tested in three separate experiments. The direction of gravity is indicated by an arrow.
[See online article for color version of this figure.]
To analyze tissue- and organ-specific expression of CRK5 during plant development, the gCRK5:GUS construct was transformed into wild-type and crk5-1 mutant plants. Whereas qRT-PCR comparison of relative transcript levels indicated that CRK5 was active in most plant organs, showing the highest transcript levels in flower buds and young flowers, characterization of gCRK5:GUS expression by histochemical GUS staining revealed that CRK5 was expressed in the root and shoot meristems; root pericycle; hypocotyl and leaf vasculature; stem vascular bundles; petals, sepals, anther filaments and pistils of flowers; and siliques (see Supplemental Figure 2 online).
Further analysis of altered developmental traits indicated that crk5-1 seedlings produced 25 to 30% more lateral roots compared with the wild type (Figure 1B). Enhancement of lateral root formation in crk5-1 suggested a possible alteration of auxin distribution leading to increased auxin transport toward the elongation zone (Benková et al., 2003). As this potential disturbance of normal auxin transport would be expected to affect the regulation of gravitropic root bending, we compared gravitropic responses of the wild type and crk5-1. Seedlings were subjected to 135° rotation after growing vertically for 1 week (Figure 1C). After 12 h of gravistimulation, 83% of the wild type but only 28% of crk5-1 roots showed a bending of 90°. After 24 h, 55% of wild-type roots were already curved by 135° but 82% of crk5-1 roots showed only 90° bending (Figure 1D). These data revealed that the crk5-1 mutation caused a marked delay in the root gravitropic response. Lugol staining of starch-containing columella cells of wild-type and crk5-1 mutant roots revealed an identical pattern, indicating that the crk5-1 mutation did not cause alteration in the accumulation of starch statoliths (Figure 1E). Surprisingly, we found that, following a 90° rotation to a horizontal position in the darkness, inflorescence stems of crk5-1 showed a maximum of 45° upward bending within 3 h, while wild-type stems were oriented vertically by bending 90° (Figure 1F). This indicated that the crk5-1 mutation also caused a delay in the shoot gravitropic response. As expected, the gCRK5:GFP complementing construct restored impaired root and shoot gravitropic responses of the crk5-1 mutant to the wild type.
CRK5 Is a PM-Associated CDPK-Related Protein Kinase
CRK5 is one of the eight members of the Arabidopsis CRK family, which share very similar structural and biochemical features with CRKs characterized in other plant species (Harper et al., 2004). N-terminal sequences of CRKs show high variability, except for two conserved domains corresponding to an MGxC myristoylation and palmitoylation motif and a downstream putative nuclear localization signal, which are also present in CRK5 (see Supplemental Figure 3 online). Ala replacements of Gly-2 and Cys-4 positions of the MGxC motive were demonstrated to abolish membrane association, leading to mixed cytoplasmic and nuclear localization of tomato (Solanum lycopersicum) CRK1 (Leclercq et al., 2005). Within their central Ser/Thr kinase domains, including the ATP binding, active-site, and activation T-loop sequences, CRKs show over 80% sequence identity (see Supplemental Figure 3 online). All CRKs undergo autoactivation by self-phosphorylation in vitro. Arabidopsis CRK3 and CRK6 autophosphorylate their T-loop Ser-311 and Ser-310 residues (Hegeman et al., 2006), which correspond to the Ser-315 position in CRK5 (see Supplemental Figure 3 online). All so far known CRKs, including CRK5, share a conserved C-terminal calmodulin (CaM) binding domain, which overlaps with the kinase autoinhibitory domain (Zhang et al., 2002). Selective CaM binding in the presence of Ca2+ simulates autophosphorylation but leads only to marginal increase of substrate phosphorylation by CRKs, which are equally active without CaM in the presence of either Ca2+ or the Ca2+ chelator EGTA (Lindzen and Choi, 1995; Furumoto et al., 1996; Lu et al., 1996; Wang et al., 2001; Zhang et al., 2002; Hua et al., 2003, 2004; Wang et al., 2004; Leclercq et al., 2005).
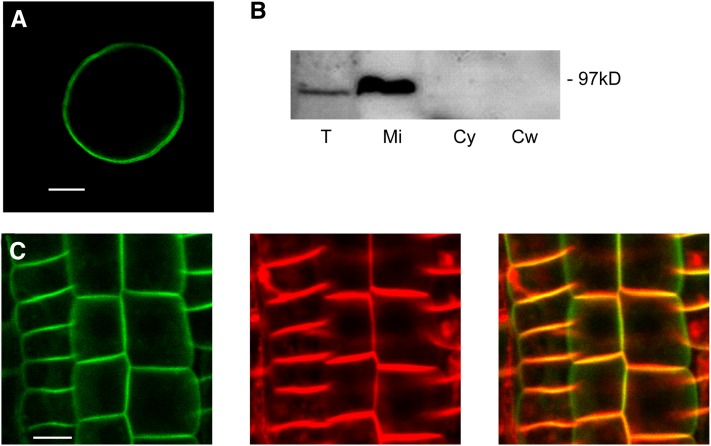
We examined several properties of CRK5, finding that they were highly similar to those of other biochemically characterized members of the CRK family. Recombinant CRK5 purified via an N-terminal His6-tag underwent autophosphorylation and phosphorylated the myelin basic protein substrate similarly in the presence of both Ca2+ and EGTA (see Supplemental Figure 4 online). Similarly to tomato CRK1, upon transient expression of the gCRK5:GFP construct in Arabidopsis protoplasts, CRK5-GFP was localized to the PM (Figure 2A). Following stable expression in an Arabidopsis root cell culture, CRK5-GFP was exclusively detected in the PM-containing purified microsomal fraction but not in the cytosolic and cell wall fractions by immunoblotting with anti-GFP antibody (Figure 2B). Furthermore, in root epidermal cells of stably transformed wild-type plants, CRK5-GFP showed colocalization with the PM-specific dye FM4-64, indicating that CRK5 is a PM-associated protein kinase (Figure 2C).
Figure 2.
PM localization of CRK5.
(A) CRK5-GFP shows association with the PM in protoplast transient expression assays. Bar = 10 µm.
(B) CRK5-GFP (97 kD) is detected in the microsomal membrane fraction. Total protein extract (T), PM-containing microsomal fraction (Mi), cytosolic fraction (Cy), and cell wall fraction (Cw) isolated from a CRK5-GFP–expressing Arabidopsis root cell culture were immunoblotted with anti-GFP antibody.
(C) In epidermal cells of wild-type plants transformed with the gCRK5:GFP construct, CRK5-GFP (left) and the membrane-specific dye FM4-64 (middle) are colocalized in PMs (merged images to the right). Bar = 25 µm.
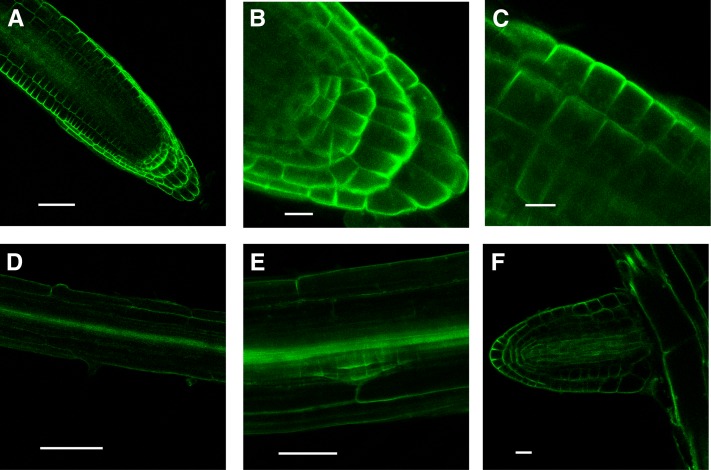
CRK5-GFP displayed particularly strong expression in the root tips. Confocal laser microscopy imaging highlighted polar localization of CRK5-GFP in U-shape patterns, which were oriented toward the lateral (surface facing) walls of epidermal, cortex, and lateral root cap cells and basal (root tip facing) membranes of columella tiers (Figures 3A to 3C). CRK5-GFP also decorated the pericycle in the lateral root initiation zone and showed epidermis-facing polar localization in dividing cells of early stage (i.e., stage I to II) root primordia. CRK5-GFP was also detectable in the lateral root vasculature and reproduced the same pattern in elongating root primordia as seen in different cell types of the main root apex (Figures 3D to 3F).
Figure 3.
Cellular Localization of CRK5-GFP in Different Cell Files of the Primary Root Apex and Lateral Root Primordia.
(A) and (B) CRK5-GFP highlights U-shape patterns, which are oriented toward outer lateral surfaces of epidermal and cortex cells, and basal (root tip–facing) walls of columella and root cap cells. Bars = 50 μm in (A) and 25 μm in (B).
(C) and (D) Details of CRK5-GFP patterns in focus of epidermis surface (C) and in the root hair differentiation zone (D). Bars = 25 μm in (C) and 100 μm in (D).
(E) CRK5-GFP expression in the stele and early stage root primordium, showing polar localization in the lateral outer membranes of dividing cells. Bar = 50 μm.
(F) CRK5-GFP is detectable in all cells of vasculature of elongating root primordia, as well as in epidermal, cortex, and root cap cells. Bar = 25 μm.
[See online article for color version of this figure.]
