Abstract
The mammalian circadian clock involves a transcriptional feedback loop in which CLOCK and BMAL1 activate the Period and Cryptochrome genes, which then feedback and repress their own transcription. We have interrogated the transcriptional architecture of the circadian transcriptional regulatory loop on a genome scale in mouse liver and find a stereotyped, time-dependent pattern of transcription factor binding, RNA polymerase II (RNAPII) recruitment, RNA expression and chromatin states. We find that the circadian transcriptional cycle of the clock consists of three distinct phases — a poised state, a coordinated de novo transcriptional activation state, and a repressed state. Interestingly only 22% of mRNA cycling genes are driven by de novo transcription, suggesting that both transcriptional and post-transcriptional mechanisms underlie the mammalian circadian clock. We also find that circadian modulation of RNAPII recruitment and chromatin remodeling occurs on a genome-wide scale far greater than that seen previously by gene expression profiling.
The circadian clock in mammals is cell autonomous and is composed of an autoregulatory transcriptional network with interlocked feedback loops (1, 2). At the core, the bHLH-PAS transcriptional activators BMAL1, CLOCK and NPAS2 activate the Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes whose transcripts and proteins slowly accumulate during the daytime (3–7). The PER and CRY proteins associate and translocate into the nucleus during the evening and physically interact with the CLOCK/NPAS2:BMAL1 complex to repress their own transcription (5, 7, 8). As the PER and CRY proteins are progressively phosphorylated during the night, they are targeted for ubiquitination by specific E3 ligases and are eventually degraded by the proteasome (9–12). The waxing and waning of this transcriptional feedback loop takes ~24 hours to complete and represents the core mechanism of the circadian clock in mammals. Biochemical analysis on a small set of target genes has shown that CLOCK, BMAL1 and CRY1 bind in a diurnal manner to regulatory regions, interact with p300 and CBP, and are accompanied by rhythmic changes in histone H3 Lys4 trimethylation (H3K4me3) and H3 Lys9 acetylation (H3K9ac) (13–19) characteristic of active promoter regions (20–23).
Although the majority of the core components of the circadian gene network are likely known (1) and many thousands of transcripts have been shown to express circadian oscillations in various tissues (24), the genome-wide architecture of the transcriptional network regulated by the core circadian clock remains to be defined (25, 26). Recent work has explored the genome-wide cis-acting targets (cistromes) of two sets of circadian transcription factors, BMAL1 (27, 28) and REV-ERBα/β (29–31). In liver, BMAL1 binds to ~2,000 genomic targets in a diurnal pattern with peak DNA binding occurring during the day (28). REV-ERBα/β bind to thousands of sites in genome and have 28% overlap with BMAL1 peaks (30). Despite these initial genome-wide views of BMAL1 and REV-ERBα/β cistromes, a comprehensive analysis of both the activators and the repressors within the core circadian transcription factor network has not been reported under constant conditions.
Circadian cistrome of the liver
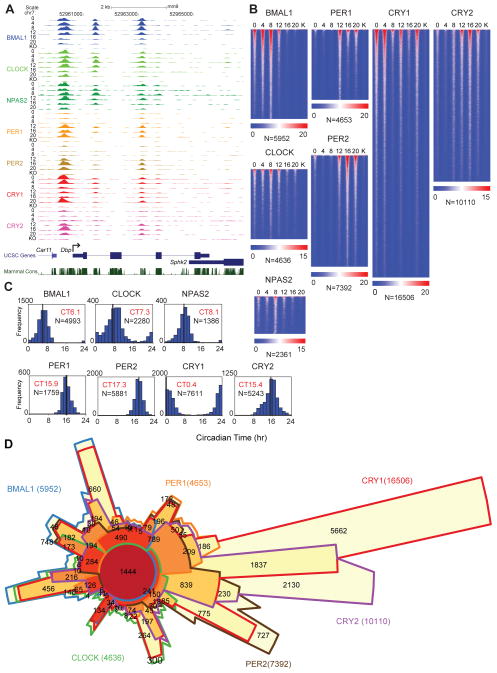
We used chromatin immunoprecipitation sequencing (ChIP-seq) to locate DNA binding sites for BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2 in vivo in murine liver. We report here the results of ~200 ChIP-seq samples encompassing more than 9 billion sequence reads (See Supporting Online Materials and Methods, fig. S1A, table S1). The cistromic landscape of the core circadian transcriptional regulators is shown at the Dbp locus as a University of California Santa Cruz (UCSC) genome browser view (32) (Fig. 1A). Similar to that reported under diurnal lighting conditions (14, 28), BMAL1 binds in a circadian manner between circadian time (CT) 0–12 to three sites in Dbp with peak binding phases occurring at CT4-8. We also confirmed that rhythmic binding of BMAL1 is sustained over two cycles in constant darkness (fig. S2). The heterodimeric partners of BMAL1, CLOCK and NPAS2, also bind in a time-dependent