This work provides biochemical evidence that ABCC transporters are directly involved in anthocyanin transport into plant vacuoles. The presence of reduced glutathione is a prerequisite for the transport. Our data support that anthocyanins and glutathione are cotransported but that no glutathione anthocyanin conjugate is formed.
Abstract
Accumulation of anthocyanins in the exocarp of red grapevine (Vitis vinifera) cultivars is one of several events that characterize the onset of grape berry ripening (véraison). Despite our thorough understanding of anthocyanin biosynthesis and regulation, little is known about the molecular aspects of their transport. The participation of ATP binding cassette (ABC) proteins in vacuolar anthocyanin transport has long been a matter of debate. Here, we present biochemical evidence that an ABC protein, ABCC1, localizes to the tonoplast and is involved in the transport of glucosylated anthocyanidins. ABCC1 is expressed in the exocarp throughout berry development and ripening, with a significant increase at véraison (i.e., the onset of ripening). Transport experiments using microsomes isolated from ABCC1-expressing yeast cells showed that ABCC1 transports malvidin 3-O-glucoside. The transport strictly depends on the presence of GSH, which is cotransported with the anthocyanins and is sensitive to inhibitors of ABC proteins. By exposing anthocyanin-producing grapevine root cultures to buthionine sulphoximine, which reduced GSH levels, a decrease in anthocyanin concentration is observed. In conclusion, we provide evidence that ABCC1 acts as an anthocyanin transporter that depends on GSH without the formation of an anthocyanin-GSH conjugate.
INTRODUCTION
Anthocyanins represent the largest class of flavonoids (Welch et al., 2008). They constitute one of the most important families of secondary metabolites and are widely distributed in plants. They have been shown to be synthesized as protective compounds in response to abiotic stresses, such as UV, cold, and drought, but also to attract pollinators. However, the importance of anthocyanins extends beyond the plant kingdom. During the last decades, anthocyanins were reported to be efficient antioxidants and regulators of signaling pathways involved in many human diseases (Winkel-Shirley, 2001; Grotewold, 2006; Wallace, 2011). The health-promoting effects mostly depend on the consumption of high levels of dietary anthocyanins. Therefore, anthocyanin-rich extracts became sought after in the food industry, and numerous attempts have been made to improve the quantity of bioactive anthocyanins in dietary products. A study directly linking anthocyanin content to beneficial effects on health was published recently, showing that feeding cancer-susceptible mice with anthocyanin-rich tomato (Solanum lycopersicum) extended their life span (Butelli et al., 2008).
Like many other secondary metabolites, anthocyanins are synthesized in the cytosol and accumulate in the vacuoles, mostly of the epidermal tissues of fruits, leaves, and flowers. It has been suggested that the structural enzymes involved in anthocyanin biosynthesis form a complex via protein–protein interaction, which is anchored to the endoplasmic reticulum membrane (Saslowsky et al., 2005). To date, more than 635 anthocyanins have been identified. The aglycone forms (anthocyanidins) differ from each other by different hydroxylation and methylation patterns. The derivatives of the most common anthocyanidins in nature (cyanidin, delphinidin, peonidin, petunidin, pelargonidin, and malvidin) represent over 90% of the anthocyanins identified to date (Wallace, 2011). Once the biosynthesis is completed, including the additional chemical modifications (glycosylation, methylation, and acylation; Winkel-Shirley, 2008), anthocyanins are transported into the vacuole. A report has shown that vacuolar accumulation of flavonoids is a prerequisite for their biosynthesis and thus fulfils a regulatory function during synthesis (Marinova et al., 2007a). The mechanisms by which anthocyanins are transported into the vacuole have long been debated. To date, two major transport models have been proposed: membrane vesicle– and membrane transporter–mediated transport (Grotewold and Davies, 2008; Zhao and Dixon, 2010). Scientific evidence supports both models. The vesicle-mediated transport system is mostly based on microscopy observations (Irani and Grotewold, 2005; Zhang et al., 2006; Gomez et al., 2011). Two distinct vesicular structures were described as having a putative role in anthocyanin transport. Cytoplasmic anthocyanin bodies, called anthocyanoplasts, are vesicular structures surrounded by membranes, which gradually fuse together. Within vacuoles, anthocyanic vacuolar inclusions have been observed. These are dynamic, nonmembrane surrounded structures that, despite being associated with proteins and membranous substances, have a role in storage rather than in transport of anthocyanins. The anthocyanoplasts have been shown to colocalize with protein storage vacuoles and to transport anthocyanins in the trans-Golgi network (Zhao and Dixon, 2010; Pourcel et al., 2010). It is likely that transporters might also be needed to load the vesicles with anthocyanins during membrane vesicle-mediated transport.
Concerning the transporter-mediated model, biochemical, molecular, and genetic evidence support the involvement of both multidrug and toxic extrusion (MATE) and the ATP binding cassette (ABC) proteins in the transport of anthocyanins (Goodman et al., 2004; Marinova et al., 2007b; Gomez et al., 2009; Zhao and Dixon, 2009; Zhao et al., 2011). MATE transporters comprise a large family of transporters, widely distributed in all living organisms. The first experimental evidence for the involvement of a MATE transporter in flavonoid transport came from a genetic screen in Arabidopsis thaliana aiming to identify the different steps of flavonoid biosynthesis. Among other identified genes, Transparent testa12 (TT12) was shown to encode a MATE-type transporter involved in the biosynthesis of flavonoids (Debeaujon et al., 2001). Further characterization demonstrated that the tonoplast-localized TT12 was able to transport cyanidin 3-O-glucoside (C3G) when heterologously expressed in yeast (Marinova et al., 2007b). When expressed in barrel medic (Medicago truncatula) hairy roots (HRs), Arabidopsis TT12 as well as a MATE transporter of barrel medic, MATE1, transport the proanthocyanidin (PA) precursor epichatechin 3-O-glucoside (Zhao and Dixon, 2009). A second barrel medic MATE transporter, MATE2, was shown to have consistently different substrate specificity despite its high similarity to MATE1; it efficiently catalyzes the vacuolar uptake of malonylated anthocyanins, while transporting glycosylated anthocyanidins at a lower rate (Zhao et al., 2011). More recently, two grapevine (Vitis vinifera) MATEs, AM1 and AM3, have been described to specifically transport acylated anthocyanins, but not glucosylated ones, which are the predominant forms that accumulate in berries of most red grapevine cultivars (Gomez et al., 2009).
Several lines of evidence also suggested the involvement of plant ABC transporters, in particular from the ABCC subfamily (formerly named multidrug resistance proteins [MRPs]) in vacuolar flavonoid sequestration (Klein et al., 2006). Complete genome sequences of several organisms revealed that genes encoding ABCC-type proteins are abundant in plants. Among the ABC transporters, unequivocal vacuolar localization has only been shown for the ABCC-type proteins. However, functionally this subfamily of ABC transporters is only marginally characterized, and ABCC transporters might be responsible for many so far unknown functions. For a long time, ABCC-type proteins were considered as the classical GSH-S conjugate pumps (Ishikawa et al., 1997). However, in the last years it became evident that their function ranges from heavy metal sequestration and detoxification to chlorophyll catabolite and inositol hexakisphosphate transport and ion channel regulation (reviewed in Kang et al., 2011). Furthermore, it was shown that rye (Secale cereale) flavones that are conjugated to glucuronate are transported into the vacuole by an ABC transporter (Klein et al., 1998). Later studies revealed that At-ABCC2 is such a glucuronide conjugate transporter (Liu et al., 2001). Moreover, genetic studies in maize (Zea mays) indicated that maize anthocyanins are delivered to the vacuole by an ABCC-type transporter, MRP3 (Goodman et al., 2004); however, this study did not provide biochemical evidence that MRP3 transports anthocyanins.
Scientific evidence also designates glutathione S-transferases (GSTs) as crucial players in the vacuolar sequestration of anthocyanins. The bronze coloration of the maize bronze-2 (bz2) mutant was shown to be correlated with the cytosolic accumulation of oxidized and cross-linked anthocyanins. A rational basis for the bz2 phenotype was obtained through the identification of Bz2 as a GST-encoding gene that might be capable of conjugating C3G with GSH (Marrs et al., 1995). Mutations in the GST-encoding genes in other plant species, such as petunia (Petunia hybrida; Alfenito et al., 1998) and Arabidopsis (Kitamura et al., 2004), also caused reduced anthocyanin accumulation and pigment mislocalization. More recently, Conn et al. (2008) showed that two grapevine GSTs are able to complement the maize bz2 phenotype. The mechanisms underlying the involvement of GSTs in anthocyanin transport are still unknown. The initial explanation for their involvement in anthocyanin transport included that GSTs might conjugate anthocyanins with GSH, thus generating the molecules that will be recognized by tonoplast-localized transporters and are imported into the vacuole. However, there is no evidence for the presence of GSH-conjugated anthocyanins in planta (Mueller et al., 2000). Alternatively, it was suggested that GSTs might act as escort proteins/ligandins of anthocyanins that deliver these compounds to the transporter (Mueller et al., 2000). Using a grapevine HR system that produces anthocyanins but is silenced for a GST known to be required for efficient anthocyanin accumulation, Gomez et al. (2011) observed that vesicles containing anthocyanins were still visible, but anthocyanin accumulation in the large vacuole was impaired. This may be interpreted in two ways: Either vacuolar anthocyanin transport depends on a GST, or GST absence forces the accumulation of anthocyanins in vesicles due to an impaired fusion of anthocyanin-containing vesicles with the large, central vacuole. Recently, it was shown that TT19, an Arabidopsis GST, directly binds C3G without conjugating it with GSH, thus reinforcing the role of GSTs as carrier proteins for vacuolar anthocyanin sequestration (Sun et al., 2012).
The recent findings on the vacuolar sequestration of anthocyanins suggest that chemical modifications of anthocyanins determine the preferential transport system. Due to the relevance of anthocyanins in grape ripening, and also because vacuolar sequestration of glucosylated anthocyanidins has only been partially characterized, we focused on elucidating whether and how ABCC-type transporters are involved in this process. Searching for Zm-MRP3 homologs in the grapevine genome database, we identified ABCC1 as having the highest protein sequence identity. Here, we report that the tonoplast-localized ABCC1 transports anthocyanidin 3-O-glucosides. The transport is preferential for malvidin 3-O-glucoside (M3G), and it is strictly GSH dependent, which assigns an important role for GSH in the current model of anthocyanin transport into the vacuole.