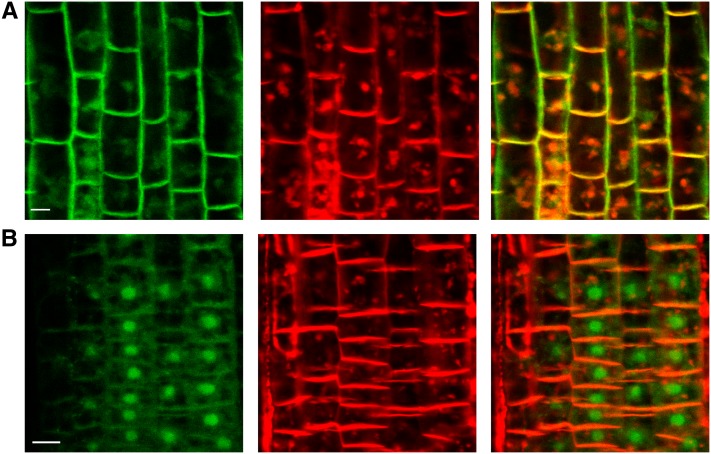
Membrane localization of CRK5-GFP confirmed by counter-staining with FM4-64 was gradually reduced in epidermal cells of roots during treatment with 50 µM BFA. Following transient accumulation of BFA bodies, CRK5-GFP showed differential localization compared with FM4-64 as the GFP signal became detectable also in the area of cell nuclei (Figure 4). Brefeldin-sensitive internalization of CRK5-GFP thus suggested that polar localization of CRK5 is likely mediated by the BFA-sensitive membrane recycling pathway.
Figure 4.
BFA Stimulates Internalization of CRK5-GFP.
Roots of wild-type CRK5-GFP–expressing plants were treated with 50 µm BFA for 40 min (A) and 90 min (B) and counterstained with 5 µm FM4-64. CRK5-GFP (left panels) and FM4-64 (middle panels) show colocalization (merged images in the right panels) in the PM and accumulating BFA bodies at 40 min (A), whereas upon 90 min of BFA treatment (B), CRK5-GFP shows differential accumulation in the area of cell nuclei. Bars = 25 µm.
The crk5-1 Mutation Delays Asymmetric Redistribution of Auxin in Gravistimulated Roots
To visualize redistribution of auxin during gravistimulation of roots (Ottenschläger et al., 2003), we introgressed the auxin-inducible DR5-GFP reporter into the crk5-1 mutant. In vertically grown roots, DR5-GFP expression levels were significantly lower in the columella tiers and root cap of the crk5-1 mutant compared with the wild type (Figure 5A), suggesting reduced auxin accumulation in the root apex. Control qRT-PCR measurements indicated that the crk5-1 mutation did not alter the transcription of a selected set of key genes acting in the regulation of auxin biosynthesis, including the Trp aminotransferase TAA1 (directing the biosynthesis of indole-3-acetic acid [IAA] precursor indole-3-pyruvate), TRP2, and TRP3 (encoding the β- and α-subunits of tryptophan synthase), YUCCA3 (involved in Trp-derived auxin biosynthesis), NIT3 (nitrilase, converting indole-3-acetonitrile to IAA), AMI1 (catalyzing the conversion of indole-3-acetamide to IAA), and CYP83B1/SUR2 (regulating the threshold of auxin glycosinolates; reviewed in Mano and Nemoto, 2012; see Supplemental Figure 5A online). Furthermore, the PIN1, PIN2, PIN3, PIN4, and PIN7 genes, which show auxin-induced transcription in the root (Vieten et al., 2005), as well as both AUX1 and LAX3 genes, displayed comparable transcription in the roots of the wild type, crk5-1 mutant, and complemented crk5-1 line carrying the gCRK5:GFP construct (see Supplemental Figure 5B online). These data indicated that the crk5-1 mutation did not result in global change of auxin-regulated gene expression, although it led to markedly reduced expression of auxin-induced DR5-GFP reporter in the root apex. However, in contrast with PID overexpression, which induces downregulation of DR5-GFP by facilitating strong shootward auxin efflux from the root apex (Friml et al., 2004), the crk5-1 mutation did not result in a collapse of root tips.
Figure 5.
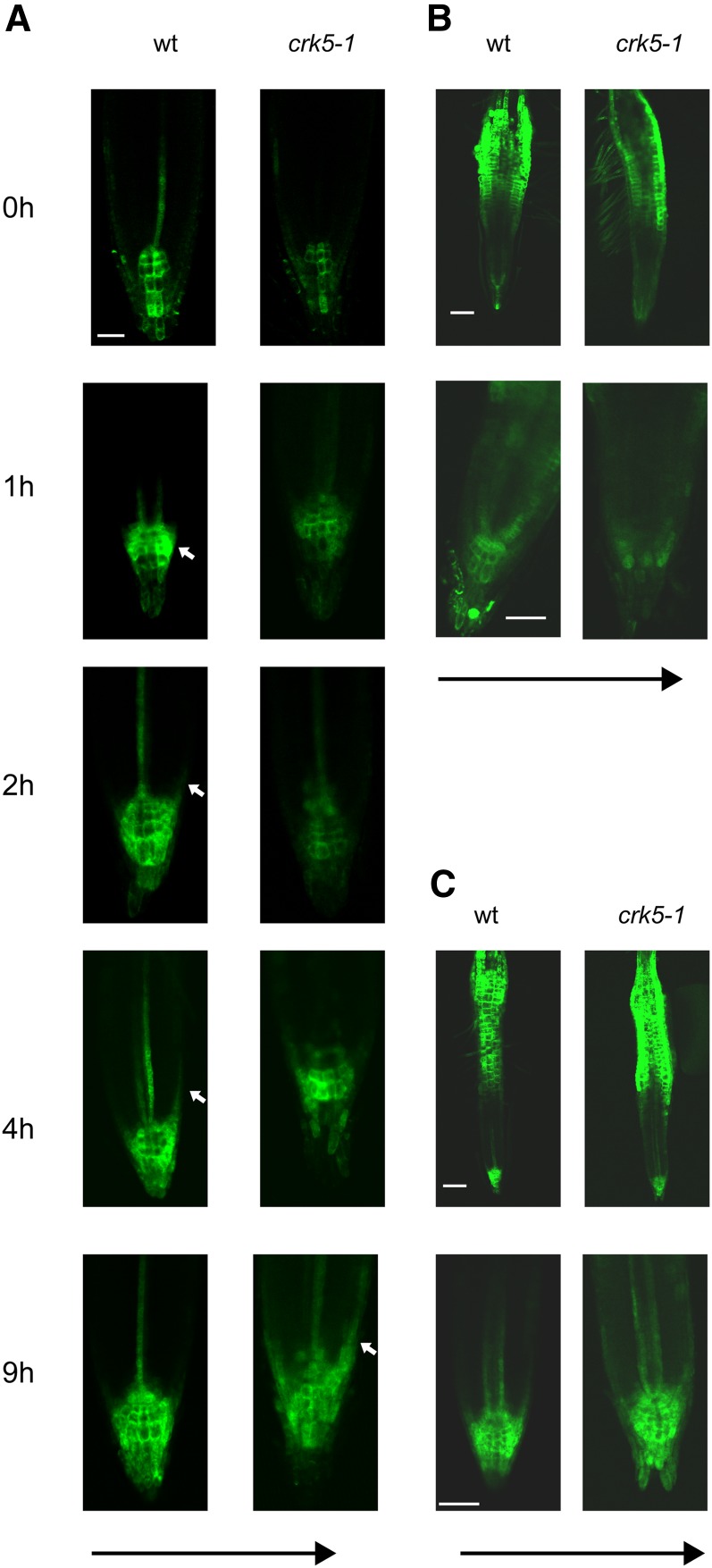
Activity of the Auxin-Induced DR5-GFP Reporter in Wild-Type and crk5-1 Roots during Gravistimulation in the Absence and Presence of NPA and NOA.
(A) Timing of asymmetric DR5-GFP activation in cells of the lower proximal lateral root cap and epidermis of wild-type (wt) and crk5-1 roots at different time points after gravistimulation. Black arrow indicates the direction of gravity, and white arrows mark the appearance of asymmetric DR5-GFP signals. Bar = 50 μm.
(B) and (C) Auxin induction of DR5-GFP is abolished by NPA (B) but enhanced by NOA (C) in the crk5-1 root apex compared with the wild type 7 h after gravistimulation.
[See online article for color version of this figure.]
Upon gravistimulation by 135° rotation, asymmetric activation of DR5-GFP in the lower proximal lateral root cap and epidermal cells of the wild type was observed within 1 to 2 h and reached a maximum in 4 h followed by a return to the symmetric vertical pattern by 9 h (Figure 5A). In the crk5-1 mutant, induction of auxin flow from the upper section of gravistimulated roots toward the root apex restored the wild-type level of DR5-GFP expression in the columella tiers 4 to 5 h after rotating the roots. A maximum of asymmetrically localized DR5-GFP expression in the lower lateral root cap and epidermis was observed only at 9 h after application of gravistimulus, indicating an ∼5-h delay in the root gravitropic response in crk5-1 (Figure 5A).
Pretreatment of wild-type roots with 10 μM 1-naphthylphtalamic acid (NPA), an inhibitor of auxin efflux, for 48 h before 6-h gravistimulation resulted in a typical upwards shift of DR5-GFP localization extending from the upper tiers of columella into the quiescent center, cortex, and endodermal files as described by Ottenschläger et al. (2003). NPA treatment further reduced weak expression of DR5-GFP in the root apex of crk5-1 mutant but did not prevent asymmetric activation of DR5-GFP in the lower epidermis of upper root hair zone (Figure 5B). By contrast, 48 h of application of auxin influx inhibitor 1-naphtoxyacetic acid (NOA) completely prevented gravitropic response and resulted in similarly high levels of DR5-GFP reporter expression in the columella, cap, vascular phloem cells, and hair zones of both wild-type and crk5-1 mutant roots (Figure 5C). Restoration of normal DR5-GFP expression in the crk5-1 root apex by NOA-mediated inhibition of AUX1 thus supported the notion that delayed gravitropic bending and enhanced differentiation of lateral roots in the crk5-1 mutant could result from enhanced shootward auxin flow from the root apex.