manner similar to BMAL1 (Fig. 1A). The repressors PER1, PER2 and CRY2 then bind the same sites between CT12 and CT20 (Fig. 1A). Interestingly, CRY1 exhibits maximal binding at CT0, which extends the repression phase into the beginning of the next cycle when BMAL1, CLOCK and NPAS2 occupancy increases again at CT0 (Fig. 1A). Heat map representations of genome-wide peak location analysis illustrate the dynamics of binding during the circadian cycle in constant darkness (Fig. 1B). Because the DNA binding of these factors is time dependent, the number of peaks detected at each time point varies. Thus we developed a method to create a master peak list for each transcription factor that compares the overlap of peaks detected by the MACS and Peaksplitter peak-finding algorithms (33, 34) among the six time samples (fig. S3, table S2). The genome-wide distributions of the peaks for each factor show that 31–36% of sites are intergenic and the remaining sites are enriched at promoter and intronic regions of genes (fig. S4). We find ~5900 BMAL1 sites, ~4600 CLOCK sites and ~2300 NPAS2 sites for the activators, and ~4600 PER1, ~7300 PER2, ~16,000 CRY1 and ~10,000 CRY2 sites for the repressors. Motif analysis shows that all 7 factors are enriched for E-box sites as well as nuclear receptor binding sites (fig. S5). Similar to that seen at the Dbp locus (Fig. 1A), the genome-wide occupancy rhythms of BMAL1, CLOCK and NPAS2 peak at CT6–8; whereas, PER1, PER2, CRY1 and CRY2 show binding rhythms that peak at CT15.9, CT17.3, CT0.4 and CT15.4, respectively (Fig. 1C). Gene ontology analysis shows that the target genes for all 7 core circadian transcriptional regulators are highly enriched for metabolic pathways, pathways in cancer and insulin signaling as seen previously (28, 30, 35, 36) (table S3, S4).
Fig. 1.
ChIP-seq analysis of the core circadian transcriptional regulators in mouse liver. (A) UCSC genome browser view of BMAL1 (blue), CLOCK (light green), NPAS2 (green), PER1 (orange), PER2 (gold), CRY1 (red) and CRY2 (purple) occupancy at the Dbp (A) locus. Each track represents the normalized ChIP-seq read coverage (wiggle plot) at a single time point. For each transcription factor, six time points every 4 hr over a circadian cycle are shown beginning at CT0 and ending at CT20. Knockout (KO) mice were used as a negative control for each factor except NPAS2. The conservation track shows 30-Way Multiz Alignment & Conservation scores (PhastCons) provided by the UCSC genome browser. Read numbers are normalized as described in the methods. The y-axis scales between tracks for Dbp are BMAL1 (0–70), CLOCK (0–40), NPAS2 (0–25), PER1 (0–35), PER2 (0–60), CRY1 (0–135), CRY2 (0–45). (B) Heat map views of genome-wide DNA binding for BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2 measured over 500 bp fragments encompassing the binding sites. Each peak in the genome is represented as a horizontal line, ordered vertically by signal strength. Six time points are shown beginning at CT0 and ending at CT20 from left to right. Knockout (K) mouse control is shown on the far right of each panel. The number of peaks in the genome is indicated at the bottom of each panel. The blue-red gradient indicates the coverage or signal strength (normalized uniquely mapped reads per 10 million reads) for all binding sites in the genome. (C) Histogram showing circadian phase distributions of BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2 binding rhythms. Cyclic binding was determined by ARSER (p<0.05) and the mean circular phases of peak binding as indicated in red are indicated using Oriana. The number of cycling peaks is indicated in black. (D) Chow-Ruskey diagram showing the 6-way overlap of BMAL1, CLOCK, PER1, PER2, CRY1 and CRY2 peaks. Master peak lists were used to determine peak overlaps and an overlap was called if two peak summits were within 120 bp of each other. The red circle in the middle represents the overlap of all six factors. Lighter shades of red, orange and yellow represent fewer overlaps of subsets. The boundaries for each protein are color coded: BMAL1 (blue), CLOCK (green), PER1 (orange), PER2 (brown), CRY1 (red) and CRY2 (purple), and the areas of each domain are proportional to the number of binding sites.