RESULTS
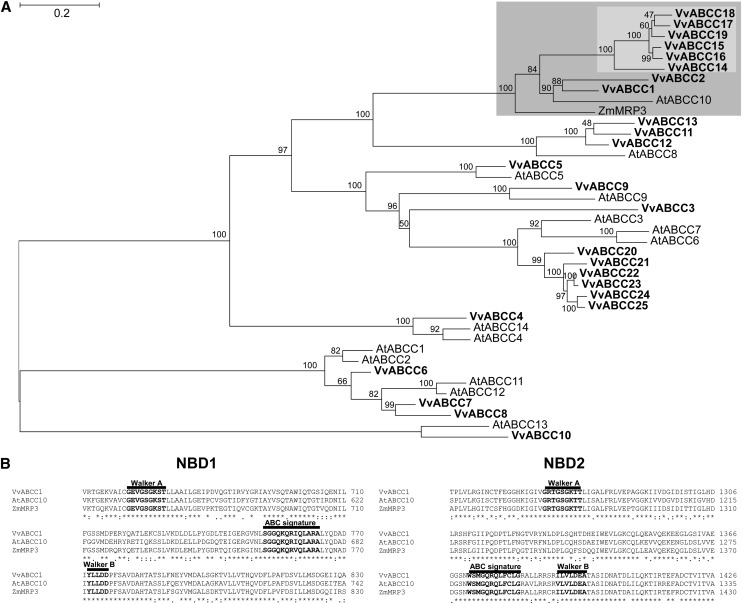
Phylogenetic Analysis of Grapevine ABCC-Type Proteins
Grapevine ABCC protein sequences from the available grapevine protein databases were chosen as candidates to carry out phylogenetic studies. The study included all Arabidopsis ABCC proteins and MRP3, the maize ABCC described as being involved in the transport of anthocyanins into the vacuole (Goodman et al., 2004). The maximum likelihood phylogenetic analysis revealed that eight grapevine ABCC proteins group within the clade containing the maize MRP3 (highlighted in dark gray, Figure 1A; see Supplemental Data Sets 1 and 2 online). Among these, Vv-ABCC1 and Vv-ABCC2 cluster with At-ABCC10, and the other six form a distinct subclade (highlighted in light gray, Figure 1A). While Vv-ABCC1 and Vv-ABCC2 are located on chromosome 16, the other six proteins all cluster on chromosome 2 in a tandem arrangement (see Supplemental Table 1 online). Vv-ABCC1/Vv-ABCC2 share a higher amino acid sequence identity with Zm-MRP3 than with any of the other six ABCCs (63%/59%), which suggests that these two proteins are candidates for anthocyanin transporters in grapevine.
Figure 1.
Phylogeny of Grapevine ABCC Proteins.
(A) Maximum likelihood phylogeny of grapevine (Vv), Arabidopsis (At) ABCC-type proteins, and Zm-MRP3/Zm-ABCC3. The clade comprising Vv-ABCC1, Vv-ABCC2, and Zm-MRP3 is shaded in dark gray, and within this clade, the subclade comprising the six other Vv-ABCCs is shaded in light gray. Branch support values indicate nonparametric bootstrap values (in percentages of 1000 replicates). The tree was estimated with PhyML 3.0 from a trimmed protein sequence alignment.
(B) Predicted ABC-type motives in Vv-ABCC1, At-ABCC10, and Zm-MRP3. The Walker A and B motifs as well the ABC signature are shown in bold letters. The three-character code is used to highlight conserved amino acids: *, full identity; :, one of the following strong groups is fully conserved (STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW); ., one of the following weaker groups is fully conserved (CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY). NBD, nucleotide binding domain. Bar = 0.2 amino acid substitutions per site.
ABCC1 Expression in Grape Berries
The phylogenetic analysis of grapevine ABCCs revealed that eight ABCCs form a distinct clade with Zm-MRP3. In this clade, ABCC1 and ABCC2 share the highest protein sequence identity with Zm-MRP3 (Figure 1A). Given the presumed role of Zm-MRP3 in anthocyanin transport (Goodman et al. 2004), these two ABCCs were chosen to verify our assumption that grapevine ABCC proteins are involved in anthocyanin transport. ABCC1 encodes a protein of 1480 amino acids with a predicted molecular mass of ∼165 kD and a calculated pI value of 6.38 (Compute pI/MW tool at http://www.expasy.ch/tools/pi_tool.html). ABCC1 exhibits the typical sequence characteristic of ABCC-type proteins, with a TMD0, two nucleotide binding domains each containing the Walker A, B, and ABC signature motifs, and two transmembrane domains (Figure 1B).
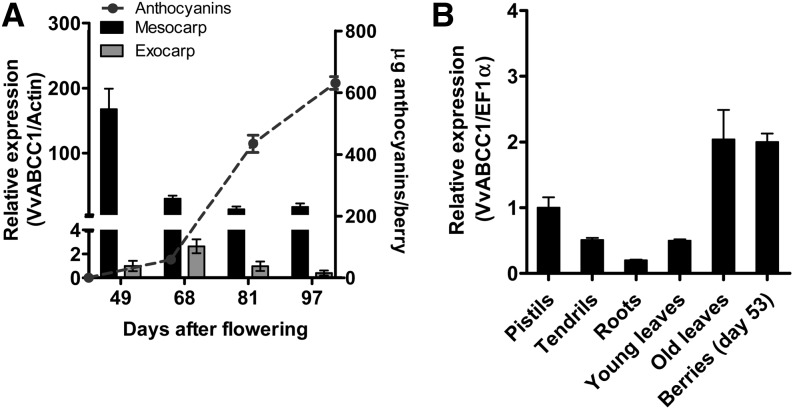
To determine when and in which berry tissues ABCC1 is expressed, quantitative RT-PCR analysis was conducted on RNA extracted from exocarp (skin) and mesocarp (pulp) at four dates, from day 49 to day 97 after flowering (DAF). ABCC1 was found to be expressed in the exocarp and mesocarp throughout berry development. However, while transcript levels decreased continuously from day 49 to 81 in the mesocarp, a strong increase could be observed in the exocarp at 68 d, during the véraison (i.e., when sugars and anthocyanins started to accumulate) (Figure 2A). The expression of ABCC1 was evaluated in several tissues and was expressed in all organs tested (Figure 2B). Higher expression was observed in old leaves and berries (Figure 2B). All attempts to amplify ABCC2 cDNA from the exocarp were unsuccessful, suggesting that it might not be expressed in detectable amounts during the ripening stage. We were also interested in determining whether members of the subclade 1 of the eight ABCCs within the Zm-MRP3–containing clade are expressed in berries. To this end, we conducted standard PCR analyses on exocarp tissues from different developmental stages. The analysis showed that some ABCC members are expressed in the exocarp during berry ripening (see Supplemental Figure 1 online).
Figure 2.
Quantitative Real-Time PCR Expression Profile of ABCC1.
(A) Expression profile of ABCC1 in grape berry tissues during fruit maturation. Transcript levels of ABCC1 were normalized to the grapevine Actin gene. Results represent the mean ± se of three replicates. Total anthocyanin content in grape berries during fruit maturation represented by the mean ± se of six biological replicates is shown.
(B) Transcript levels of ABCC1 in various grapevine organs. Young leaves correspond to leaves with a fresh weight of 0.3 g, and old leaves are fully expanded and had a mean fresh weight of 2.8 g. Gene expression was normalized to the grapevine EF1α reference gene. Results represent the mean ± se of three replicates.
ABCC1 Localizes to the Tonoplast
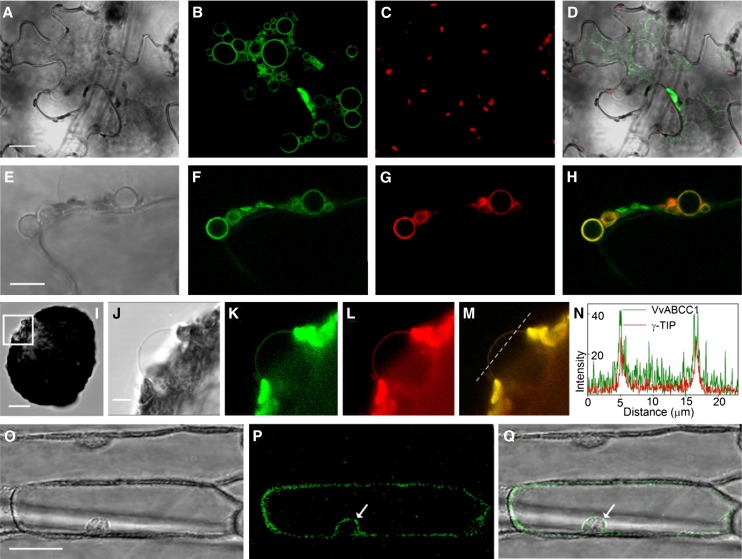
To determine the subcellular localization of ABCC1, full-length ABCC1 cDNA was C-terminally fused to green fluorescent protein (GFP) and expressed under the control of the cauliflower mosaic virus 35S promoter. A γ-TIP-RFP (for red fluorescent protein) was used as a control for tonoplast localization (Nelson et al., 2007).
Agrobacterium tumefaciens carrying the P35S:VvABC1-GFP construct was infiltrated into tobacco (Nicotiana benthamiana) leaves (Figures 3A to 3H). Four days after infiltration, we observed the formation of small vacuoles (Figures 3A and 3E), with the GFP signal encircling these structures (Figures 3B and 3F). To confirm that these small structures indeed corresponded to vacuoles, we performed coinfiltration with the tonoplast marker γ-TIP-RFP (Figure 3G). Figure 3H shows colocalization of both recombinant proteins around the edge of the small vacuoles. To further elucidate VvABCC1-GFP localization, protoplasts of transformed N. benthamiana leaves were subjected to gentle lysis (Figure 3I). A bright-field close-up of the released small vacuole in Figure 3I is shown in Figure 3J. VvABCC1-GFP and γ-TIP-RFP fluorescence is observed around the released small vacuole (Figures 3K and 3L). Both signals colocalize (Figure 3M) at the tonoplast. Colocalization was further confirmed by fluorescence intensity over distance analysis (Figure 3N) along the dotted line (Figure 3M).
Figure 3.
ABCC1-GFP Localizes to the Tonoplast.