To determine how asymmetric redistribution of auxin during gravistimulation affects polar PM localization of CRK5, similar root gravitropism assays were performed with wild-type plants expressing the gCRK5:GFP construct. Polar localization patterns of CRK5-GFP remained unaltered, indicating that alterations in the auxin flow during gravistimulation of roots did not affect polar PM localization of CRK5 in the columella, root cap, epidermis, and cortex cells (see Supplemental Figure 6 online).
Localization of PINs and AUX1 in the crk5-1 Mutant
To compare polar cellular localization of AUX1 and PIN auxin influx and efflux carriers, respectively, in wild-type and crk5-1 roots, gene constructs for expression of AUX1-YFP (for yellow fluorescent protein; Swarup et al., 2001), PIN1-GFP (Benková et al., 2003), PIN2-GFP (Xu and Scheres, 2005), PIN3-GFP (Zádníková et al., 2010), PIN4-GFP, and PIN7-GFP (Blilou et al., 2005) were introgressed into the crk5-1 mutant. PIN1 showed similar basal membrane localization in cells of the vasculature in both vertically grown and gravistimulated wild-type and crk5-1 roots, respectively (see Supplemental Figure 7A online). PIN4-GFP also displayed identical localization in the first columella tiers, quiescent center, and upper adjacent layers of dividing cells of wild-type and crk5-1 roots independently of gravistimulation (see Supplemental Figure 7B online). Although the level of PIN7-GFP protein was somewhat lower compared with wild-type in crk5-1 roots, its basal localization in stele cells and apolar pattern in the columella did not differ in vertically grown and gravistimulated wild-type and crk5-1 roots (see Supplemental Figure 7C online). Using color-coded heat maps for PIN3-GFP localization in columella cells according to Kleine-Vehn et al. (2010), we found that PIN3-GFP showed apolar PM localization in 90% of vertically positioned roots of both wild-type and crk5-1 plants. After 30 min of gravistimulation by 90° reorientation, translocation of PIN3-GFP to basal PM was observed only in 10% of both wild-type and crk5-1 roots, in which PIN3-GFP showed previous partial polarization also in the absence of gravistimulus (see Supplemental Figure 8A online) as reported by Kleine-Vehn et al. (2010). A subsequent time course indicated that PIN3-GFP was nonpolarized and partially polarized in 90% versus 10% of vertically grown roots equally in both the wild type and crk5-1 mutant, and this ratio did not change during subsequent gravistimulation for 30, 60, or 120 min (see Supplemental Figure 8B online). Analogously, AUX1-YFP localization in the basal and inner lateral membranes of root cap and basal PM of epidermal cells proved to be identical in wild-type and crk5-1 roots (see Supplemental Figure 9 online).
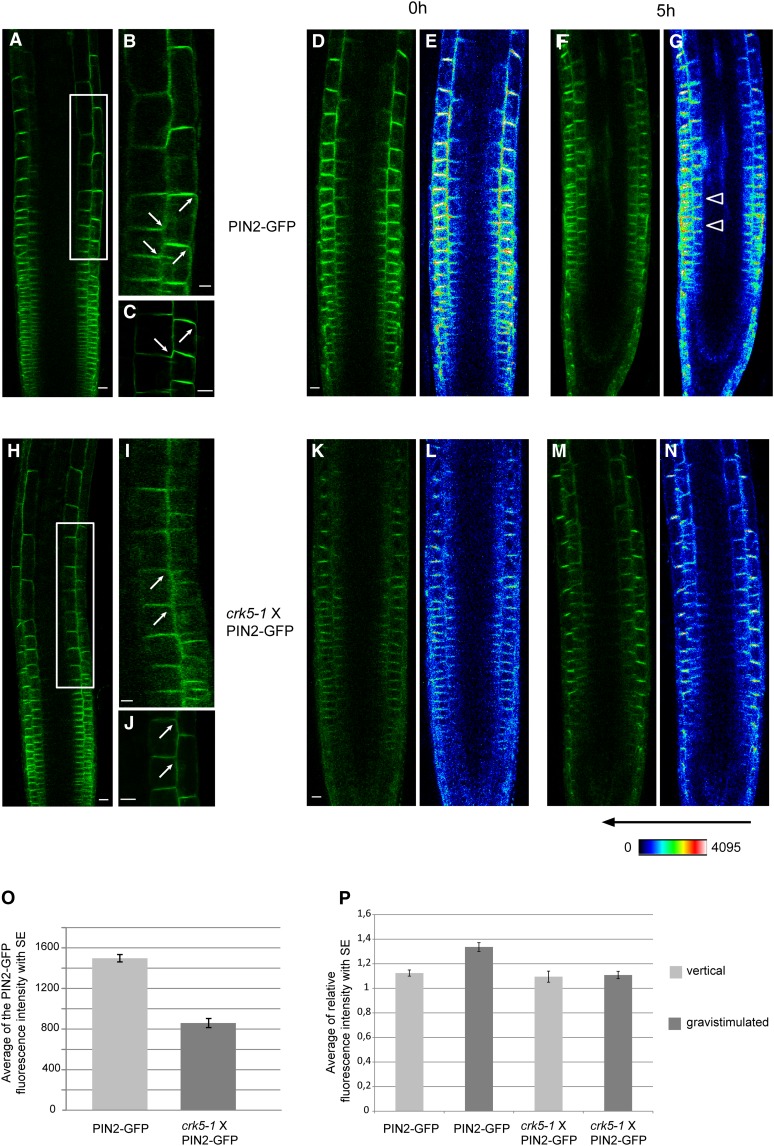
Whereas the crk5-1 mutation appeared to have no effect on polar localization of AUX1, PIN1, PIN3, PIN4, and PIN7, we found that the amount of PIN2-GFP reporter measured by heat maps was about twofold lower in the transition zone between the root cap and elongation zones (Verbelen et al., 2006) of vertically grown crk5-1 roots compared with the wild type (Figures 6A to 6E, 6H to 6L, and 6O). Moreover, in the transition zone, the amount of PIN2-GFP was markedly reduced in the upper PM of epidermal cells, whereas PIN2-GFP showed a basal-to-apical shift of PM localization in the adjacent cortex cells of crk5-1 compared with the wild type (Figures 6A to 6C and 6H to 6J). In contrast with previous reports (Kleine-Vehn et al., 2008b, Baster et al., 2013), however, we failed to detect enhanced vacuolar accumulation, suggesting facilitated endocytosis and degradation of PIN2-GFP in the light- or dark-grown crk5-1 mutant. Following 5 h of gravistimulation by 90° rotation, PIN2-GFP showed a typical asymmetric distribution in wild-type roots due to its reduction in the upper and accumulation in the lower section of epidermal and cortex cell files (Figures 6F, 6G, and 6P). At the same time, equal signal intensities were detected in the upper and lower sections of horizontally placed crk5-1 roots, indicating that symmetric distribution of PIN2-GFP remained unaltered upon 5 h of gravistimulation (Figures 6M, 6N, and 6P).
Figure 6.
Comparison of Polar PIN2-GFP Localization in the Transition Zones of Wild-Type and crk5-1 Roots before and after Gravistimulation.
(A) to (C) PIN2-GFP localization in cells of transient zone of wild type roots. Root section framed in (A) is enlarged in (B). Bars = 25 µm.
(D) to (G) Confocal GFP ([D] and [F]) and fluorescence intensity heat map ([E] and [G]) images of PIN2-GFP localization in wild-type roots grown vertically for 5 d (0 h) ([D] and [E]) and then subjected to gravistimulation for 5 h ([F] and [G]). Arrows in (G) indicate asymmetric distribution of PIN2-GFP signal accumulating at the lower section of gravistimulated wild-type root. Bar = 25 µm.
(H) to (J) PIN2-GFP localization in cells of transient zone of crk5-1 roots. Arrows indicate altered PIN2-GFP localization. PIN2-GFP is expressed at a lower level in crk5-1, it shows depletion from the apical membranes of epidermal cells, and a basal-to-apical shift of localization in the cortex cells of the transition zone of vertically grown roots. Root section framed in (H) is enlarged in (I). Bars = 25 µm.
(K) to (N) Confocal GFP ([K] and [M]) and fluorescence intensity heat map ([L] and [N]) images of PIN2-GFP localization in crk5-1 roots grown vertically for 5 d (0 h) ([K] and [L]) and then gravistimulated for 5 h ([M] and [N]).
(O) Comparison of average PIN2-GFP fluorescent intensities in vertically grown wild-type and crk5-1 roots calculated from quantitative data of a series of heat maps (n > 20 in two repeats).
(P) Average GFP intensities measured in the transient zones of lower and upper sections of vertical and gravistimulated wild-type and crk5-1 mutant roots using a series of heat map images from at least 20 plants in each of two biological repeats. Error bars in (O) and (P) indicate sd.