To analyze the overlap of the binding sites of the circadian regulators genome-wide, we compared the 6-way overlap of the factors BMAL1, CLOCK, PER1, PER2, CRY1 and CRY2 and show the results in a Chow-Ruskey diagram (Fig. 1D). At the core, 1444 sites in the genome are bound by all 6 factors. Binding site coverage plots of these common sites are shown in fig. S6A. Motif analysis of the common binding sites shows a clear enrichment for the canonical CLOCK:BMAL1 E-box motif (CACGTG) as well as CEBPA and a number of nuclear receptor motifs (fig. S7). Of particular interest are the large number of CRY1, CRY2 and PER2 sites that are uniquely bound by each factor and appear to be independent of BMAL1:CLOCK sites (Fig. 1D). Motif analysis of these unique sites shows a reduction in E-box sites and enrichment for nuclear hormone receptor sites (fig. S7) consistent with recent reports that PER2 and CRY1 interact with nuclear receptors (37, 38). To visualize the dynamics of activator and repressor binding on a genome-wide level, we analyzed the overlap of the activators, BMAL1, CLOCK and NPAS2, in comparison to the repressors, PER1, PER2, CRY1 and CRY2 during the circadian cycle and see a reciprocal binding pattern between activators and repressors (fig. S6B, C).
Whole transcriptome analysis of circadian gene expression
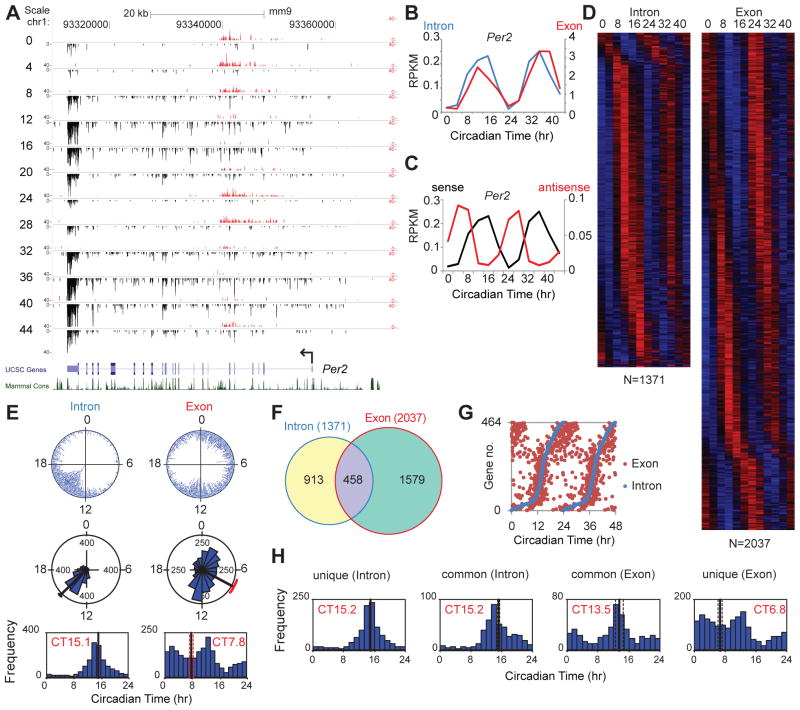
In order to assess transcriptional readouts on a genome level, we used whole transcriptome RNA-seq to measure circadian gene expression in the liver over a 48-hr interval sampled every 4 hrs in constant darkness. A high-amplitude RNA rhythm can be seen at the Per2 locus with a peak at CT12 and CT36 (Fig. 2A, B). In addition, there is a long noncoding antisense RNA transcript that oscillates in antiphase in a manner similar to that seen at the Neurospora frequency locus (39) (Fig. 2C, table S5). To quantitate RNA expression levels across the genome, we calculated the RNA-seq read coverage in reads per kilobase per million reads (RPKM) in exons and introns separately for the UCSC canonical gene set (28,661 total; 21,789 with introns). We interpret the intron signal as a representation of pre-mRNA expression or nascent transcription (40) and the exon signal as a representation of mRNA expression, which can reflect not only transcriptional activity but also post-transcriptional processing events. At the Per2 locus, the intron and exon signals both oscillate and their phases are similar (Fig. 2B) as seen previously using qPCR measurements of pre-mRNA and mRNA (12, 14) (fig. S8). In genome-wide analyses, we detected 1371 intron and 2037 exon RNA cycling transcripts (table S6, S7), which are shown as heat maps ordered by the phase of RNA expression (Fig. 2D). Gene ontology analysis of both classes of cycling RNAs shows highly significant enrichment for metabolic pathways, pathways in cancer and many other pathways (table S8, S9). A comparison of the phase distribution of the intron and exon cycling transcripts reveals a clear difference: the intron cycling transcripts are clustered and peak at CT15.1; whereas, the exon cycling transcripts have