(A) Bright-field image of a tobacco epidermis cell transiently expressing ABCC1-GFP. Bar = 20 µm.
(B) GFP signal encircling small vacuoles, which only occur in tobacco epidermis cells overexpressing VvABCC1-GFP.
(C) Chlorophyll autofluorescence corresponding to the image in (E).
(D) Bright-field, chlorophyll, and GFP fluorescence overlay image.
(E) Bright-field image of the VvABCC1-GFP overexpression-induced small vacuoles in a tobacco epidermis cell. Bar = 10 µm.
(F) VvABCC1-GFP signal encircling small vacuoles.
(G) Signal of the tonoplast marker γ-TIP-RFP encircling small vacuoles.
(H) Overlay image of VvABCC1-GFP and γ-TIP-RFP fluorescence showing colocalization of both dyes in small vacuole membranes.
(I) Bright-field image of a transiently transformed tobacco protoplast releasing a small vacuole (box). Bar = 20 µm.
(J) Close-up bright-field image of the small vacuole in (I). Bar = 5 µm.
(K) VvABCC1-GFP signal at the vacuolar membrane.
(L) Tonoplast marker γ-TIP-RFP at the vacuolar membrane.
(M) Overlay image of VvABCC1-GFP and γ-TIP-RFP fluorescence at the membrane encircling the released small vacuole.
(N) Fluorescence intensity over distance plot of VvABCC1-GFP and γ-TIP-RFP fluorescence along the dotted line in M.
(O) Bright-field image of onion epidermis cells transiently expressing VvABCC1-GFP. Bar = 50 µm.
(P) VvABCC1-GFP signal.
(Q) Bright-field and GFP fluorescence overlay image showing a signal around the nucleus indicative of tonoplast localization (arrows in [P] and [Q]).
Vacuolar fragmentation is a phenomenon described to occur in guard cells during different stages of stomata opening and closing (Gao et al., 2005), as a response to osmotic stress (Chang et al., 1996), or during senescence events (Kasaras et al., 2012). In this study, the small vacuoles may reflect a physiological adaptation to any VvABCC1-GFP overexpression-induced changes in the cellular and/or vacuolar equilibrium. Therefore, we extended our VvABCC1 localization analysis by performing transient expression in onion (Allium cepa) bulb epidermis cells by biolistic particle delivery. Figure 3O shows the bright field image of onion epidermis cell with the corresponding VvABCC1-GFP signal (Figure 3P). The fluorescence of the VvABCC1-GFP fusion protein showed a signal cytoplasmically fitting against, but not surrounding, the nucleus (arrows in Figures 3P and 3Q), which is indicative of tonoplast localization. Taken together, our results show that VvABCC1-GFP localizes to the vacuolar membrane in N. benthamiana plants, a finding that is further supported by VvABCC1-GFP tonoplast localization in onion epidermis cells.
ABCC1 Mediates M3G Transport
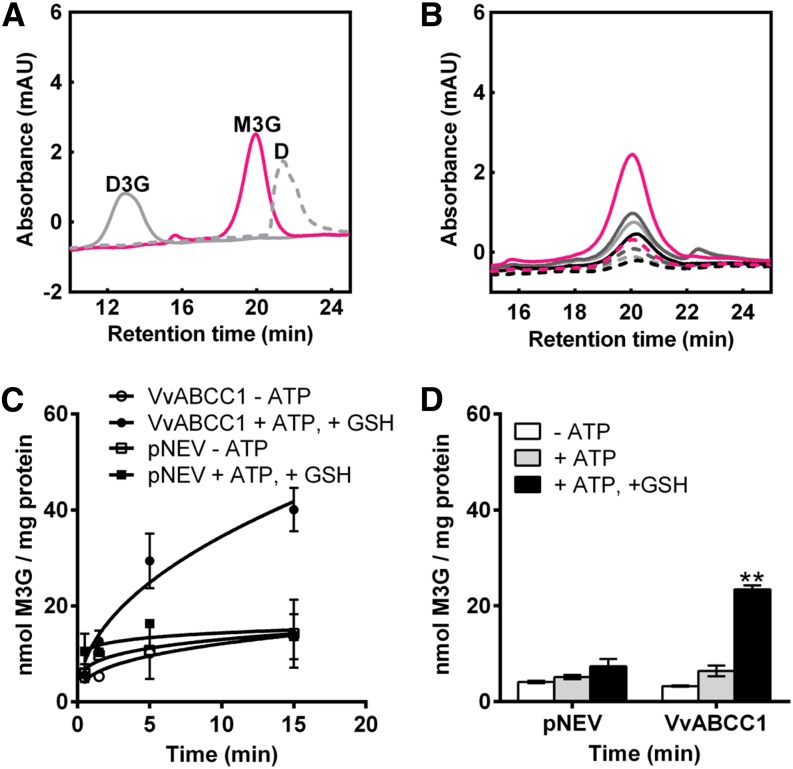
It is known that ABCC transporters can transport a large number of chemically unrelated compounds (Kang et al., 2011). The observation that ABCC1 is expressed in the exocarp of ripening berries and its close relation to Zm-MRP3 encouraged us to evaluate whether ABCC1 is an anthocyanin transporter. For this purpose, we cloned the full-length cDNA encoding ABCC1 (see Supplemental Table 1 online) into the pNEV-Ura (Sauer and Stolz, 1994) plasmid and expressed it in Saccharomyces cerevisiae defective in the ABCC transporter Yeast Bile Transporter1 (YBT1) (ybt1 strain). Yeast cells transformed with the empty vector were used as control (pNEV). Total microsomal membrane vesicles were isolated from both yeast transformants. To assess the physiological intactness of the isolated vesicles, we decided to use the vesicles lumen acidification method. Acidification occurs only when vesicles are intact and is mainly the result of the activity of the plasma membrane and tonoplast H+-ATPases. Acidification leads to the accumulation of the weak base 9-amino-6-chloro-2-methoxyacridin (ACMA) and a concomitant quenching of ACMA fluorescence (Casadio, 1991). The addition of (NH4)2SO4 restored the basal fluorescence, indicating that the proton gradient formed across the membranes collapsed, further proof that the vesicles were intact. To elucidate whether and in what relative proportion vacuolar or plasma membrane vesicles were present in the mixture, and whether both were sealed, we performed the ACMA-quenching experiment in the presence of either vanadate (inhibitor of the plasma membrane H+-ATPases), bafilomycin A1 (inhibitor of vacuolar H+-ATPases), or both. The results showed that both vanadate and bafilomycin reduced the ATP-dependent quenching to a similar extent, indicating that both plasma and vacuolar membrane vesicles were present and intact (see Supplemental Figure 2 online).
We conducted the initial transport studies with M3G, the major anthocyanin of grape berries (see Supplemental Table 2 online). The transport experiments were performed at room temperature, with a standard substrate concentration of 0.5 mM, using the rapid filtration technique as reported by Tommasini et al. (1996). HPLC was used to quantify the anthocyanins taken up into the vesicles and to confirm that there is no alteration in the structure of the substrates during the transport experiments (Figure 4A). Significant M3G uptake into yeast microsomes could not be observed in the absence or presence of ATP. Knowing that ABCCs mainly transport organic anions and that for some animal ABC transporters GSH was shown to be cotransported with their substrates (König et al., 2003), we included the negatively charged GSH into our transport assays at a 5 mM final concentration. Under these conditions, M3G was taken up in a time-dependent manner by the yeast vesicles expressing ABCC1 and uptake of M3G was ATP and GSH dependent (Figures 4B to 4D). To validate whether the transport was strictly ABC transporter driven, we included known ABC-type transporter inhibitors in the transport assays. In the presence of the sulphonylurea glibenclamide or probenecid, M3G transport was strongly reduced to 1 and 4%, respectively, of the transport activity observed without inhibitors (Table 1). Vanadate completely abolished M3G transport, and the nonhydrolyzable ATP analog, adenyl-5′-yl imidodiphosphate (AMPPNP), could not replace ATP, demonstrating that ATP hydrolysis is a prerequisite for the transport (Table 1). These results further confirm that ABCC1 mediates M3G transport in an ABC-type fashion. M3G transport was not detectable when experiments were performed at 0°C. These observations are further proof that the transport data presented in Table 1 are not due to nonspecific permeability of yeast microsomes or to the binding of M3G to yeast vesicles.
Figure 4.
ABCC1 Transports M3G in Yeast Microsomal Vesicles.
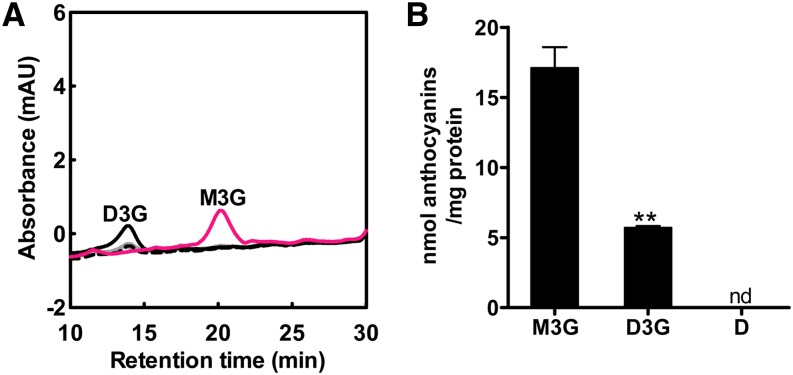
(A) HPLC profiles of M3G (pink line), D3G (gray solid line), and delphinidin (D; gray dotted line) standards; mAU, mili absorbance units.
(B) HPLC analysis of M3G taken up into microsomal vesicles isolated from yeast cells expressing Vv-ABCC1. Light-gray dotted line, −ATP, 5 s of incubation; light-gray solid line, −ATP, 15 min; dark-gray dotted line, +MgATP, 5 s; dark-gray solid line, +MgATP, 15 min; black dotted line, −ATP, +GSH, 5 s; black solid line, −ATP, +GSH, 15 min; pink dotted line, +ATP, +GSH, 5 s; pink solid line, +ATP, +GSH, 15 min.
(C) Time-dependent uptake of M3G into vesicles isolated from yeast cells transformed either with the empty vector (pNEV) or with Vv-ABCC1. Results are presented as mean values ± se of two independent uptake experiments.