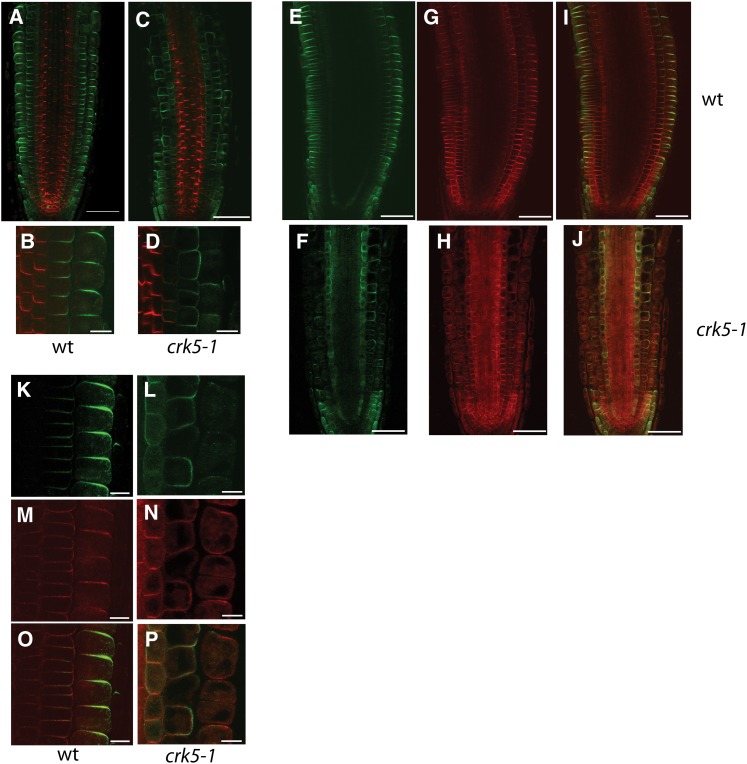
To confirm altered localization of PIN2 in the crk5-1 mutant by an independent means, we performed dual immunolocalization of PIN1 (Figures 7A to 7D, red) and PIN2 (Figures 7A to 7D, green). Whereas PIN1 similarly decorated the basal membranes of vascular cells in both wild-type and crk5-1 roots, immunolocalization detected similar changes in PIN2 localization as those seen with the PIN2-GFP reporter in crk5-1 roots (Figures 6A to 6C, 6H to 6J, and 7A to 7D). To exclude potential artifacts, we performed an alternative dual-labeling approach, in which PIN2 was colocalized with the PM H+-ATPase (Figures 7E to 7P). Both series of immunolocalization experiments demonstrated that in cells below the transition zone, PIN2 was properly localized in the apical and basal membranes of epidermal and cortex files, respectively, as expected. However, in epidermal cells of the crk5-1 root transition zone, the apical PIN2 membrane localization was markedly diminished along with a change in PIN2 localization to apolar or apical in the adjacent cortex cells and accompanied by appearance of some PIN2 signal also in membranes of endodermal cells (Figures 7C, 7D, 7F, and 7L). Upwards of the crk5-1 transition zone, PIN2 was detected, if at all, as weak internalized signal in the epidermis, and its localization was shifted to apical in the cortex. Since such apical localization of PIN2 in the cortex induced by low concentration BFA treatment was previously demonstrated to abolish root gravitropic responses (Rahman et al., 2010), these data suggested that aberrant polar localization of PIN2 in both epidermis and cortex was likely responsible for the delayed gravitropic response of crk5-1 mutant roots.
Figure 7.
Immunolocalization of PIN2 in Wild-Type and crk5-1 Roots.
(A) to (D) Dual immunolocalization of PIN1 (red) in stele and PIN2 (green) in epidermal and cortex cells of wild-type (wt; [A] and [B]) and crk5-1 roots ([C] and [D]). PIN1 shows identical patterns in basal membranes of root vascular cells of wild-type ([A] and [B]) and crk5-1 ([C] and [D]) roots.
(E) to (P) Dual immunolocalization of PIN2 (green) and H+-ATPase (red) in wild-type ([E], [G], [I], [K], [M], and [O]) and crk5-1 ([F], [H], [J], [L], [N], and [P]) roots. Immunolocalization detects PIN2 (green) in the apical membrane of epidermis and basal membranes of cortex cells in the wild type ([A], [B], [E], [I], [K], and [O]). In the crk5-1 mutant ([C], [D], [F], [J], [L], and [P]), epidermal signal of PIN2 is strongly reduced, whereas PIN2 polarity is changed gradually to apolar/apical in the cortex. Compared with the wild type, PIN2 is also observed in the endodermis of crk5-1 roots, where it decorates the inner lateral membrane with some portion at the apical and basal membranes ([F] and [L]). Dual immunolocalization with anti-PIN2 and anti-H+-ATPase antibodies ([E] to [P]) demonstrates that changes observed in PIN2 localization are not due to aberrant tissue penetration of antibody. Ubiquitous expression of H+-ATPase is only mildly affected by the crk5-1 mutation ([H] and [N]).
Bars = 50 µm in (A), (C), and (E) to (J) and 10 µm in (B), (D), and (K) to (P).
The crk5-1 Mutation Decelerates PIN2 Exocytosis and PIN2 Is a Candidate CRK5 Substrate
The altered polar localization of PIN2 in cells of the transition zone and the generally reduced amount of PIN2 in epidermal and cortex cell membranes of crk5-1 roots compared with the wild type suggested a potential defect in membrane recycling leading to destabilization of PIN2 (Michniewicz et al., 2007; Dhonukshe et al., 2010). Similarly to the effects of phosphatidylinositol monophosphate 5-kinase 2 (Mei et al., 2012), we found that BFA treatment stimulated faster internalization of PIN2-GFP from epidermal cell membranes of crk5-1 roots compared with the wild type (Figures 8A to 8H). Upon 30 min of treatment with 50 µM BFA, the number of BFA bodies was approximately threefold higher in crk5-1 compared with the wild type, and this difference was not balanced by prolonging the BFA treatment up to 2 h (Figure 8I). To exclude possible impact of de novo protein synthesis on enhanced accumulation of PIN2-GFP in BFA bodies, in control experiments, wild-type and crk5-1 seedlings were treated with the protein biosynthesis inhibitor cycloheximide (CHX; 50 µM) for 30 min and then incubated with BFA (50 µM) and CHX for 30 min before removal of BFA and further treatment with CHX for 1 h (Figures 8J to 8P). As without CHX, PIN2-GFP showed approximately threefold higher accumulation in brefeldin bodies in BFA+CHX but not in CHX-treated crk5-1 root cells compared with the wild type, indicating that the crk5-1 mutation decelerated BFA-sensitive exocytosis of PIN2 without affecting its de novo synthesis.
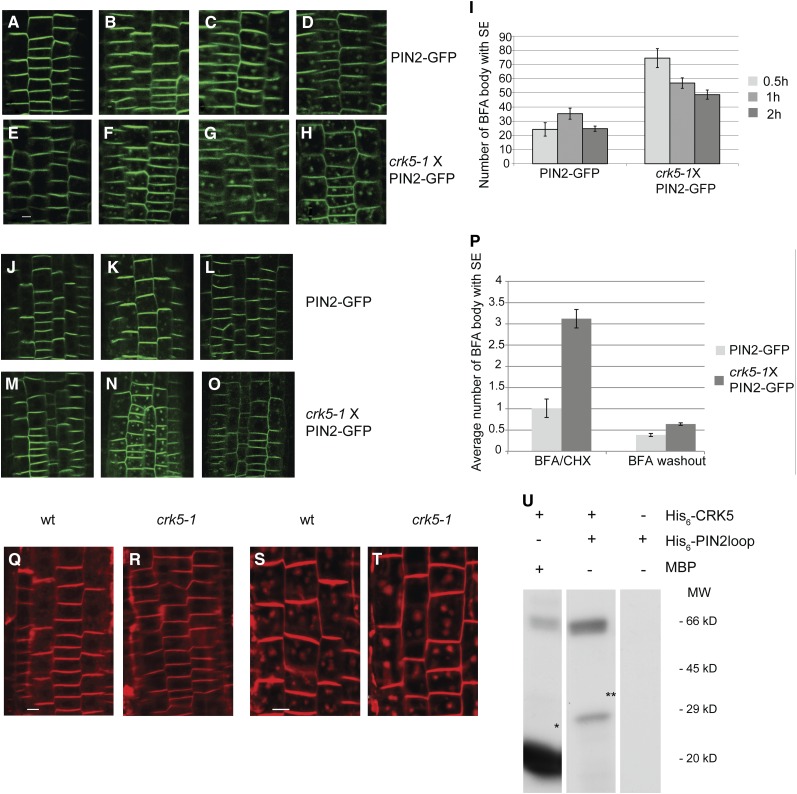
Figure 8.
The crk5-1 Mutation Decelerates Membrane Recycling of PIN2.
(A) to (H) Comparison of PIN2-GFP internalization in root epidermal cells of 5-d-old wild-type ([A] to [D]) and crk5-1 ([E] to [H]) seedlings at 0 ([A] and [E]), 0.5 ([B] and [F]), 1 ([C] and [G]), and 2 h ([D] and [H]) treatment with 50 μM BFA indicates enhanced inhibitory effect of BFA on PIN2-GFP cycling in crk5-1. Bar = 25 μm.
(I) Number of PIN2-GFP BFA bodies at 0.5, 1, and 2 h after BFA treatment in epidermal cells of wild-type and crk5-1 roots. Error bars indicate se of measurements by inspecting at least 10 roots in three biological replicates.
(J) to (O) Wild-type ([J] to [L]) and crk5-1 ([M] to [O]) roots were treated with CHX for 30 min ([J] and [M]) and then with CHX+BFA for 30 min ([K] and [N]) before BFA removal (washout) and further incubation with CHX for 1 h ([L] and [O]).
(P) Comparison of average number of BFA bodies in wild-type (PIN2-GFP) and crk5-1 (crk5-1 × PIN2-GFP) root cells upon combined BFA+CHX treatment and after BFA washout in the presence of CHX. Error bars indicate se of measurements performed with at least 10 roots in three biological replicates.