three peak phases and are distributed across all phases (Fig. 2E) as seen in previous microarray experiments (35, 41–43). Comparing the genes in the intron and exon cycling transcript classes shows that only 458 genes (22% of the exon cycling class) are in common (Fig. 2F), and this set of common genes is enriched for known circadian clock genes and high amplitude cycling target genes reported previously (28, 43). The phases of the common intron and exon cycling transcripts are correlated suggesting that transcriptional cycles primarily drive these mRNA rhythms (Fig. 2G). In the intron cycling/exon not cycling class, the cycling pre-mRNA transcripts are clustered at the same phase as the overall intron cycling class, but the steady-state mRNAs are likely to have long half-lives (>24 hrs) which would damp oscillations generated at the transcriptional level as described previously (44, 45) (Fig. 2H). By contrast, in the intron not cycling/exon cycling class, the phases are widely distributed as seen in the overall exon cycling class and these rhythms likely arise from post-transcriptional regulatory processes such as rhythms in RNA splicing, polyadenylation or mRNA stability (Fig. 2H) (46, 47). Taken together, the analysis of intron- and exon-specific cycling transcripts shows that circadian regulation of gene expression can occur at both transcriptional and post-transcriptional levels. The genome-wide phase clustering of the intron cycling transcripts is unexpected and suggests that a global circadian regulation of de novo transcription may exist.
Fig. 2.
Whole-transcriptome RNA-seq analysis of circadian gene expression. (A) UCSC genome browser views of RNA-seq data at the Per2 locus. Sense strand reads are shown in black and reverse strand reads in red as normalized average reads per 50 million total reads in 10 bp bins. The y-axis scales are: Per2 (0–40 for the positive strand, −40–0 for the negative strand). (B) RNA-seq read coverage in reads per kilobase per million (RPKM) reads in exons and introns. The intron (blue) and exon (red) RNA expression of Per2 is circadian (ARSER: p<0.01). (C) Cyclic expression of Per2 sense and antisense transcripts. The Per2 antisense transcript is circadian (ARSER; p<0.05) and antiphasic (phase of sense: CT14.4; phase of antisense: CT7.1). (D) Heat map view of intron (left) and exon (right) cycling genes. Each gene is represented as a horizontal line, ordered vertically by phase determined by ARSER. (E) The phase distribution of cycling genes. The phase of each transcript rhythm is represented in a dot plot (top), rose plot (middle) and histogram plot (bottom). The mean circular phase of the rhythms is indicated in red. (F) Venn diagram of intron and exon cycling genes. (G) Comparison of the phase of intron and exon cycling transcripts for common genes. The RNA peak phase of intron (blue) and exon (red) is double plotted and ordered by intron phase. (H) Histogram of intron phase in intron-only cycling genes, intron phase in common genes, exon phase in common genes, and exon phase in exon-only cycling genes (mean circular phase is shown in red).