(D) Net M3G uptake by ABCC1 and its vector control. Net transport was calculated by subtracting the 5-s values from the values measured after 15 min. Results are presented as mean values ± se of three independent uptake experiments; asterisks indicate statistical significance (Student’s t test; P < 0.01).
Table 1. Uptake of M3G by ABCC1 in the Presence of the Nonhydrolyzable ATP Analog AMPPNP, Glibenclamide, Probenecid, and Vanadate on Ice (0°C).
| Conditions | M3G Uptake (% Control) |
|---|---|
| ATP | 100 |
| AMPPNP (4 mM) | nd |
| Glibenclamide (150 µM) | 1.3 ± 2.74** |
| Probenecid (1 mM) | 4 ± 4.89** |
| Vanadate (1 mM) | nd |
| 0°C | nd |
With the exception of the 0°C condition, transport experiments were performed at room temperature. Results represent the means ± se of three independent uptake experiments from three independent microsome preparations. nd, not detectable. Asterisks indicate significance differences between conditions (Student’s t test; P < 0.01).
Substrate Specificity of ABCC1
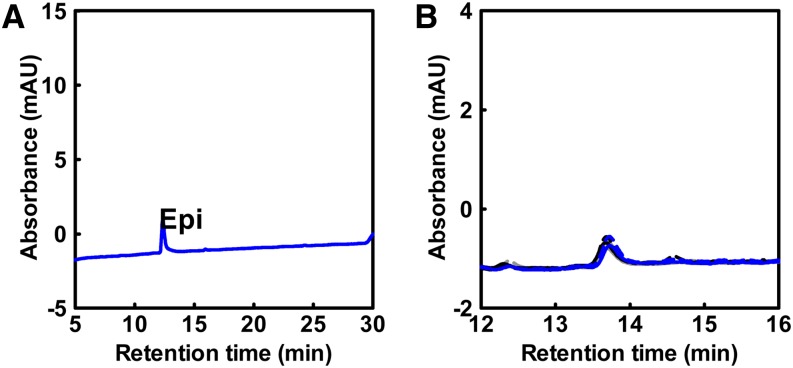
To verify the substrate specificity of ABCC1, additional transport experiments were performed with delphinidin-3-O-glucoside (D3G) and the aglycon delphinidin. D3G is one of the five major anthocyanins present in grape berries and is structurally the most distinct compared with M3G. Transport experiments were conducted as described for M3G. No uptake was observed when delphinidin was added as a substrate (Figure 5A). By contrast, ABCC1 transported the 3-O-glucoside of delphinidin, although at a significantly lower rate compared with M3G (Figure 5B). As for M3G, D3G uptake was dependent on both ATP and GSH (Figure 5A). To investigate whether ABCC1 could transport non-anthocyanin-type flavonoids, we performed a set of transport experiments with the flavan-3-ol epicatechin (Figure 6A). However, no transport was observed with epicatechin as substrate (Figure 6B).
Figure 5.
ABCC1 Transports Specifically Glucosylated Anthocyanins.
(A) HPLC profile of D3G taken up into microsomes isolated from Vv-ABCC1–transformed yeast cells. Light-gray dotted line, −ATP, 5 s; light-gray solid line: −ATP, 15 min; black dotted line, +ATP, +GSH 5 s; black solid line, +ATP, + GSH, 15 min; pink solid line, M3G uptake in the presence of both ATP and GSH, 15 min. No absorbance peak corresponding to delphinidin was observed. Transport experiments were performed at room temperature. The data represent the means ± se of two independent uptake experiments performed with microsomes from two independent preparations; mAU, mili absorbance units.
(B) Microsomes isolated from yeast cells transformed with Vv-ABCC1 transport preferentially M3G and to a lesser extent D3G, but not delphinidin (D). The data represent the means ± se of two independent uptake experiments performed with microsomes from two independent preparations. Asterisks indicate statistical significance (Student’s t test; P < 0.01); nd, not detectable.
Figure 6.
ABCC1 Does Not Transport Flavan-3-ols (Epicatechin).
(A) HPLC profile of an epicatechin (Epi) standard; mAU, mili absorbance units.
(B) HPLC analysis of epicatechin transport by microsomal vesicles isolated from yeast cells expressing ABCC1. Gray dotted line, −ATP, 30 s; gray solid line, −ATP, 15 min; black dotted line, +ATP, 30 s; black solid line, +ATP, 15 min; blue dotted line, +ATP, +GSH, 30 s; blue solid line, +ATP, +GSH, 15 min.
Uptake of Radiolabeled GSH in the Presence of M3G
Transport of M3G and D3G was only observed in the presence of GSH (Figures 4 and 5), but the mechanism behind the requirement for GSH was still unclear. Although it was suggested that GST would conjugate GSH with anthocyanins, the HPLC analyses following our transport assays did not reveal changes in the retention time of the substrates or additional peaks that could point to GSH-conjugated or modified M3G or D3G (Figures 4 and 5). These results led us to hypothesize a similar mechanism as postulated in animals (i.e., cotransport of a GSH substrate) (König et al., 2003). To test this hypothesis, cotransport experiments with M3G and radiolabeled GSH were performed. All transport experiments were performed in the presence of 5 mM NH4Cl to dissipate the proton gradient, thus inhibiting the endogenous yeast GSH transporters, which were described to depend on a proton gradient (Bourbouloux et al., 2000). No ATP-dependent GSH uptake into microsomes was detected when M3G was absent from the transport mixtures (Table 2). However, with 0.5 mM M3G in the transport reaction, we observed a time and ATP-dependent uptake of GSH in Vv-ABCC1–carrying yeast microsomes. By contrast, microsomes isolated from yeast cells carrying the empty vector did not take up GSH (Table 2).
Table 2. ABCC1 Cotransports [3H]GSH and M3G.
Microsomes isolated from yeast cells expressing ABCC1 transport GSH only in the presence of M3G. Empty vector–transformed yeast cells were used as a negative control. Transport assays were performed at room temperature for 15 min. To exclude the endogenous proton gradient-driven GSH transport, NH4Cl was added to the transport mix. The data represent the mean of three independent uptake experiments ± se; nd, not detectable. Asterisk indicates statistical significance (Student’s t test; P < 0.05).
Decrease in GSH Levels Alters the Anthocyanin Concentration in HRs
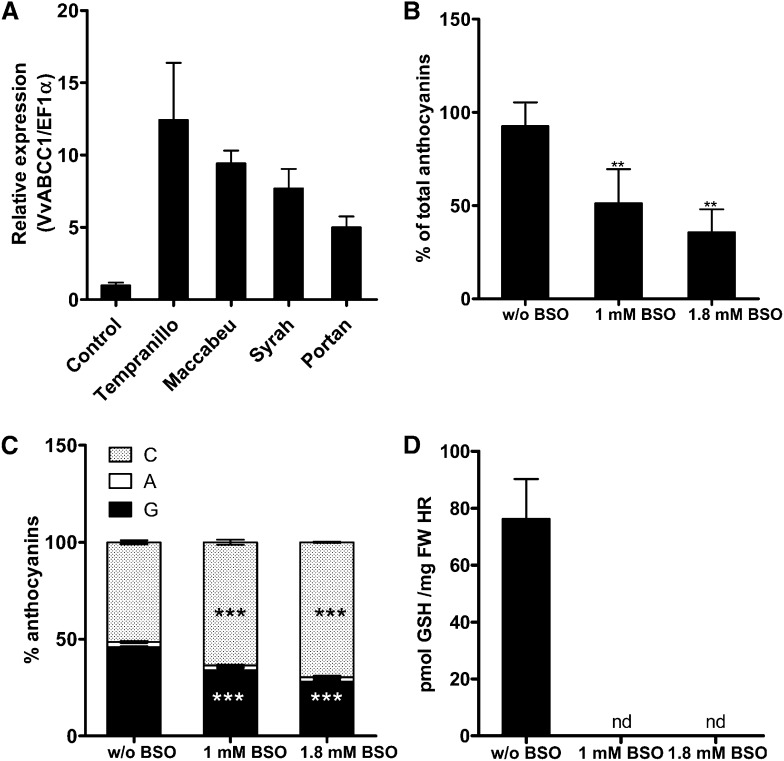
To further investigate and validate the mechanism indicating that GSH is required for anthocyanin transport, we used a transgenic anthocyanin-producing HR system (Cutanda-Perez et al., 2009). In this system, grapevine HRs transformed with the transcription factor Vv-MYBA1 exhibit a constitutive synthesis and accumulation of anthocyanins. In addition, Vv-ABCC1 is highly expressed in transformed HRs from different grapevine cultivars compared with the HRs obtained with a wild-type Agrobacterium rhizogenes (Figure 7A). Transgenic MYBA1-HRs from Tempranillo cultivar grown on LGO medium (Torregrosa and Bouquet, 1997) were transferred to fresh medium supplemented with different concentrations of buthionine sulfoximine (BSO), a compound that inhibits glutamyl-Cys synthetase, the enzyme responsible for the first step in GSH synthesis. The growth rate of HRs was similar in control and BSO-containing media; however, 2 weeks after exposure to BSO, we observed a significant decrease in anthocyanin concentration in HRs grown on BSO medium (Figure 7B). An HPLC profile of the extracted anthocyanins revealed that there was a preferential decrease in the amount of glucosylated anthocyanidins compared with the acylated fraction (Figure 7C). When comparing Tempranillo to other cultivars (i.e., Portan and Syrah), we observed that BSO mostly affected the anthocyanin concentration and profile in HRs of Portan (see Supplemental Figure 3 online). As expected, BSO affected the GSH concentration in HRs; that is, GSH concentration was significantly lower in HRs growing in BSO-containing medium compared with the control. The trend in the reduction of GSH concentrations (Figure 7D) was similar to that observed for anthocyanins (Figure 7B).
Figure 7.
Effect of BSO on Anthocyanin Concentration and Profile in Tempranillo HRs.
(A) Quantitative real-time PCR expression profile of ABCC1 in HRs. Expression was tested in red HR of different cultivars. Control experiment: expression level in HR obtained with wild-type A. rhizogenes.
(B) Anthocyanin-producing Tempranillo HRs were grown on increasing concentrations of BSO, a compound that inhibits glutamyl-Cys synthetase, the enzyme responsible for the first step of GSH biosynthesis. Root samples were harvested 14 d after BSO treatment. Anthocyanin concentration was determined by HPLC. Increasing BSO concentration led to a decrease of the total anthocyanin concentration. The concentration of anthocyanins in roots grown on media without BSO was set as 100% (corresponding to 0.92 mg anthocyanins/g fresh weight HR). The data represent the means ± se of three independent experiments. Asterisks indicate statistical significance (Student’s t test; P < 0.01).