(Q) and (R) Compared with the wild type (wt) (Q), endocytosis of the membrane dye FM4-64 is slower in the crk5-1 mutant (R). Internalization of FM4-64 was monitored by staining of roots of 4-d-old seedlings (n > 20) with 5 μM FM4-64 for 30 min. Bar = 25 μm.
(S) and (T) Internalization of FM4-64 into BFA compartments is accelerated by the crk5-1 mutation. Roots of 4-d-old wild-type (S) and crk5-1 (T) seedlings (n > 20) were treated with FM4-64 (5 μM) and BFA (50 μM) for 30 min to compare the size of BFA bodies over time. Bar = 25 μm.
(U) In vitro kinase assay with purified His6-CRK5 and His6-PIN2loop and myelin basic protein (MBP) substrates. Asterisks indicate the phosphorylated myelin basic protein and His6-PIN2loop substrates.
[See online article for color version of this figure.]
In support of the observations of Teh and Moore (2007), we found that endocytosis of the membrane-specific dye FM4-64 was significantly slower in epidermal cells of crk5-1 roots compared with the wild type. However, FM4-64 showed slower internalization in the wild type compared with crk5-1 in the presence of BFA, as was indicated by the size of accumulating BFA bodies, which increased with time during BFA treatment (Figures 8Q to 8T).
As polar membrane recycling was demonstrated to be dependent on phosphorylation of central hydrophilic loops of PIN proteins, we tested whether PIN2 was a candidate substrate of CRK5. The hydrophilic T-loop domain of PIN2 expressed in Escherichia coli and purified via N-terminal His6-tag was phosphorylated by CRK5 in vitro analogously to the control myelin basic protein (Figure 8U).
DISCUSSION
Based on pharmacological studies, Ca2+/calmodulin signaling was suggested early on to play an important role in the regulation of root gravitropic responses (Sinclair and Trewavas, 1997). For example, KN-93, an inhibitor of animal calmodulin-activated kinase II, was found to abolish light-regulated root gravitropism in maize (Zea mays) and to compete effectively with CaM binding of the maize CRK MCK1 (Lu et al., 1996). Nonetheless, specific roles of CRKs in the control of root gravitropism remained thus far hidden because the available inhibitors and agonists could target more than one classes of signaling factors. The CaM antagonist KM-93 might thus interfere with the activities of CCaMK Ca2+ and calmodulin-activated kinases, which carry visinin-like C-terminal Ca2+ binding motives (Zhang and Lu, 2003), in addition to inhibiting CRKs that harbor defective Ca2+ binding EF-hand motives, and are thus active also in the absence of Ca2+ (Harper et al., 2004). Unlike some other plant species, intriguingly, no genuine CCaMK kinase is encoded by the Arabidopsis genome (Hrabak et al., 2003). Therefore, CRKs probably represent the major class of CaM-activated kinases in this model plant. Although biochemical studies of Arabidopsis CRK1 and CRK3 indicate that CaM increases by ∼10-fold, the autophosphorylation of these CRKs in the presence of Ca2+, CaM, and Ca2+ together stimulate only a marginal two- to threefold increase in their substrate phosphorylation activities (Wang et al., 2004; Du et al., 2005; Jeong et al., 2007). Consequently, it is still unclear whether CRKs are genuine CaM-activated kinases, and it is suggested that their CaM induction requires some yet undefined Ca2+ binding partners in vivo (Harmon, 2003).
The Arabidopsis CRK family includes eight highly similar members, which feature significant sequence divergence only in their heterologous N-terminal domains. All Arabidopsis CRKs carry the same N-terminal myristoylation/palmitolylation motif, mutations of which are known to prevent PM localization of tomato CRK1 and the related Ca2+-activated tobacco (Nicotiana tabacum) kinase Nt-CPK5 (Leclercq et al., 2005; Wang et al., 2005). Consequently, all CRKs carrying the same conserved N-terminal motif are predicted to be membrane-associated kinases. Our data confirm this prediction and show that CRK5, which shares all distinguishing sequence and structural features of biochemically characterized CRKs in Arabidopsis and other plant species, is a PM-associated protein kinase. Similarly to BFA-stimulated internalization of CRK5-GFP (Figure 4), mutation of the myristoylation motif leads to nuclear accumulation of tomato CRK1 (Leclercq et al., 2005). This suggests that some members of the CRK family might perform nuclear regulatory functions under specific conditions stimulating their internalization or that, in the case of CRK5-GFP, selective degradation of the N-terminal CRK5 domain allows some nuclear import of the potentially more stable GFP reporter protein. In any case, further research is required to test these possibilities.
Under normal growth conditions, CRK5 displays characteristic polar U-shape localization patterns facing the root tip in columella and root cap cells but decorates primarily the lateral outer membranes of epidermal and cortex cells in the meristematic, transition, and elongation zones of roots. PM localization domains of CRK5 thus partially overlap in different root cell types with those of examined PINs (Figures 3 and 7; see Supplemental Figures 6 to 8 online). In the epidermis, CRK5 localization closely resembles that of the boron efflux transporter BOR4 (Miwa et al., 2007) but clearly differs from those of ABCB/PGP1, 4, and 19 auxin transporters, which appear to selectively modulate the activities of specific PINs (Geisler et al., 2005; Terasaka et al., 2005; Titapiwatanakun et al., 2009; Titapiwatanakun and Murphy, 2009). Similarly, the cell type–specific polar localization of CRK5 also completely differs from that of AUX1 (see Supplemental Figure 9 online), which appears to act independently of so far studied ABCB/PGPs (Blakeslee et al., 2007).
Recently, Langowski et al. (2010) found that BFA treatment stimulates partial internalization and accumulation of BOR4 and the similarly localized ABCG37/PIS1 and PEN3 proteins in intracellular agglomerates without abolishing outer lateral membrane localization of these membrane transporters. Consequently, BFA appears to inhibit polar lateral secretion (i.e., exocytosis) of BOR4, ABCG37/PIS1, and PEN3, which show similar localization as CRK5. In comparison, we found that CRK5 accumulates in typical BFA compartments within 30 min and is completely depleted from the membrane during 90 min of BFA treatment. Despite this apparent difference in BFA sensitivity, it will be interesting to determine whether inactivation of CRK5 affects polar lateral membrane localization and phosphorylation of BOR4, ABCG37/PIS1, and PEN3. In addition to a potential role facilitating outer lateral targeting of some membrane transporters, CRK5 could also play a role in recycling proteins from the outer lateral membranes and thereby contribute to their targeting to the BFA-sensitive membrane recycling pathway. As polar localization of PIN proteins appears to be mediated though nonpolar secretion followed by their internalization and recycling-dependent polarization (Dhonukshe et al., 2008), current models propose that sequential phosphorylation of hydrophilic T-loops of PINs by AGCs and possibly other protein kinases (Zhang et al., 2010) directs their stabilization in basal membranes and subsequent apical transcytosis (Rademacher and Offringa, 2012; Löfke et al., 2013). As most evidence supporting these models is derived from the analysis of polar localization of PIN1 and PIN2 in vertically grown and gravistimulated roots, in this work, we examined in detail how inactivation of CRK5 affects the root gravitropic response. In addition to PIN2 and AUX1, which play pivotal roles in the root gravitropic response, we explored how the crk5-1 mutation affects polar localization of other PIN proteins, including PIN1, PIN3, PIN4, and PIN7.
We have found that the crk5-1 null mutation inhibits positive and negative gravitropic bending of roots and shoots, respectively. By examining how inactivation of CRK5 affects asymmetric activation of auxin-stimulated DR5-GFP reporter in gravistimulated roots, we found that in the area of quiescent center, columella, and root cap initials, the expression level of DR5-GFP is markedly lower in the crk5-1 mutant compared with the wild type. However, upon gravistimulation for 4 to 5 h, the auxin-induced DR5-GFP maximum in these areas of root tip is restored to the wild type by redirection of auxin flow from the upper toward the lower root section. This suggests that the root tip area of crk5-1 has lower auxin content, which is also reflected by ∼30% reduction of crk5-1 root growth compared with the wild type. Furthermore, restoration of wild-type DR5-GFP expression in crk5-1 root tip by NOA inhibition of AUX1 influx carrier, but not by NPA-mediated inhibition of shootward epidermal auxin efflux, strongly suggests that the crk5-1 mutation enhances shootward auxin flow from the root tip through the cortex toward the elongation zone. This is also consistent with our finding that the number of lateral roots is increased by at least 30% in the crk5-1 mutant.
We excluded that reduced DR5-GFP activity in crk5-1 was a consequence of impaired auxin signaling by showing that the crk5-1 mutation does not alter the expression of numerous genes regulating auxin biosynthesis and the auxin-induced PIN genes in the root. In addition, the crk5-1 mutation did not lead to alteration in the polar localization of AUX1 or the PIN auxin efflux facilitators PIN1, PIN3, PIN4, and PIN7 with or without gravistimulation. By contrast, consistent with a major function of PIN2 in the control of root gravitropic responses, we found that in the root transition zone, which plays a critical role in the bending response to gravistimulus (Verbelen et al., 2006; Muraro et al., 2013), PIN2 is largely depleted from apical membranes of epidermal cells. PIN2 shows either apolar or apical localization in the neighboring cortical cells, consistent with a possible defect of backward auxin recycling from the cortex to the root tip. The effect of crk5-1 mutation thus appears to be very similar to the consequence of treating roots with low concentrations of BFA, which stimulates basal-to-apical relocation of PIN2 in the cortex inhibiting root gravitropic response (Rahman et al., 2010). In fact, we observed that the crk5-1 mutation not only reduces the amount of PM-associated PIN2 in the epidermis of transition zone, but also markedly accelerates brefeldin-sensitive internalization of PIN2 in epidermal cells. Subsequently, we found that CRK5 phosphorylates in vitro the central hydrophilic T-loop of PIN2, which modulates its PM association and stability in the epidermis (Michniewicz et al., 2007; Dhonukshe et al., 2010). In conclusion, these results suggest that CRK5 phosphorylation is specifically involved in the regulation of brefeldin-sensitive membrane recycling of PIN2 in the root transition zone.