Circadian expression of RNAPII and co-activator occupancy
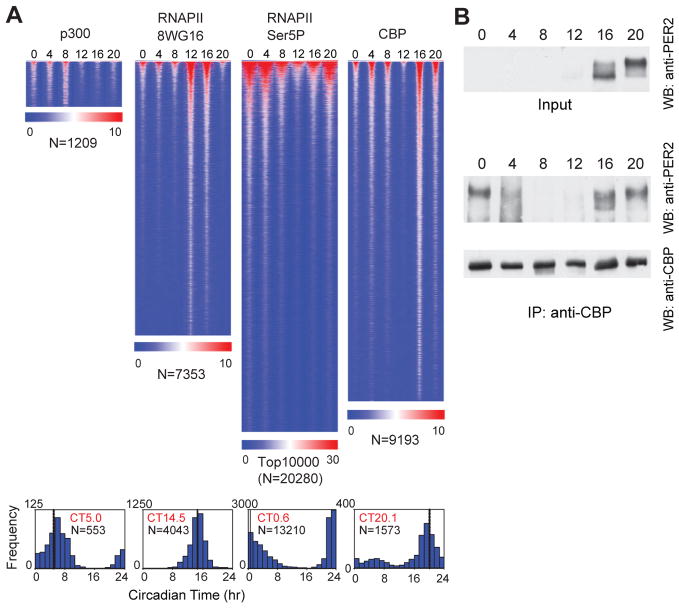
To explore the origins of the global rhythms in nascent transcription, we analyzed the genome-wide occupancy of RNA polymerase II as well as p300 and CBP co-activators as a function of the circadian cycle. The C-terminal domain (CTD) of the large subunit of RNAPII is modified at various stages of transcription (48, 49). RNAPII is recruited into the pre-initiation complex with a hypophosphorylated CTD that is recognized by the 8WG16 antibody (50). The CTD is then phosphorylated on serine 5 (Ser5P) during initiation and is recognized by the 3E8 Ser5P antibody (51). We used ChIP-seq with these two antibodies to measure RNAPII occupancy and to estimate RNAPII recruitment and initiation (Fig. 3, S1B). On a genome-wide basis, we detect over 7,300 RNAPII-8WG16 peaks and 20,000 RNAPII-Ser5P peaks, and strikingly, RNAPII occupancy is circadian with 8WG16 signal peaking at CT14.5 and Ser5P signal peaking at CT0.6 (Fig. 3A). The peak of RNAPII-8WG16 at CT14.5 coincides closely with the peak of intron cycling transcripts at CT15.1 (Fig. 2E) further supporting the idea that intron cycling transcripts represent nascent or de novo transcription events. On the other hand, the peak of RNAPII-Ser5P occupancy at CT0.6 coincides with the peak of CRY1 occupancy at CT0.4. At this time, CLOCK and BMAL1 are beginning a new cycle of binding, but are transcriptionally silent likely because CRY1 is bound at the same sites. One possible scenario is that RNAPII can be recruited by CLOCK:BMAL1 and that RNAPII initiation occurs, but then pauses or stalls, and accumulates at CT0. An example of this can be seen at the Nr1d1 locus where RNAPII-Ser5P occupancy initiates at CT0, but RNA expression and RNAPII-8WG16 occupancy begins later at CT4 (fig. S9A, a 4-hr lag between RNAPII-Ser5P and transcription can be seen at many CLOCK:BMAL1 targets). Alternatively, RNAPII-Ser5P occupancy at CT0 could be independent of CLOCK:BMAL1 and could reflect a peak of transcription at CT0. For example, RNAPII-Ser5P occupancy at CT0 overlaps with REV-ERBα/β peaks (30) in the Nfil3, Adck3 and Ppp1r3c genes (along with ~300 intron cycling genes) in which peak RNA expression occurs between CT20-CT4 (fig. S9B–D). In these cases, CLOCK:BMAL1 and CRY1 sites are present nearby, but do not appear to overlap with RNAPII-Ser5P. Interestingly, in these cases, RNAPII-8WG16 occupancy tends to occur in antiphase to RNAPII-Ser5P.
Fig. 3.
Circadian expression of RNAPII and co-activator occupancy. (A) Heat map view of genome-wide DNA binding for p300, RNAPII-8WG16, RNAPII-Ser5P and CBP as described in Fig. 1B. Histograms of the phase of the rhythms are shown below the heat map (mean circular phase is shown in red and number of cycling peaks is indicated in black). (B) PER2 and CBP protein interaction using co-immunoprecipitation of native extracts during the circadian cycle. Co-immunoprecipitation was carried out with anti-CBP antibody and mouse cerebellum protein lysate. The immunoprecipitates were visualized on western blots with anti-PER2 or anti-CBP antibodies.