(C) Analysis of the anthocyanin profile in the root samples described in (B) shows that BSO affects mostly the glucosylated fraction. The data represent the means ± se of three independent experiments. Asterisks indicate statistical significance (Student’s t test; P < 0.001). C, coumaroylated; A, acylated; G, glucosylated anthocyanins.
(D) GSH concentration in red HRs subjected to various BSO concentrations. FW, fresh weight; nd, not detectable.
DISCUSSION
Grapevine is one of the most important fruit crops worldwide. Apart from their use for wine production, grape berries can also be consumed as fresh or dried fruit, serve for juice production, and, more recently, used in nutraceuticals and cosmetics industries. The grape berry is composed of a pericarp tissue and up to four seeds; the pericarp is divided into exocarp, mesocarp, and endocarp (Kanellis and Roubelakis-Angelakis, 1993). The exocarp cells have an active metabolism, potentially regulating the other pericarp tissues (Coombe and Hale, 1973). Most of the color, aroma, and flavor compounds accumulate in the exocarp. Mesocarp cells accumulate primarily high amounts of organic acids and sugars in the vacuole. The endocarp is the tissue that surrounds the seeds and is hardly distinguishable from the mesocarp (Kanellis and Roubelakis-Angelakis, 1993).
The grapevine genome has recently been sequenced. This, together with the multitude of flavonoids accumulating in the various tissues, assigns grapevine as a suitable model plant for studying flavonoid biosynthesis and transport (Boss and Davies, 2009; Zhao et al., 2010). Qualitative differences in berry characteristics produce unique organoleptic traits most relevant to the wine making industry. Still, much remains to be learned about the mechanisms underlying the profound changes that occur during fruit development and ripening, regardless of the significant progress made over the past few years (Conde et al., 2007; Grimplet et al., 2009).
Grape anthocyanin biosynthesis is developmentally triggered at the onset of fruit ripening and lasts until harvest (Boss et al., 1996). The basic aglycone forms of anthocyanins (i.e., cyanidin, delphinidin, peonidin, petunidin, and malvidin) exist in red grapes in hundreds of forms, through glucosylation or acylation at different sites of the anthocyanidins. The 3-O-monoglucosides and corresponding acylated forms are the most common derivatives within grape (see Supplemental Table 2 online), while the 3,5-O-diglucosides are marker compounds of nongrape and red hybrid species (He et al., 2010). In recent years, the steps of the biosynthetic pathway, regulatory mechanisms, as well as intracellular transport and accumulation of anthocyanins started to be unveiled (reviewed in Boss and Davies, 2009; He et al., 2010). Furthermore, due to their importance for the quality of wine grapes, the effects of environmental factors and viticulture practices on anthocyanin accumulation are also increasingly investigated (Castellarin et al., 2007a, 2007b; Chaves et al., 2010). Thorough analysis revealed that anthocyanin biosynthesis is regulated by a transcriptional complex that is well conserved in different plant species. Among these transcriptional regulators, the MYB family, which has been largely investigated in grape, constitutes the most abundant group. MYBA1 and MYBA2 are able to induce gene expression of UDP Glc flavonoid 3-O-glucosyltransferase that catalyzes one of the last steps in the anthocyanin biosynthetic pathway (Kobayashi et al., 2002; Ageorges et al., 2006; Walker et al., 2007). Moreover, the white phenotype in grapes has been related to the presence of a transposable element, Gret1, in the promoter of the MYBA1 locus (Kobayashi et al., 2004), showing the relevance of MYBA1 as a regulator of anthocyanins biosynthesis.
Direct biochemical evidence proving the involvement of transporters in the delivery of anthocyanins into the vacuole was only recently published. Two tonoplast-localized grapevine MATEs, AM1 and AM3, were described to specifically transport acylated anthocyanins but not glucosylated ones (Gomez et al., 2009). The ectopic expression of MYBA1 in a grapevine HR system caused the induction of several genes specifically related to the last steps of anthocyanin metabolism, including a MATE encoding gene and, to a lesser extent, a gene encoding an ABCC transporter homologous to At-ABCC10 (Cutanda-Perez et al., 2009). These results and the genetic studies on vacuolar sequestration of anthocyanins in maize (Goodman et al., 2004) strongly suggested that ABCC proteins are involved in the transport of grape berry anthocyanins and raised the question whether ABC proteins act directly as anthocyanin transporters.
Our phylogenetic analysis revealed that eight ABCCs cluster with Zm-MRP3 and that two of them, which form a subclade, exhibit the highest protein sequence identity with Zm-MRP3 (Figure 1A). These findings were the rationale for choosing ABCC1 and ABCC2 as candidates to verify our assumption that ABCC proteins are directly involved in anthocyanin transport.
Due to unsuccessful attempts to amplify ABCC2, our attention focused on the characterization of ABCC1. While AM1 and AM3 had an expression pattern that correlated with anthocyanin accumulation in grape berries (Gomez et al., 2009), ABCC1, which is localized in the vacuolar membrane as the two MATE-type transporters, was shown to be expressed in exocarp throughout berry development and ripening (i.e., even before the onset of véraison). However, a significant increase in expression was observed at véraison, when anthocyanin synthesis starts (Figure 2). A strict correlation between ABCC1 expression and anthocyanin accumulation during the entire time of berry ripening was not observed. It should be mentioned, though, that transporters of the ABCC class are known to exhibit broad substrate specificity and, in contrast with ABCGs, are in most cases not upregulated by their substrates (Song et al., 2010). Therefore, the observation that ABCC1 expression is not strictly correlated with anthocyanin accumulation is not surprising (Figure 2). Although grape berries accumulate several chemical derivatives of five anthocyanidins, M3G is the predominant anthocyanin in the exocarp of red grape berries (see Supplemental Table 2 online). Therefore, our initial studies focused on M3G. Our findings demonstrate that ABCC1 has the ability to transport anthocyanidin 3-O-glucosides in vitro in an ATP- and GSH-dependent manner, suggesting that ABCC1 is part of the mechanism underlying vacuolar anthocyanin accumulation. Reduction of transport activity in the presence of inhibitors or a nonhydrolyzable ATP analog confirms that ABCC1 fulfils the pharmacologically described characteristics for ABC transporters. Our results also show that ABCC1 preferentially transports M3G over D3G (Figure 5). It is tempting to hypothesize that proteins with the closest homology to ABCC1 could be involved in the transport of other forms of anthocyanins present in grape berry. The expression the closest ABCC1/2 homologs in the berry exocarp (Figure 1) favors this hypothesis (see Supplemental Figure 1 online).
In grape berry exocarp, the expression of genes encoding enzymes involved in flavonoid biosynthesis was detected up to 4 weeks after flowering, followed by a decrease and finally a peak of expression again close to véraison (Boss et al., 1996). It was proposed that this expression pattern may be related to the overlapping activity of some of these enzymes in other branched pathways of flavonoids biosynthesis, namely, PAs, which are colorless polymers of flavan-3-ols (Boss et al., 1996). In grape berries, the synthesis of flavan-3-ol monomers, such as epicatechin, a common building block of PA, is completed by véraison. Although ABCC1 was expressed in exocarp tissues even before the onset of véraison, ABCC1 did not show epicatechin transport. This indicated that this transporter is not involved in PA precursor transport, but further work is needed to elucidate whether ABCC1 may transport other flavonoids that accumulate in grape berries mostly during pre-véraison.
In this study, ABCC1-mediated transport of M3G and D3G was strictly GSH dependent. Glutathionated compounds are high-affinity substrates for ABCCs from different species (Ishikawa et al., 1997; Rea et al., 1998). The close functional resemblance of plant ABCCs to mammalian GSH-conjugated Mg+-ATPases (termed GS-X pumps) was recognized through the uptake of two model GS conjugates and the glutathionated herbicide metolachlor-GS by isolated vacuoles or membrane vesicles (Martinoia et al., 1993; Li et al., 1995). Anthocyanins are positively charged molecules and therefore are not typical substrates of ABCC transporters, which often catalyze the transfer of organic anions. Thus, GS-conjugated anthocyanins were suggested to be ideal substrates for these vacuolar transporters (Marrs et al., 1995); nevertheless, HPLC analysis of the products we recovered after transport assays (Figures 4 and 5) suggests that there is no structural alteration of the anthocyanins during transport. In addition, formation of M3G- or D3G-GSs has not been reported, and it is also unlikely to occur due to chemical constraints. Therefore, GS conjugation of anthocyanins is not a prerequisite for the ABCC-driven transport. Animal ABCCs were reported to transport positively charged compounds in the presence of free GSH (König et al., 2003). The cotransport of labeled GSH and M3G (Table 2) suggests that a similar transport mechanism exists in plants. Although GSTs have been shown to be required for anthocyanin transport (Alfenito et al., 1998), in vitro vacuolar anthocyanin transport mediated by different classes of transporters was independent of GST (Zhao and Dixon, 2009). Therefore, the putative role of GSTs is either to keep cellular anthocyanin concentrations low or to ensure an efficient delivery of these compounds to the transporter.
The rapid and high accumulation of anthocyanins during grape berry ripening raises the question whether this could have an impact on the redox state of the cell. The overall GSH concentration of a cell is around 1 mM. However, only a minor part is localized within the vacuole; therefore, the concentration in the cytosol, which makes up ∼20% of the cell volume, can exceed 5 mM. In the exocarp, anthocyanins accumulate up to 3.5 mg/g fresh weight, corresponding to an anthocyanin concentration of ∼7 mM (based on present data). Our results indicate a 1:1 stoichiometry of transport of GSH and anthocyanin (i.e., ∼7 µmoles of GSH are transported per gram of exocarp). Since anthocyanin accumulation takes place over at least 20 d, the daily GSH consumption would not exceed 0.35 µmoles GSH per gram of exocarp. This rough calculation suggests that in the exocarp, anthocyanin transport has indeed an impact on the GSH budget, since ∼35% of the GSH would be used for the delivery of anthocyanins to the vacuole.