Based on the remarkable conservation of kinase domains in the CRK family, it is tempting to speculate that in analogy to the AGC kinases PID, WAG1, WAG2, and D6PK (reviewed in Ganguly et al., 2012), CRKs might contribute to cell type–specific phosphorylation of specific residues in hydrophilic loops of certain PINs and thereby regulate their polar membrane recycling. Thus, homologous members of the CRK family might perform overlapping cell type–specific functions, which could explain the relatively mild developmental effects of the crk5-1 mutation. In addition to further study of CRK-mediated PIN phosphorylation, it remains important to determine whether CRKs contribute to the regulation of activity and membrane localization of any of the PIN-specific AGC kinases, and vice versa, in order to establish the position of CRKs in the gravistimulation signaling pathway. A further intriguing question is whether CRKs also contribute to regulation of ABCB/PGP auxin transporters. In this respect, it is intriguing that according to Blakeslee et al. (2007) PIN2 is colocalized in epidermal and cortex cells of the root transition zone with ABCB/PGP1 and 19. Although the pgp19 mutant does not show altered root gravitropic response (Lewis et al., 2007), the pgp1 and pgp19 mutations are reported to enhance synergistically the gravitropic defect of the pin2 mutant (Blakeslee et al., 2007). Nevertheless, PGP19 was found not to be expressed in the root, but to play a major role in the regulation of shoot gravitropic responses (Rojas-Pierce et al., 2007; Nagashima et al., 2008). By contrast, the pgp4 mutation impairs both acro- and basipetal auxin transport only in lateral roots without affecting the elongation and gravitropic response of primary root (Wu et al., 2007). Despite these apparently contradictory observations, based on the finding that some ABCB/PGPs, such as PGP1 (Henrichs et al., 2012), are phosphorylated by the AGC PIN kinase PID, it will be interesting to determine whether CRK5 or other members of the Arabidopsis CRK family contribute by phosphorylation to the modulation of regulatory interactions between ABCB/PGPs and PINs (Bandyopadhyay et al., 2007; Titapiwatanakun and Murphy, 2009). Definition of the role of CRK5 in the regulation of root gravitropic response now opens the way to answer these important questions by combined genetic analysis of members of the CRK family.
METHODS
Plant Materials, Growth Conditions, and Gravitropism Assays
Seeds of wild-type Arabidopsis thaliana (Columbia-0) and mutant lines were germinated after 1 d of stratification at 4°C on half-strength Murashige and Skoog (MS) medium containing 0.5% Suc in a growth chamber (22°C with 120 mol/m2s light intensity, constant light) or in soil under controlled greenhouse conditions (16 h light/8 h dark period; 22 to 24°C day temperature and 18°C night temperature, 80 to 120 mol/m2s light intensity) as described (Koncz et al., 1994). Polyethylene glycol–mediated transformation of cell suspension–derived protoplasts and transient expression studies were performed 18 h after protoplast transformation as described (Rigó et al., 2008). Root and shoot gravitropism assays were performed in growth chambers using continuous light. Five-day-old vertically grown seedlings were reoriented by 135°, and the degree of root bending was recorded by scanning 12 and 24 h after rotation. The rate of root bending was determined by measuring the angle formed between the growth direction of root tip and horizontal baseline. At least 300 wild-type and crk5-1 seedlings were tested in four separate experiments. Gravitropic response of inflorescence stems was similarly assayed using 4-week-old greenhouse grown plants carrying inflorescence stems of 10 cm after turning them to a horizontal position for 3 h in darkness.
Identification of T-DNA Insertion Mutants, Plasmid Construction, and Plant Transformation
Identification of the crk5-1 (MPIZ38225) and crk5-2 (Salk_003774) T-DNA insertion mutants, construction of CRK5 and PIN2 loop E. coli expression vectors, assembly of gCRK5:GFP and gCRK5:GUS gene constructs in Agrobacterium tumefaciens binary vectors, and generation of transgenic Arabidopsis plants are described in the Supplemental Methods and References 1 and Supplemental Table 1 online.
RNA Isolation, RT-PCR, and qRT-PCR Analyses
Total RNA was isolated from leaf or root tissues of 3-week-old seedlings using the TRI reagent (Sigma-Aldrich). Transcript levels were monitored by qRT-PCR using oligonucleotide primers (see Supplemental Table 1 online). cDNA templates were generated from DNase-treated (Fermentas) total RNA (1 μg) by RevertAid M-MuLV reverse transcriptase (Fermentas). qRT-PCR reactions were performed with SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) using an ABIPRISM 7700 sequence detection system (Applied Biosystems) and the following protocol: denaturation 95°C for 10 min, 40 cycles of 95°C for 10 s, and 60°C for 60 s.
CRK5 and PIN2loop Substrate Purification and in Vitro Kinase Assays
His6-CRK5 and His6-Pin2loop proteins were purified by Ni-nitrilotriacetic acid agarose affinity chromatography (Novagen) according to the manufacturer’s instructions. Following gradient elution with 50 to 500 mM imidazole and size separation by 10% SDS-PAGE, fractions containing apparently homogeneous His6-CRK5 or His6-PIN2loop were identified, pooled, dialyzed with storage buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10% glycerin, and 5 mM 2-mercaptoethanol) at 4°C, and stored at −80°C. In vitro phosphorylation assays were performed with 1 μg His6-CRK5 in 20 μL kinase buffer (20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM DTT, and 5 µCi [γ-32P]ATP) containing either 2 μg myelin basic protein (Sigma-Aldrich) or 2 μg His6-PIN2 loop as substrate at 24°C for 30 min. CaCl2 was added, at a final concentration of 0.1 mM with or without 1 mM Ca2+-chelator EGTA where indicated. The kinase assays were size separated by 12% SDS-PAGE; the gels were stained with Coomassie Brilliant Blue dye, dried, and subjected to autoradiography using x-ray film.
Microsomal Membrane and Cell Wall Preparations and Immunoblotting
From seedlings expressing the gCRK5:GFP construct, root cultures were established as described (Mathur and Koncz, 1998). Root samples (15 g) were homogenized in liquid nitrogen, solubilized in homogenization buffer (100 mM Tris-HCl, pH 7.5, 300 mM Suc, 10 mM EDTA, 2 mM EGTA, and 0.1% protease inhibitor cocktail Sigma-Aldrich P9599), and filtered through Miracloth (Calbiochem) to prepare total cell extract by centrifugation at 2000g for 15 min at 4°C as described (Dammann et al., 2003). Microsomal membranes were collected by centrifugation 100,000g for 1 h; the supernatant represented the cytoplasmic fraction. Cell wall fraction was purified as described (Feiz et al., 2006) by extraction of 15 g of root sample with extraction buffer (5 mM acetate, pH 4.6, 0.4 M Suc, and 0.1% protease inhibitor cocktail Sigma-Aldrich P9599) followed by centrifugation at 1000g at 4°C for 15 min. The pellet was washed successively with extraction buffer containing 0.4, 0.6, and 1 M Suc and then with 5 mM acetate buffer before lyophilization of purified sample. Following standardization of protein concentrations using Bio-Rad protein assay, 25 μg of each sample was size separated by 8% SDS-PAGE, transferred onto Immobilon Membrane (Millipore), and incubated for 2 h in 1× TBST blocking buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20, and 5% dry skimmed milk) and for 2 h with anti-GFP antibody (Roche; 1:1000 dilution) in blocking buffer. After washing with 1× TBST three times, the membranes were incubated for 1.5 h with an anti-mouse-POD secondary antibody (Pierce; dilution 1:5000), washed with 1× TBST, and overlaid with Immobilon Western Chemiluminescent horseradish peroxidase substrate (Millipore) to detect CRK5-GFP by autoradiography.
Histochemical GUS and Starch Staining and Immunolocalization of PIN1 and PIN2
Selected T3 lines carrying the gCRK5-GUS construct in wild-type and crk5-1 mutant backgrounds were germinated and subjected to GUS staining according to Gallagher (1992). Starch staining of roots was performed with Lugol solution (Fluka) for 10 min, and then the samples were destained in water. Immunolocalization of PIN1 and PIN2 was performed with wild-type and crk5-1 mutant seedlings grown vertically on 0.5× MS medium supplemented with 0.5% Suc for 7 d in constant light as described (Friml et al., 2003). Immunolocalization of PIN1 was performed using an affinity-purified rabbit anti-PIN1 antibody as described by Gälweiler et al. (1998). Dilutions of primary anti-PIN2 guinea pig antibody (Ditengou et al., 2008) was 1:600 and for the anti-PM H+-ATPase rabbit antibody (Langhans et al., 2001; Agrisera AS07 260) 1:1000. As secondary antibodies, ALEXA Fluor 488 goat anti-guinea pig (Invitrogen) and ALEXA Fluor 555 goat anti-rabbit (Invitrogen) diluted to 1:600 were used.