Following this CRY1-repressed phase at CT0, CLOCK and BMAL1 occupancy increases at CT4-8 along with p300 occupancy (peak at CT5) (Fig. 3A), and global nascent transcription begins at CT8 and peaks at CT15. Examination of the time-dependent pattern of CBP occupancy reveals that CBP can have either a 24-hr or a 12-hr binding rhythm with the 24-hr rhythm peaking at CT16-20 and the 12-hr rhythm peaking at CT4 and CT16 (Fig. 3A, S10). Because CBP can interact at E-box sites either at CT4 or CT16, this suggests that CBP may be able to function as either a co-activator at CT4 or a co-repressor at CT16. In support of this hypothesis, we find that CBP can interact with the repressor, PER2, in native extracts in a time-dependent manner with maximal interaction occurring at CT16–20 when PER2 levels are elevated (Fig. 3B).
Circadian regulation of chromatin remodeling
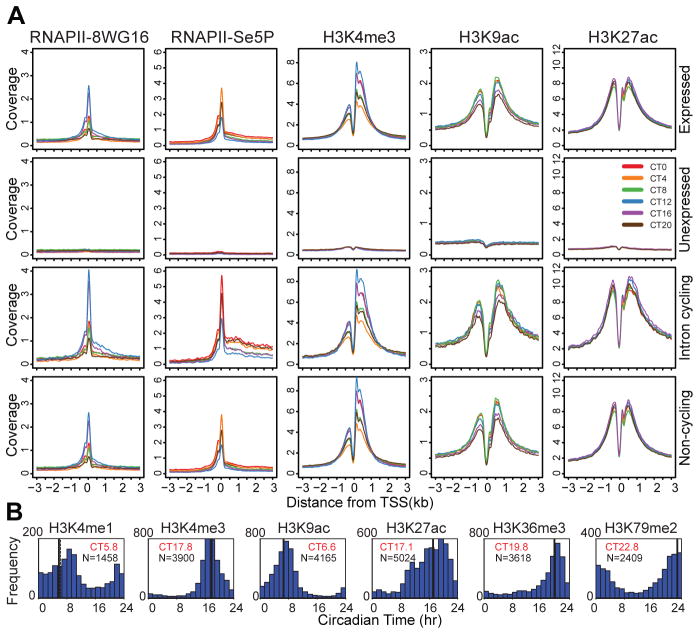
Given the genome-wide circadian rhythms of RNAPII and co-activator occupancy, we next assessed chromatin states associated with transcription initiation and elongation using ChIP-seq during the circadian cycle (20, 22, 23, 52–55). Histone H3K4me3, H3K9ac and H3K27ac are enriched at promoters and show robust circadian rhythms in occupancy at transcription start sites (TSS) (Fig. 4A, S1C). Histone H3K4me1, a mark that is characteristic of enhancer sites and gene bodies, exhibits a very subtle circadian modulation (Fig. 4B). Noticeably, there is an antiphase rhythm in H3K4me1 and H3K4me3 occupancy at the Dbp intron 1 site (fig. S1C a, b). Consistent with recent reports (54, 55), histone H3K27ac is also highly enriched at both intragenic and intergenic enhancer sites. The elongation marks, H3K36me3 and H3K79me2, also express very low amplitude circadian modulation (Fig. 4B). On a genome-wide level, clear circadian rhythms in RNAPII-8WG16, RNAPII-Ser5P, H3K4me3, H3K9ac and H3K27ac occupancy at TSS’s can be seen in all classes of expressed genes as compared to unexpressed genes (Fig. 4A, B). These rhythms in RNAPII occupancy and histone modifications are stronger in intron RNA cycling genes (Fig. 4A), and rhythmic RNAPII-Ser5P elongation past the TSS can be seen prominently in cycling genes relative to all expressed genes (Fig. 4A, S11). Curiously, non-cycling intron expressed genes also show circadian modulation of RNAPII occupancy and histone modifications similar to that see in all expressed genes (Fig. 4A bottom). In contrast to the observation reported from the analysis of human embryonic stem cells that inactive genes harbor paused RNAPII (22, 56, 57), we see no evidence of RNAPII (8WG16 and Ser5P) pausing at the promoters of unexpressed genes (Fig. 4A, S11). Genome-wide analysis of the periodicity and phase of these histone marks reveals that a large number of genes exhibit circadian rhythms in histone modifications (Fig. 4B). The overall number of histone modification sites does not appear to vary on a circadian basis; rather, the recruitment (and initiation) of RNAPII appears to underlie the variation in the amplitude of histone marks (fig. S12A). Thus, our analysis reveals that the majority of expressed genes are undergoing circadian histone modifications irrespective of whether RNA cycling can be detected.