As generally observed for ABCCs, reduced GSH alone is not recognized as a substrate by ABCC1. It may be speculated that GSH and anthocyanins are brought closely together in the cavity of ABCC1 via an unknown mechanism to allow their cotransport. In the future, structural investigations of this type of ABC transporters will be helpful to elucidate the transport mechanism. Transformation of grapevine plants is a long procedure, and it takes several years until fruits can be collected (Martínez-Zapater et al., 2009). Therefore, to assess the in vivo influence of GSH on vacuolar sequestration of anthocyanins, the HR system established by Cutanda-Perez et al. (2009) was used. We observed that ABCC1 expression was highly upregulated in these roots and that the presence of GSH is a prerequisite for anthocyanin accumulation in vivo. Moreover, impaired GSH content led to a reduction of anthocyanin levels and a modified anthocyanin profile in HRs. This suggests that a feedback mechanism for the regulation of anthocyanin biosynthesis may be activated, which preferentially affects the biosynthesis of glucosylated anthocyanins.
Taken together, the presented results allow us to conclude that ABCC-type proteins can actively transport anthocyanidin 3-O-glucosides and that GSH is essential for this ABCC-type mediated transport. This study paves the way for investigating vacuolar transport of other positively charged compounds, which are not typical substrates of ABCC transporters. Reduction and modification of the anthocyanin pattern in HRs upon BSO treatment suggest that the influence of GSH is not restricted to the transport processes but also affects the biosynthetic pathway. However, the mechanism(s) underlying its role needs further elucidation. It will also be interesting to biochemically test if homologs within the ABCC1 clade of ABCCs in grapevine (Figure 1A) are involved in the vacuolar transport of other anthocyanins, to evaluate their transport efficiency and substrate specificity, and to elucidate whether this overrepresentation of ABCC proteins in this clade is due to functional redundancy or rather to developmental/environmental adaptations.
METHODS
Chemicals and Reagents
M3G, D3G, and delphinidin were purchased from PhytoLab, epicatechin from Extrasynthèse, and radiolabeled [3H]GSH from Sigma-Aldrich. Other chemicals were purchased from Sigma-Aldrich.
Plant Material
Berry samples (Vitis vinifera cv Tempranillo syn. Aragonez) were collected from an experimental plot at the commercial vineyard Monte dos Seis Reis (South of Portugal, Estremoz, Portugal). Berries were collected during the summer season of 2007 at four representative stages of development and maturation: pre-véraison at pea size (49 DAF), véraison (68 DAF) when 50% berries were colored, maturation (81 DAF), and full maturation (97 DAF). Collected berries were immediately frozen in liquid nitrogen and stored at −80°C before use.
Plant material (roots, shoots, leaves, and berries) were also collected from grapevine plants grown in Sup Agro-Institut National de la Recherche Agronomique as previously described by Gomez et al. (2009).
Grapevine HRs producing anthocyanins were generated from Tempranillo, Portan, and Syrah plantlets and are described by Cutanda-Perez et al. (2009). Culturing of the transgenic HRs was performed as described by Torregrosa and Bouquet (1997) with the modifications reported by Cutanda-Perez et al. (2009). Stable HR lines were grown on solidified LGO medium (Cutanda-Perez et al., 2009; LGO as described in Torregrosa and Bouquet, 1997) and subcultured every 4 to 5 weeks until use. Prior to phenotyping anthocyanin concentration and profile, 4- to 5-week-old HRs were exposed to LGO medium containing various concentrations of BSO for 14 d.
Phylogenetic Analysis of ABCC Proteins
The sequence of ABCC1 was experimentally determined during this work. The other grapevine ABCC protein sequences were identified by BLASTp searches against the V. vinifera National Center for Biotechnology Information (NCBI) RefSeq database (http://blast.ncbi.nlm.nih.gov) and the GENOSCOPE V. vinifera 12X March 2010 release database (retrieved via Phytozome v7.0; http://www.phytozome.net/grape; Goodstein et al., 2012) on the June 2, 2012. BLASTP searches were performed with standard parameters using the Arabidopsis thaliana ABCC13, ABCC5, and ABCC3 sequences as BLAST seeds (see below). NCBI models/sequences were generally more complete compared with corresponding GENOSCOPE models and were therefore used, when available, to obtain the protein and cDNA sequences used in this work, except Vv-ABCC18/22/25, which were manually annotated. Manual annotations were based on alignments of corresponding NCBI/Genocope models with other Vv-ABCCs and Arabidopsis ABCCs (see below) and on GENESCAN predictions on corresponding genomic sequences (http://genes.mit.edu/GENSCAN.html). The GENOSCOPE model GSVIVG01016879001 comprises two distinct adjacent full-size ABCC genes, which are annotated as distinct genes in the NCBI database. GSVIVG01016879001-A represents the first and GSVIVG01016879001-B the second full-size ABCC gene of this model. The GENOSCOPE models GSVIVT01021593001 and GSVIVT01021594001 encode partial ABCC protein sequences, which are annotated in the NCBI database as being distinct parts of the same ABCC gene. Vv-ABCC21/22/25/26 were only found in the GENOSCOPE database. The model for Vv-ABCC26 has a length of only 287 amino acids. On the chromosomal location of Vv-ABCC26, additional partial gene models corresponding to distinct regions of an ABCC gene were identified; however, it was not possible to predict any new model comprising these partial models. Assembly of this chromosomal location may be erroneous, or this gene is degenerated and might not encode a full-size ABCC gene. Vv-ABCC26 was not included in the phylogenetic analysis. All sequences used for subsequent analysis are presented in Supplemental Data Set 1 online. Sequences of Arabidopsis ABCC proteins were obtained from The Arabidopsis Information Resource (version TAIR10; http://www.Arabidopsis.org) using the Arabidopsis ABCC inventory (Verrier et al., 2008). The maize (Zea mays) MRP3 sequence (gene ID: GRMZM2G361256_P01) was obtained from the Maize Genome Sequencing Project (release 5b.60; http://www.maizesequence.org).
Protein sequences were aligned using MAFFT with the E-INS-i algorithm (Katoh and Toh, 2008) as implemented in the GUIDANCE Web server (Penn et al., 2010), which additionally estimates alignment confidence scores. Alignment columns with a GUIDANCE score below 0.93 were removed, and additional columns with >20% gaps were deleted using TrimAl (Capella-Gutiérrez et al., 2009). The obtained alignment (see Supplemental Data Set 1 online) was used to estimate the unrooted maximum likelihood phylogenetic tree with PhyML 3.0 (Guindon et al., 2010) using the LG+G4+I+F model, which was selected by a ProtTest 3 analysis (Darriba et al., 2011). Branch support values represent nonparametric bootstrap values in percentages of 1000 bootstrap samples. The phylogenetic tree was visualized with MEGA5 (Tamura et al., 2011) and graphically adapted in Adobe Illustrator.
Gene Expression Analysis
Total RNA was extracted from frozen grapevine tissues (roots, shoots, and leaves) and from berry tissues (exocarp and mesocarp) according to Reid et al. (2006). Total RNA was purified using an RNeasy kit (Qiagen) with the addition of an on-column DNase I digestion step. Total RNA (0.5 µg) was reverse transcribed using M-MLV reverse transcriptase (Promega) and priming with oligo(dT)18 according to the manufacturer’s recommendations. Transcript levels were determined by quantitative RT-PCR using the 7500 Fast Real-Time PCR system (Applied Biosystems) with 7500 Software version 2.0.4. Reactions were performed in a final reaction mixture of 20 μL of cDNA (diluted 1:10), 0.25 μM gene-specific primers, and SYBR Green PCR Master Mix (Applied Biosystems). Reaction conditions for the thermal cycling were as follows: after enzyme activation at 95°C for 10 min, amplification was performed in a two-step PCR procedure with 40 cycles of 15 s at 95°C for denaturation and 1 min at 60°C for annealing/extension. All reactions were performed in triplicate with three biological replicates. Gene primer sequences used in the quantitative RT-PCR analyses were as follows: ABCC1 forward, 5′-AATTCAAAGATTGGAAGC-3′ and ABCC1 reverse 5′-GCACTGATTTTGAATAGAA-3′. Dissociation curves were analyzed to verify the specificity of each amplification reaction; the dissociation curve was obtained by heating the amplicon from 60 to 95°C. Transcript levels were calculated using the standard curve method and normalized against the grapevine Actin gene or the grapevine EF1-α gene as described by Pfaffl (2001).
Gene-specific primer sequences used for the standard PCR analyses (see Supplemental Figure 1 online) were as follows: Vv-ABCC18 reverse 5′-CAATAGTAGGCCAACTGAC-3′, Vv-ABCC19 forward 3′-ATCGTATGGCCTTTCAGTG-5′, and Vv-ABCC19 reverse 3′-CCAATAGTAGGCCAACTGGG-5′. Expression levels were evaluated after 20, 25, and 30 cycles. Results after 25 cycles are depicted.
Molecular Cloning of ABCC1
RNA was extracted from berries collected at the véraison stage. Total RNA was isolated as described above. The full-length cDNA of ABCC1 was amplified using the high-fidelity PCR Extender polymerase mix (5PRIME) according to the manufacturer’s instructions. The following primers were used: ABCC1 forward (5′-ATAAGAATGCGGCCGCATGGGGGATCTGTGGACTATGTTTTGTGGG-3′) and ABCC1 reverse (5′-ATAAGAATGCGGCCGCTCAATGTGATTCCGCTGAGTGAAAATGGGACC-3′) (NotI restriction sites are underlined). The resulting PCR product was cloned into the yeast expression vector pNEV-Ura (Sauer and Stolz, 1994) using the NotI sites, thereby producing pNEV-VvABCC1.
The CaMV35S:VvABCC1-GFP construct was generated by amplifying the full length of Vv-ABCC1 cDNA without the stop codon by PCR using the primers Vv1 PacI forward, 5′-CTTAATTAAATGGGGGATCTGTGGACTATG-3′, and Vv1 AscI reverse, 5′-GGCGCGCCAATGTGATTCCGCTGAGTGAAAAT-3′ (PacI and AscI restriction sites are underlined, respectively), followed by digestion of the PCR product with PacI and AscI restriction enzymes and ligation into the PacI-AscI–digested binary vector pMDC83 (Curtis and Grossniklaus, 2003).