Confocal Laser Scanning Microscopy
To compare spatial and temporal expression patterns of CRK5-GFP reporter construct, at least 20 offspring derived originally from five independent wild-type and crk5-1 lines were examined. For monitoring DR5-GFP localization in gravitropism assays, stratified sterile seeds carrying the reporter construct were placed onto microscope slides covered with a thin layer of 0.5× MS medium containing 0.8% phytoagar (Duchefa) and germinated in vertical position for 5 d in continuous light. Upon gravistimulation by rotating the slides with 135°, the DR5-GFP expression pattern in 50 seedlings from each independent line was monitored at different time points using an Olympus FV1000 laser scanning microscope with the following configuration: objective lenses, UPLSAPO ×20 (dry, numerical aperture [NA] of 0.75), UPLFLN ×40 (oil, NA of 1.3), and UPLSAPO ×60 (oil, NA of 1.35); sampling speed, 4 µs/pixel; line averaging, 2×; scanning mode, sequential unidirectional; excitation, 405 nm (4′,6-diamidino-2-phenylindole) and 488 nm (GFP); laser transmissivity, <10%; main dichroic beam splitter, DM405/488; intermediate dichroic beam splitter, SDM 490; and GFP fluorescence detection between 505 and 530 nm. Roots of seedlings grown in continuous light vertically for 4 d were stained with 5 μM FM4-64 (Invitrogen T3166) for 30 min, and then images were recorded using an excitation wavelength of 543 nm. For monitoring PIN2-GFP in cells of the root transient zone, wild-type and crk5-1 seedlings were grown either in the light or dark vertically for 5 d and subjected to gravistimulation by 90° rotation. Fluorescence intensities were measured by construction of color-coded heat maps as described (Kleine-Vehn et al., 2008b; Baster et al., 2013), and the data derived from examination of at least 20 seedlings from each of two biological repeats were evaluated using the Student’s t test (P < 0.001). The vesicle trafficking inhibitor BFA (Sigma-Aldrich B-7651) was applied at 50 µM concentration at various time points (0, 0.5, 1, and 2 h) without or parallel with the FM4-64 staining. Combined BFA+CHX treatments were performed as described (Kakar et al., 2013). Five-day-old seedlings were treated in 0.5× MS medium with 50 µM CHX for 30 min, and then with CHX plus 50 µM BFA for 30 min, and finally with CHX alone for 1 h. Seedlings sampled from each phase of treatment were subjected to confocal imaging to visualize and count BFA bodies. Roots of AUX1-YFP–expressing plants were counterstained with propidium iodine (5 µg/mL in water) for 5 to 20 min. Differential interference contrast images were captured with a 488-nm laser line for FM4-64: excitation, 543 nm; emission, long-pass 560 nm. PIN immunolocalization images were recorded with a 510 Confocal Microscope from Zeiss equipped with ×20, ×40, and ×63 lenses. Fluorophores were excited with 488 and 561 nm. Emitted light was collected with 505- to 550-nm and 575- to 615-nm band-pass filters. Composite images were prepared using the Adobe Photoshop and Illustrator software.
Bioinformatics Analyses
Amino acid and nucleotide sequences of the CRK family were retrieved from The Arabidopsis Information Resource (http://www.Arabidopsis.org/). Searches for putative protein targeting signals were performed using the TargetP, Myristoylation Predictor, and cNLS Mapper databases (http://www.cbs.dtu.dk/services/TargetP, http://plantsp.genomics.purdue.edu/myrist.html, and http://nls-mapper.iab.keio.ac.jp). BLAST searches based on multiple sequence alignments were performed using the National Center for Biotechnology Information COBALT tool with standard settings, and the alignments were colored using GeneDoc software (http://www.nrbsc.org/gfx/genedoc/). Statistical analyses were performed using the SPSS Statistic software version 14.0.
Accession numbers
Sequence data from this article can be found in The Arabidopsis Information Resource and GenBank (National Center for Biotechnology Information) databases under the following accession numbers: CRK1 (At2g41140), CRK2 (At3g19100), CRK3 (At2g46700), CRK4 (At5g24430), CRK5 (At3g50530)/PIP-D (GenBank Y09418.1), CRK6 (At3g49370), CRK7 (At3g56760), CRK8 (At1g49580), Sl-CRK1 (AY079049), Nt-CBK1 (AF435450), Nt-CBK2 (AF435452), Dc-CRK1 (CAA58750), Zm-MCK1 (AAB47181), Zm-MCK2 (AF289237), Os-CBK1 (AF368282), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At3g70940), PIN4 (At2g01420), PIN7 (At1g23080), AUX1 (At2g38120), LAX3 (At1g77690), TRP2 (At5g54810),TRP3 (At3g54640), YUCCA3 (At1g04610), AMI (At1g08980), TAA1 (At1g70560), CYP83B1 (At4g31500), NIT3 (At3g44320), and GAPDH2 (At1g13440).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Positions of T-DNA Insertion in CRK5 and qRT-PCR Analysis of Transcription of crk5 Mutant Alleles.
Supplemental Figure 2. qRT-PCR Measurement of CRK5 Transcript Levels and Detection of CRK5-GUS Expression in Different Organs.
Supplemental Figure 3. Cobalt Sequence Alignment of Conserved Domains of CRK Family Members from Different Plant Species.
Supplemental Figure 4. In Vitro Kinase Assays with Purified His6-CRK5.
Supplemental Figure 5. qRT-PCR Comparison of mRNA Levels of Genes Involved in the Regulation of Auxin Biosynthesis and Encoding PIN Auxin Efflux and AUX/LAX Influx Carriers in Roots of Wild-Type, crk5-1 Mutant, and Genetically Complemented crk5-1/gCRK5-GFP Seedlings.
Supplemental Figure 6. Polar Localization of CRK5-GFP Is Not Changed in Root Cell Files in Response to Gravistimulation.
Supplemental Figure 7. Localization of PIN1-GFP, PIN4-GFP, and PIN7-GFP in Vertically Grown and Gravistimulated Roots of Wild-Type and crk5-1 Mutant Plants.
Supplemental Figure 8. Comparison of PIN3-GFP Localization in Vertically Grown and Gravistimulated Roots of the Wild Type and crk5-1 Mutant.
Supplemental Figure 9. Cellular Localization of AUX1-YFP in Wild-Type and crk5-1 Mutant Roots.
Supplemental Table 1. List of PCR Oligonucleotide Primers.
Supplemental Methods and References 1. Plasmid Constructs and Identification of T-DNA Insertion Mutants.
Supplementary Material
Acknowledgments
We thank Anna-Mária Király and Zsuzsa Kószó for excellent technical assistance and Jiří Friml, Malcolm J. Bennett, and Xugang Li for providing the PIN-GFP, AUX1-YFP, and pET28a-PIN2loop constructs, respectively. This work was supported by the OTKA Grant K81765 to G.R., TET_12_RO-1-2013-0010 to Á.C., and OTKA K68226 and NKFP 4-038-04 Grants to L.S.; by the Excellence Initiative of the German Federal and State Governments (EXC 294), SFB 592, Graduiertenkolleg 1305, European Space Agency, Bundesministerium für Forschung und Technik, Deutsches Zentrum für Luft und Raumfahrt, the Freiburg Initiative for Systems Biology, and FP6 European Union (Autoscreen) to K.P.; and by SFB635 and AFGN (KO 1438/12-1) Grants from the Deutsche Forschungsgemeinschaft to C.K.
AUTHOR CONTRIBUTIONS
G.R. performed genetic and biochemical studies and prepared all expression constructs and transgenic plants. O.T., F.A., and A.C. performed confocal microscopy studies. L.Z. carried out PCR analyses and root growth assays. H.K. contributed statistical analyses. Sequence analyses were done by A.P. K.S. performed yeast two-hybrid studies. C.K., A.C., L.S., and K.P. designed the research. Z.D. and K.F.M. performed matrix-assisted laser desorption/ionization (MALDI) analysis of PIN2loop CRK5 phosphorylation sites. C.K. prepared the article, which was critically commented on by K.P., A.C., L.S., and O.T.