Fig. 4.
Circadian regulation of histone modifications. (A) Binding profile of RNAPII-8WG16, RNAPII-Ser5P, H3K4me3, H3K9ac and H3K27ac at the TSS +/− 3kb in 12,680 expressed genes (top row), 8,945 unexpressed genes (second row), 1371 intron cycling genes (third row) and 5,839 non-cycling genes (bottom row). The y-axis represents the average signal in a 10 bp bin (normalized uniquely mapped reads per 10 million reads). (B) Histograms showing circadian phase distributions of histone modification rhythms (ARSER: p<0.05). Mean circular phase is shown in red. The signal in TSS +/− 1 kb was used to determine H3K4me1, H3K4me3, H3K9ac and H3K27ac binding rhythm phases. The signal in the gene body was used for H3K36me3 and H3K79me2. Genes without MACS peaks in the corresponding area (TSS or gene body) were filtered out from the analysis.
Circadian regulation at intergenic enhancers
To determine whether circadian regulation of RNAPII occupancy and histone modifications also occurs at enhancer sites, we identified intergenic enhancer sites bound by either BMAL1 or CBP and compared these sites to BMAL1 or CBP promoter sites, respectively (fig. S12B, C). Coverage profiles for BMAL1 show circadian occupancy that is similar at both promoter and enhancer sites with peaks at CT8. RNAPII-8WG16 profiles show clear circadian modulation at BMAL1- and CBP-bound promoter sites as well as enhancer sites with a peak at CT12 except that occupancy was much lower at enhancer sites. H3K4me1 showed clear enrichment at enhancer sites and depletion at promoter sites as expected and also showed a subtle circadian modulation in occupancy that peaked at CT8. In contrast, H3K4me3 was enriched at promoter sites and was lower at enhancer sites, but in this case the timing of the circadian modulation was different where promoter sites peaked at CT12 and enhancer sites peaked at CT20. H3K9ac was enriched at promoter sites but a clear circadian modulation can be seen at both promoter and enhancer sites with peaks at CT8. Finally, H3K27ac was equally enriched at both promoter and enhancer sites and showed low amplitude circadian modulation with peaks at CT16. Thus, overall we observe circadian modulation of BMAL1 occupancy, RNAPII occupancy and histone modifications at both promoter proximal sites as well as distal intergenic enhancer sites.
Discussion
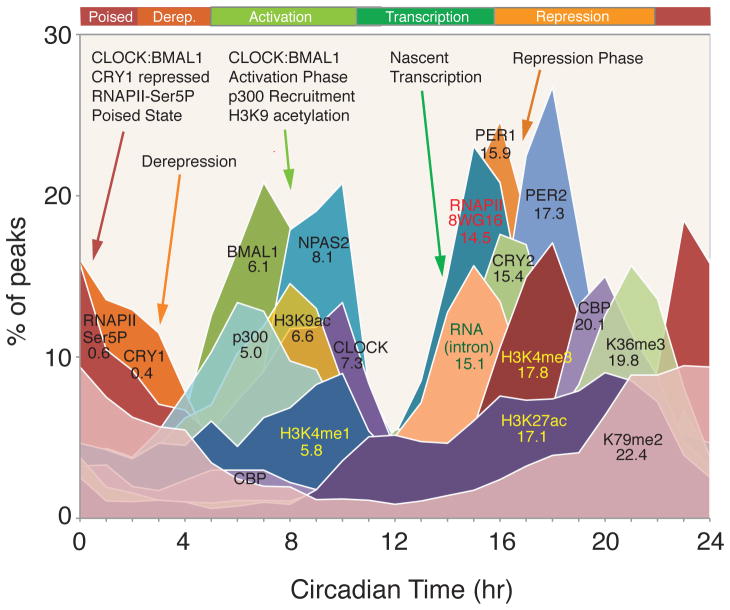
We have defined the cis-regulatory network of the entire core circadian transcriptional feedback loop under constant conditions coupled with whole transcriptome RNA expression, RNAPII recruitment and initiation as well as chromatin states on a genome-wide basis. We find three distinct phases of the circadian transcriptional cycle of the clock in the murine liver: 1) a transcriptionally poised state; 2) a global peak of de novo transcription that peaks at CT15; and 3) a repressed state in which CLOCK:BMAL1 occupancy decreases at CT16-20 (Fig. 5). The poised state of CLOCK:BMAL1 bound with CRY1 is consistent with that seen in vitro (58) and in vivo (19), and is followed by derepression (59) as the occupancy of CRY1 declines and recruitment of the co-activator, p300, occurs. The details of this cis-regulatory cycle of circadian transcriptional regulators differ from that reported in Drosophila in which PER sequesters CLOCK both on and off DNA and RNAPII appears to be paused and is regulated by elongation at the period locus and by recruitment at the timeless locus (60).