Subcellular Localization
To investigate the subcellular localization of ABBC1 in plant cells, Vv-ABCC1-GFP and γ-TIP-RFP constructs were transiently expressed in Nicotiana benthamiana leaves. Briefly, Agrobacterium tumefaciens (GV3101) was transformed by electroporation with the corresponding plasmids and subsequently infiltrated into N. benthamiana leaves as described previously (Voinnet et al., 2003). To reduce the silencing of the transgenes, all constructs were coinfiltrated with the silencing suppressor RK19.
Mesophyll protoplasts and vacuoles of N. benthamiana leaves were prepared as described by Kovermann et al. (2007).
Transient expression in onion (Allium cepa) bulb epidermal cells was performed by biolistic particle delivery of P35S:VvABCC1-GFP using DNA-coated 0.6-μm gold particles as microcarriers and the Bio-Rad He Biolistic PDS-1000/He system following the manufacturer's instructions.
Tissue samples were examined 2 to 6 d after transformation using a Leica TCS SP5 confocal microscope. GFP fluorescence was excited using a 488-nm laser, with GFP and chlorophyll fluorescence being collected between 500 and 600 nm and 698 and 787 nm, respectively. For colocalization studies, Vv-ABCC1-GFP and γ-TIP-RFP fluorescence was excited simultaneously at 458 and 561 nm, respectively. GFP and RFP emission was separated and collected between 468 and 558 nm and 567 and 665 nm, respectively, with crosstalk between detection channels being controlled for and excluded in each experiment. Colocalization analysis was conducted using the Leica LAS AF Lite software.
Functional Analysis in Yeast Vesicles
The ybt1 yeast mutant (MATa; ura3Δ::HIS3; leu2-3, 112; his3-Δ200; bat1Δ1::URA3) was transformed as described by Gietz and Woods (2002) with either the empty vector pNEV-Ura (pNEV) or pNEV-VvABCC1. Transformants were selected on minimal synthetic dropout medium lacking uracil. For in vitro transport studies, yeast microsomes were isolated as described by Tommasini et al. (1996), and intactness of the vesicle fractions was evaluated by quenching of the fluorescent dye ACMA (Gomez et al. 2009). Briefly, vesicle extracts (300 µg) were added to a reaction solution (1 mL) containing 2 µM ACMA, 0.4 M glycerol, 6 mM MgSO4, 1 mM DTT, 0.1 M KCl, 20 mM Tris-MES, pH 7.4, and 5 mM MgATP. The fluorescence intensities were measured by spectrofluorimetry, and the ACMA signal was acquired at 485 nm after excitation at 425 nm. Evaluation of proton gradient-dependent uptake of ACMA into yeast vesicles was performed through addition of 25 mM (NH4)2SO4 to the reaction solution.
To study the transport of M3G, D3G, and delphinidin into membrane microsomes, uptake experiments were performed using the rapid filtration technique (Tommasini et al., 1996) with nitrocellulose filters (0.45-mm pore size; Millipore). Anthocyanin transport assays were performed with 0.2 mL of vesicles mixed with ice-cold transport buffer (0.4 M glycerol, 0.1 M KCl, and 20 mM Tris-MES, pH 7.4) and freshly added 1 mM DTT, 6 mM or 1 mM MgSO4 (in the presence or absence of MgATP, respectively), 100 µg/mL creatine kinase, and 10 mM creatine-phosphate. M3G, D3G, or delphinidin (0.5 mM) was assayed either in the presence or absence of 4 mM MgATP and 5 mM GSH in a total reaction volume of 0.8 mL. The transport assay was performed at room temperature. At the time points indicated in the figures, 0.35 mL of the reaction mixture was immediately loaded on a prewetted filter and rapidly washed with 3 × 2 mL of ice-cold transport buffer. The filter-bound anthocyanins were dissolved by adding 50% (v/v) methanol and 0.1% (v/v) HCl at 37°C and extracting for 10 min. The eluted anthocyanins were then quantified by HPLC. Briefly, HPLC separation was performed on a CC 250/4 Hypersil 100-5 C18HD column (Macherey-Nagel) with 100% acetonitrile (A) and 0.1% phosphoric acid (B) at a total flow rate of 1 mL/min in the following gradient: 0 to 25 min from 90 to 75% B; 25 to 35 min 75 to 30% B; 35 to 40 min 30 to 0% B, and 40 to 50 min from 0 to 90% B. Parameters were controlled by a Gynkotek liquid chromatography system (Dionex) equipped with a UVD340S diode array detector set at 520, 310, and 280 nm for anthocyanin identification. The transport experiments with epicatechin (0.5 mM) were performed as described for M3G. Inhibition assays were performed in the presence of the following potential inhibitors: 4 mM AMPPNP, 1 mM probenecid, 150 µM glibenclamide, and 1 mM vanadate. For the inhibition experiments, yeast microsomes were incubated at room temperature in the reaction mix containing the indicated potential inhibitors. After 10 min, anthocyanins were added and the uptake experiment performed as described above. For experiments on ice, the tubes containing the reaction mix and M3G were placed on ice immediately after the addition of yeast microsomes. Experiments were repeated three times with independent vesicle preparations unless stated otherwise.
For experiments with labeled GSH, 5 mM NH4Cl was added to the transport mixes with the purpose of dissipating the proton gradient, thus repressing endogenous yeast GSH transport. A 0.1-µCi/nitrocellulose filter of [3H]GSH was used. The experiments were performed at room temperature in the presence and absence of M3G. Labeled GSH was quantified in a scintillation counter.
Intactness of yeast microsomes vesicles was assessed by measuring their capacity to generate a pH gradient. Briefly, vesicle isolated from pNEV and pNEV-VvABCC1 (300 µg) were added to a reaction solution (1 mL) containing 2 µM ACMA, 0.4 M glycerol, 6 mM MgSO4, 1 mM DTT, 0.1 M KCl, 20 mM Tris-MES, pH 7.4, and 5 mM MgATP. A final concentration of 1 mM vanadate and 5 µM bafilomycin was used for the inhibition of ATPases. The fluorescence intensities were measured by spectrofluorimetry at 15-s intervals, and the ACMA signal was acquired at 485 nm after excitation at 425 nm. Evaluation of proton gradient–dependent uptake of ACMA into yeast vesicles was performed through the addition of 25 mM (NH4)2SO4 to the reaction solution.
Quantitative Analysis of Anthocyanins
Anthocyanin extraction from grape berries was performed twice using acidified methanol. Briefly, 600 μL of acidified methanol (1% [w/v] HCl) was added to 100 mg of grounded tissue and centrifuged at 4°C for 15 min at 16,100g. Supernatants were collected and anthocyanins quantified as described by Zarrouk et al. (2012). Anthocyanin contents are expressed as milligrams of malvidin equivalent per berry fresh weight (Zarrouk et al., 2012). For anthocyanin determination from HRs, 200 mg of ground, frozen tissues was used. Determination of anthocyanin composition was performed by HPLC as described by Ageorges et al. (2006).
Determination of GSH Concentration in HRs
GSH was analyzed by liquid chromatography–tandem mass spectrometry (Agilent 6460) and quantified by stable isotope dilution assay using labeled GSH as internal standard. In practice, the analysis consisted of a derivatization step using a maleimide derivative and then a direct injection. The detection was performed in multiple reaction monitoring mode as previously reported (Shuford et al., 2012).
Accession Numbers
Sequence data for Vv-ABCC1 have been deposited at GenBank/EMBL data libraries under accession number JX245004. Other accession numbers of genes mentioned in this article are as follows: GRMZM2G361256_P01, GU585869, BQ799343, At3g13080, At1g04120, and At2g07680.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Analyses of the Closest Homologous of Vv ABCC1/2.
Supplemental Figure 2. Assessment of Physiological Intactness of Yeast Microsomes.
Supplemental Figure 3. The BSO Effect on Anthocyanin Concentration and Profile in Hairy Roots from Two Other Cultivars: Portan and Syrah.
Supplemental Table 1. Vitis vinifera ABCC Protein Inventory.
Supplemental Table 2. Structure and Distribution of the Five Major Anthocyanins of Grape Berries.
Supplemental Data Set 1. FASTA File of Amino Acid Sequences of Vitis vinifera ABCCs Used for the Construction of the Phylogenetic Tree.
Supplemental Data Set 2. Alignment Used to Generate the Phylogenetic Tree Shown in Figure 1A.
Supplementary Material
Acknowledgments
We acknowledge the financial support from Fundação para a Ciência e a Tecnologia of Ministério da Ciência, Tecnologia e do Ensino Superior (Portugal) through Grants SFRH/BD/30344/2006 (R.M.F.), SFRH/BPD/74210/2010 (R.M.F.), SFRH/BPD/34986/2007 (A.R.), and SFRH/BPD/34488/2006 (O.Z.) and from the Swiss National Foundation (3100AO-11-7780/1; E.M.). We thank Aurèlie Roland-Vialaret from Nysèos Montpellier for her analytical help in GSH analysis. We also thank Stefan Hoertensteiner for valuable comments and suggestions on the article and Steffi Pfrunder for helping with the article preparation. E.M. thanks Charles and Annemarie Steiner Schernelz for initially providing plant material and all the discussions around the wine.
AUTHOR CONTRIBUTIONS
A.R., M.M.C., and E.M. raised the hypothesis underlying this work. R.M.F., R.N., and E.M. designed experiments. R.M.F., R.N., B.J.B., B.B., C.E., A.A., S.V., T.M., and O.Z. performed the experiments and analyzed the data. R.M.F., E.M., A.R., and R.N. wrote the article. E.M. and R.N. directed the study.
Glossary
- C3G
cyanidin 3-O-glucoside
- PA
proanthocyanidin
- GST
glutathione S-transferase
- HR
hairy root
- M3G
malvidin 3-O-glucoside
- DAF
days after flowering
- GFP
green fluorescent protein
- ACMA
9-amino-6-chloro-2-methoxyacridin
- D3G
delphinidin-3-O-glucoside
- LGO
a growing medium
- BSO
buthionine sulfoximine
- NCBI
National Center for Biotechnology Information
- AMPPNP
adenyl-5′-yl imidodiphosphate
References
- Ageorges A., Fernandez L., Vialet S., Merdinoglu D., Terrier N., Romieu C. (2006). Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 170: 372–383 [Google Scholar]
- Alfenito M.R., Souer E., Goodman C.D., Buell R., Mol J.N.M., Koes R., Walbot V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss, P.K., and Davies, C. (2009). Molecular biology of anthocyanin accumulation in grape berries. In Grapevine Molecular Physiology and Biotechnology, K.A. Roubelakis-Angelakis, ed (Dordrecht, The Netherlands: Springer), pp. 263–292. [Google Scholar]
- Boss P.K., Davies C., Robinson S.P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbouloux A., Shahi P., Chakladar A., Delrot S., Bachhawat A.K. (2000). Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275: 13259–13265 [DOI] [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H.P., Matros A., Peterek S., Schijlen E.G.W.M., Hall R.D., Bovy A.G., Luo J., Martin C. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio, R. (1991). Measurements of transmembrane pH differences of low extents in bacterial chromatophores. Eur. Biophys. J. 19: 189–201.