Glossary
- BFA
brefeldin A
- PM
plasma membrane
- qRT-PCR
quantitative RT-PCR
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- CaM
C-terminal calmodulin
- IAA
indole-3-acetic acid
- NPA
1-naphthylphtalamic acid
- NOA
1-naphtoxyacetic acid
- CHX
cycloheximide
- MS
Murashige and Skoog
- NA
numerical aperture
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 Erratum. Nat. Cell Biol. 8: 424. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baldwin K.L., Strohm A.K., Masson P.H. (2013). Gravity sensing and signal transduction in vascular plant primary roots. Am. J. Bot. 100: 126–142 [DOI] [PubMed] [Google Scholar]
- Baster P., Robert S., Kleine-Vehn J., Vanneste S., Kania U., Grunewald W., De Rybel B., Beeckman T., Friml J. (2013). SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32: 260–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Ampudia C.S., Hooykaas P.J., Offringa R. (2003). PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 132: 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor E.B. (2013). Regulation of plant gravity sensing and signaling by the actin cytoskeleton. Am. J. Bot. 100: 143–152 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., et al. (2007). Interactions of PIN and PGP auxin transport mechanisms. Biochem. Soc. Trans. 35: 137–141 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Dammann C., Ichida A., Hong B., Romanowsky S.M., Hrabak E.M., Harmon A.C., Pickard B.G., Harper J.F. (2003). Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski L., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Ditengou F.A., Teale W.D., Kochersperger P., Flittner K.A., Kneuper I., van der Graaff E., Nziengui H., Pinosa F., Li X., Nitschke R., Laux T., Palme K. (2008). Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 18818–18823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Wang Y., Liang S., And Lu Y.-T. (2005). Biochemical and expression analysis of an Arabidopsis calcium-dependent protein kinase-related kinase. Plant Sci. 168: 1181–1192 [Google Scholar]
- Estelle M. (1996). Plant tropisms: The ins and outs of auxin. Curr. Biol. 6: 1589–1591 [DOI] [PubMed] [Google Scholar]
- Feiz L., Irshad M., Pont-Lezica R.F., Canut H., Jamet E. (2006). Evaluation of cell wall preparations for proteomics: A new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. (2010). Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235 [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Mayer U., Palme K., Muster G. (2003). Automated whole mount localisation techniques for plant seedlings. Plant J. 34: 115–124 [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Furumoto T., Ogawa N., Hata S., Izui K. (1996). Plant calcium-dependent protein kinase-related kinases (CRKs) do not require calcium for their activities. FEBS Lett. 396: 147–151 [DOI] [PubMed] [Google Scholar]
- Gallagher, S.R. (1992). GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. (London: Academic Press). [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Ganguly A., Sasayama D., Cho H.T. (2012). Regulation of the polarity of protein trafficking by phosphorylation. Mol. Cells 33: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon A.C. (2003). Calcium-regulated protein kinases of plants. Gravit. Space Biol. Bull. 16: 83–90 [PubMed] [Google Scholar]
- Harper J.F., Breton G., Harmon A. (2004). Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Hegeman A.D., Rodriguez M., Han B.W., Uno Y., Phillips G.N., Jr, Hrabak E.M., Cushman J.C., Harper J.F., Harmon A.C., Sussman M.R. (2006). A phyloproteomic characterization of in vitro autophosphorylation in calcium-dependent protein kinases. Proteomics 6: 3649–3664 [DOI] [PubMed] [Google Scholar]
- Henrichs S., et al. (2012). Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J. 31: 2965–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak E.M., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua W., Liang S., Lu Y.T. (2003). A tobacco (Nicotiana tabacum) calmodulin-binding protein kinase, NtCBK2, is regulated differentially by calmodulin isoforms. Biochem. J. 376: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua W., Zhang L., Liang S., Jones R.L., Lu Y.T. (2004). A tobacco calcium/calmodulin-binding protein kinase functions as a negative regulator of flowering. J. Biol. Chem. 279: 31483–31494 [DOI] [PubMed] [Google Scholar]
- Jeong J.C., Shin D., Lee J., Kang C.H., Baek D., Cho M.J., Kim M.C., Yun D.J. (2007). Isolation and characterization of a novel calcium/calmodulin-dependent protein kinase, AtCK, from Arabidopsis. Mol. Cells 24: 276–282 [PubMed] [Google Scholar]
- Kakar K., Zhang H., Scheres B., Dhonukshe P. (2013). CLASP-mediated cortical microtubule organization guides PIN polarization axis. Nature 495: 529–533 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Ding Z., Jones A.R., Tasaka M., Morita M.T., Friml J. (2010). Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. USA 107: 22344–22349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Langowski L., Wisniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008a). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008b). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z. (2012). The spliceosome-activating complex: Molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, S. Gelvin and B. Schilperoort, eds (Dordrecht-Boston-London: Kluwer Academic Publishers), pp. 1–22. [Google Scholar]
- Kramer E.M. (2004). PIN and AUX/LAX proteins: Their role in auxin accumulation. Trends Plant Sci. 9: 578–582 [DOI] [PubMed] [Google Scholar]
- Kurusu, T., Kuchitsu, K., Nakano, M., Nakayama, Y., and Iida, H. (2013). Plant mechanosensing and Ca(2+) transport. Trends Plant Sci. 18: 227–233. [DOI] [PubMed]
- Langhans M., Ratajczak R., Lützelschwab M., Michalke W., Wächter R., Fischer-Schliebs E., Ullrich C.I. (2001). Immunolocalization of plasma-membrane H+-ATPase and tonoplast-type pyrophosphatase in the plasma membrane of the sieve element-companion cell complex in the stem of Ricinus communis L. Planta 213: 11–19 [DOI] [PubMed] [Google Scholar]
- Langowski L., Růzicka K., Naramoto S., Kleine-Vehn J., Friml J. (2010). Trafficking to the outer polar domain defines the root-soil interface. Curr. Biol. 20: 904–908 [DOI] [PubMed] [Google Scholar]
- Leclercq J., Ranty B., Sanchez-Ballesta M.T., Li Z., Jones B., Jauneau A., Pech J.C., Latché A., Ranjeva R., Bouzayen M. (2005). Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J. Exp. Bot. 56: 25–35 [DOI] [PubMed] [Google Scholar]
- Lewis D.R., Miller N.D., Splitt B.L., Wu G., Spalding E.P. (2007). Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell 19: 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindzen E., Choi J.H. (1995). A carrot cDNA encoding an atypical protein kinase homologous to plant calcium-dependent protein kinases. Plant Mol. Biol. 28: 785–797 [DOI] [PubMed] [Google Scholar]
- Löfke C., Luschnig C., Kleine-Vehn J. (2013). Posttranslational modification and trafficking of PIN auxin efflux carriers. Mech. Dev. 130: 82–94 [DOI] [PubMed] [Google Scholar]
- Lu Y.T., Hidaka H., Feldman L.J. (1996). Characterization of a calcium/calmodulin-dependent protein kinase homolog from maize roots showing light-regulated gravitropism. Planta 199: 18–24 [DOI] [PubMed] [Google Scholar]
- Mano Y., Nemoto K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63: 2853–2872 [DOI] [PubMed] [Google Scholar]
- Mathur J., Koncz C. (1998). Callus culture and regeneration. Methods Mol. Biol. 82: 31–34 [DOI] [PubMed] [Google Scholar]
- Mei Y., Jia W.J., Chu Y.J., Xue H.W. (2012). Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 22: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Miwa K., Takano J., Omori H., Seki M., Shinozaki K., Fujiwara T. (2007). Plants tolerant of high boron levels. Science 318: 1417. [DOI] [PubMed] [Google Scholar]
- Morita M.T. (2010). Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61: 705–720 [DOI] [PubMed] [Google Scholar]
- Muraro D., Byrne H., King J., Bennett M. (2013). The role of auxin and cytokinin signalling in specifying the root architecture of Arabidopsis thaliana. J. Theor. Biol. 317: 71–86 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A., Uehara Y., Sakai T. (2008). The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant Cell Physiol. 49: 1250–1255 [DOI] [PubMed] [Google Scholar]
- Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., Scheres B., Fu Y., Yang Z. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E.H., Offringa R. (2012). Evolutionary adaptations of plant AGC kinases: From light signaling to cell polarity regulation. Front. Plant Sci. 3: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Takahashi M., Shibasaki K., Wu S., Inaba T., Tsurumi S., Baskin T.I. (2010). Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell 22: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H., Gallego-Bartolomé J., Vanstraelen M., Robert H.S., Alabadí D., Blázquez M.A., Benková E., Friml J. (2011). Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 67: 817–826 [DOI] [PubMed] [Google Scholar]
- Rigó G., Ayaydin F., Szabados L., Koncz C., Cséplő A. (2008). Suspension protoplasts as useful experimental tool to study localization of GFP-tagged proteins in Arabidopsis thaliana. Acta Biol. Szeged. 52: 59–61 [Google Scholar]
- Ríos G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 32: 243–253 [DOI] [PubMed] [Google Scholar]
- Rojas-Pierce M., Titapiwatanakun B., Sohn E.J., Fang F., Larive C.K., Blakeslee J., Cheng Y., Cutler S.R., Peer W.A., Murphy A.S., Raikhel N.V. (2007). Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chem. Biol. 14: 1366–1376 Erratum. Chem. Biol. 15: 87. [DOI] [PubMed] [Google Scholar]
- Sinclair W., Trewavas A.J. (1997). Calcium in gravitropism. A re-examination. Planta 203 (suppl. 1): S85–S90 [DOI] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kramer E.M., Perry P., Knox K., Leyser H.M., Haseloff J., Beemster G.T., Bhalerao R., Bennett M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Teh O.K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496 [DOI] [PubMed] [Google Scholar]
- Terasaka K., Blakeslee J.J., Titapiwatanakun B., Peer W.A., Bandyopadhyay A., Makam S.N., Lee O.R., Richards E.L., Murphy A.S., Sato F., Yazaki K. (2005). PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B., et al. (2009). ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 57: 27–44 [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B., Murphy A.S. (2009). Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J. Exp. Bot. 60: 1093–1107 [DOI] [PubMed] [Google Scholar]
- Verbelen J.P., De Cnodder T., Le J., Vissenberg K., Baluska F. (2006). The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: Meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signal. Behav. 1: 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wisniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C., Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Wang L., Liang S., Lu Y.T. (2001). Characterization, physical location and expression of the genes encoding calcium/calmodulin-dependent protein kinases in maize (Zea mays L.). Planta 213: 556–564 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liang S., Xie Q.G., Lu Y.T. (2004). Characterization of a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis. Biochem. J. 383: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang M., Ke K., Lu Y.T. (2005). Cellular localization and biochemical characterization of a novel calcium-dependent protein kinase from tobacco. Cell Res. 15: 604–612 [DOI] [PubMed] [Google Scholar]
- Went F.W. (1974). Reflections and speculations. Annu. Rev. Plant Physiol. 25: 1–26 [Google Scholar]
- Wu G., Lewis D.R., Spalding E.P. (2007). Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zegzouti H., Anthony R.G., Jahchan N., Bögre L., Christensen S.K. (2006). Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 6404–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2011). Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev. Cell 20: 855–866 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu B.F., Liang S., Jones R.L., Lu Y.T. (2002). Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem. J. 368: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lu Y.T. (2003). Calmodulin-binding protein kinases in plants. Trends Plant Sci. 8: 123–127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.