- Castellarin S.D., Matthews M.A., Di Gaspero G., Gambetta G.A. (2007a). Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 227: 101–112 [DOI] [PubMed] [Google Scholar]
- Castellarin S.D., Pfeiffer A., Sivilotti P., Degan M., Peterlunger E., DI Gaspero G. (2007b). Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 30: 1381–1399 [DOI] [PubMed] [Google Scholar]
- Chang P.-F.L., et al. (1996). Alterations in cell membrane structure and expression of a membrane-associated protein after adaptation to osmotic stress. Physiol. Plant. 98: 505–516 [Google Scholar]
- Chaves M.M., Zarrouk O., Francisco R., Costa J.M., Santos T., Regalado A.P., Rodrigues M.L., Lopes C.M. (2010). Grapevine under deficit irrigation: Hints from physiological and molecular data. Ann. Bot. (Lond.) 105: 661–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C., Silva P., Fontes N., Dias A.C.P., Tavares R.M., Sousa M.J., Agasse A., Delrot S., Gerós H. (2007). Biochemical changes throughout grape berry development and fruit and wine quality. Food 1: 1–22 [Google Scholar]
- Conn S., Curtin C., Bézier A., Franco C., Zhang W. (2008). Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 59: 3621–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe B.G., Hale C.R. (1973). The hormone content of ripening grape berries and the effects of growth substance treatments. Plant Physiol. 51: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutanda-Perez M.C., Ageorges A., Gomez C., Vialet S., Terrier N., Romieu C., Torregrosa L. (2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69: 633–648 [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. (2011). ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Peeters A.J., Léon-Kloosterziel K.M., Koornneef M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.Q., Li C.G., Wei P.C., Zhang X.Y., Chen J., Wang X.-C. (2005). The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol. 139: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Gomez C., Conejero G., Torregrosa L., Cheynier V., Terrier N., Ageorges A. (2011). In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 67: 960–970 [DOI] [PubMed] [Google Scholar]
- Gomez C., Terrier N., Torregrosa L., Vialet S., Fournier-Level A., Verriès C., Souquet J.M., Mazauric J.P., Klein M., Cheynier V., Ageorges A. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150: 402–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C.D., Casati P., Walbot V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16: 1812–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40 (Database issue): D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J., Cramer G.R., Dickerson J.A., Mathiason K., Van Hemert J., Fennell A.Y. (2009). VitisNet: “Omics” integration through grapevine molecular networks. PLoS ONE 4: e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Grotewold E., Davies K. (2008). Trafficking and sequestration of anthocyanins. Nat. Prod. Comm. 3: 1251–1258 [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321 [DOI] [PubMed] [Google Scholar]
- He F., Mu L., Yan G.L., Liang N.N., Pan Q.H., Wang J., Reeves M.J., Duan C.Q. (2010). Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 15: 9057–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani N.G., Grotewold E. (2005). Light-induced morphological alteration in anthocyanin-accumulating vacuoles of maize cells. BMC Plant Biol. 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Li Z.S., Lu Y.P., Rea P.A. (1997). The GS-X pump in plant, yeast, and animal cells: structure, function, and gene expression. Biosci. Rep. 17: 189–207 [DOI] [PubMed] [Google Scholar]
- Kanellis, A.K., and Roubelakis-Angelakis, K.A. (1993). Grape. In Biochemistry of Fruit Ripening, G. Seymour, J. Taylor, and G. Tucker, eds (London: Chapman and Hall), pp. 189–234. [Google Scholar]
- Kang J., Park J., Choi H., Burla B., Kretzschmar T., Lee Y., Martinoia E. (2011). Plant ABC transporters. The Arabidopsis Book 9: e0153, doi/10.1199/tab.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaras A., Melzer M., Kunze R. (2012). Arabidopsis senescence-associated protein DMP1 is involved in membrane remodeling of the ER and tonoplast. BMC Plant Biol. 12: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kitamura S., Shikazono N., Tanaka A. (2004). TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Klein M., Burla B., Martinoia E. (2006). The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 580: 1112–1122 [DOI] [PubMed] [Google Scholar]
- Klein M., Martinoia E., Weissenböck G. (1998). Directly energized uptake of beta-estradiol 17-(beta-D-glucuronide) in plant vacuoles is strongly stimulated by glutathione conjugates. J. Biol. Chem. 273: 262–270 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Goto-Yamamoto N., Hirochika H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Ishimaru M., Hiraoka K., Honda C. (2002). Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215: 924–933 [DOI] [PubMed] [Google Scholar]
- König, J., Nies, A.T., Cui, Y., and Keppler, D. (2003). MRP2, the apical export pump for anionic conjugates. In ABC Proteins: From Bacteria to Man, I.B. Holland, K. Kuchler, C. Higgins, and S.P.C. Cole, eds (London: Academic Press), pp. 423–443. [Google Scholar]
- Kovermann P., Meyer S., Hörtensteiner S., Picco C., Scholz-Starke J., Ravera S., Lee Y., Martinoia E. (2007). The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 52: 1169–1180 [DOI] [PubMed] [Google Scholar]
- Li Z.-S., Zhao Y., Rea P.A. (1995). Magnesium adenosine 5′-trisphosphate-energized transport of glutathione S-conjugates by plant vacuolar membrane vesicles. Plant Physiol. 107: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Sánchez-Fernández R., Li Z.S., Rea P.A. (2001). Enhanced multispecificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J. Biol. Chem. 276: 8648–8656 [DOI] [PubMed] [Google Scholar]
- Marinova K., Kleinschmidt K., Weissenböck G., Klein M. (2007a). Flavonoid biosynthesis in barley primary leaves requires the presence of the vacuole and controls the activity of vacuolar flavonoid transport. Plant Physiol. 144: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova K., Pourcel L., Weder B., Schwarz M., Barron D., Routaboul J.M., Debeaujon I., Klein M. (2007b). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs K.A., Alfenito M.R., Lloyd A.M., Walbot V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Martínez-Zapater J.M., Carmona M.J., Díaz-Riquelme J., Fernández L., Lijavetzky D. (2009). Grapevine genetics after the genome sequence: Challenges and limitations. Aust. J. Grape Wine Res. 16: 33–46 [Google Scholar]
- Martinoia E., Grill E., Tommasini R., Kreuz K., Amrhein N. (1993). ATP-dependent glutathione S-conjugate 'export' pump in the vacuolar membrane of plants. Nature 364: 247–249 [Google Scholar]
- Mueller L.A., Goodman C.D., Silady R.A., Walbot V. (2000). AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 123: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Penn O., Privman E., Ashkenazy H., Landan G., Graur D., Pupko T. (2010). GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 38 (Web Server issue): W23–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L., Irani N.G., Lu Y., Riedl K., Schwartz S., Grotewold E. (2010). The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant 3: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea P.A., Li Z.S., Lu Y.P., Drozdowicz Y.M., Martinoia E. (1998). From vacuolar GS-X pumps to multispecific ABC transporters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 727–760 [DOI] [PubMed] [Google Scholar]
- Reid K.E., Olsson N., Schlosser J., Peng F., Lund S.T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky D.E., Warek U., Winkel B.S. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280: 23735–23740 [DOI] [PubMed] [Google Scholar]
- Sauer N., Stolz J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 6: 67–77 [DOI] [PubMed] [Google Scholar]
- Shuford C.M., Poteat M.D., Buchwalter D.B., Muddiman D.C. (2012). Absolute quantification of free glutathione and cysteine in aquatic insects using isotope dilution and selected reaction monitoring. Anal. Bioanal. Chem. 402: 357–366 [DOI] [PubMed] [Google Scholar]
- Song W.-Y., et al. (2010). Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 107: 21187–21192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li H., Huang J.R. (2012). Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 5: 387–400 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R., Evers R., Vogt E., Mornet C., Zaman G.J.R., Schinkel A.H., Borst P., Martinoia E. (1996). The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor 1. Proc. Natl. Acad. Sci. USA 93: 6743–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrosa L., Bouquet A. (1997). Agrobacterium rhizogenes and A. tumefaciens co-transformation to obtain grapevine hairy roots producing the coat protein of grapevine chrome mosaic nepovirus. Plant Cell Tissue Organ Cult. 49: 53–62 [Google Scholar]
- Verrier P.J., et al. (2008). Plant ABC proteins—A unified nomenclature and updated inventory. Trends Plant Sci. 13: 151–159 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walker A.R., Lee E., Bogs J., McDavid D.A.J., Thomas M.R., Robinson S.P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wallace T.C. (2011). Anthocyanins in cardiovascular disease. Adv. Nutr. 2: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C.R., Wu Q., Simon J.E. (2008). Recent advances in anthocyanin analysis and characterization. Curr. Anal. Chem. 4: 75–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 127: 1399–1404 [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (2008). The biosynthesis of flavonoids. In The Science of Flavonoids, E. Grotewold, ed (New York: Springer Science), pp.71–95. [Google Scholar]
- Zarrouk O., Francisco R., Pintó-Marijuan M., Brossa R., Santos R.R., Pinheiro C., Costa J.M., Lopes C., Chaves M.M. (2012). Impact of irrigation regime on berry development and flavonoids composition in Aragonez (Syn.Tempranillo) grapevine. Agric. Water Manage. 114: 18–29 [Google Scholar]
- Zhang H., Wang L., Deroles S., Bennett R., Davies K. (2006). New insight into the structures and formation of anthocyanic vacuolar inclusions in flower petals. BMC Plant Biol. 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Dixon R.A. (2009). MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21: 2323–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Dixon R.A. (2010). The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 15: 72–80 [DOI] [PubMed] [Google Scholar]
- Zhao J., Huhman D., Shadle G., He X.Z., Sumner L.W., Tang Y., Dixon R.A. (2011). MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23: 1536–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Pang Y., Dixon R.A. (2010). The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 153: 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.