Fig. 5.
Circadian landscape of the cistrome and epigenome of the liver. Phase distributions of circadian transcriptional regulators, intron cycling RNA transcripts and histone modifications as shown in Fig. 1C, 2E, 3A and 4B. The mean circular phase of peak binding is indicated under the name.
The dynamics of nascent transcription, RNAPII occupancy and histone modifications provide important insights into the relationships among these events. While co-activator recruitment of p300 by CLOCK:BMAL1 and histone H3K9ac precede nascent transcription, there is a clear lag in histone H3K4me3 relative to RNAPII recruitment as measured by 8WG16 (Fig. 5). Histone H3K27ac also peaks well after nascent transcription as do the elongation marks H3K36me3 and H3K79me2 and thus may reflect processes involved in the consequences of elongation rather than elongation events themselves (Fig. 5). Finally, it will be of interest to explore the role of HDACs and co-repressor complexes in circadian regulation. Both SIN3A/HDAC1 (61) and HDAC3 (29) have been found in PER2 and REV-ERBα complexes, respectively, as well as RNAPII accumulation at the termination sites of Per and Cry genes (62).
Although global circadian rhythms in steady state mRNA levels have been known for a decade (35, 41, 42), here we find that only ~22% of cycling mRNA transcripts are driven by transcription (Fig. 2). Therefore, post-transcriptional regulatory events must contribute significantly to the generation of steady state cycling mRNA levels (46, 47). Surprisingly the most pervasive circadian regulation observed on a genome scale are rhythms in RNAPII recruitment and initiation, H3K4me3, H3K9ac and H3K27ac, which occur at thousands of expressed genes whether or not RNA cycling was detectable. What accounts for these genome-wide circadian rhythms in RNAPII occupancy and histone modifications? Examination of the relationship between circadian transcription factor occupancy and gene expression shows that approximately 90% of genes bound by these factors are expressed whereas only 1–5% of unexpressed genes are similarly bound (Table S10). These results demonstrate that gene expression per se rather than rhythmicity of gene expression is tightly correlated with circadian transcription factor binding. Rhythmic circadian transcription factor occupancy in turn could then be responsible for RNAPII recruitment and initiation on a genome-wide basis, which would then lead to the global rhythmic histone modifications seen here. Thus, circadian transcriptional regulators appear to be involved in the initial stages of RNAPII recruitment and initiation and the histone modifications associated with these events to set the stage for gene expression on a global scale, but additional control steps must then determine the ultimate transcriptional outputs from these sites.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (J.S.T.), Whitehall Foundation grant (T.-K.K), NIH grant NS053616 (C.L.). We thank Helen Hobbs and David Mangelsdorf for helpful discussions and support; Billy Crider, Vanessa Schmid, Aaron Guillory, Sara Hildebrand, John Knepper, Julia Denton, Sijing Niu and the McDermott Next Generation Sequencing Facility for outstanding technical and computational support; and Steven L. McKnight for NPAS2 antibody. J.S.T. is an Investigator, S.H.Y. and V.K. were Associates in the Howard Hughes Medical Institute. J.S.T. is a co-founder of, a Scientific Advisory Board member of, and a paid consultant for Reset Therapeutics, Inc., a biotechnology company aimed at discovering small-molecule therapies that modulate circadian activity for a variety of disease indications.
Footnotes
ChIP-seq and RNA-seq data sets have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE39860 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39860).
References and Notes
- 1.Lowrey PL, Takahashi JS. Adv Genet. 2011;74:175. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass J, Takahashi JS. Science. 2010;330:1349. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King DP, et al. Cell. 1997;89:641. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gekakis N, et al. Science. 1998;280:1564. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 5.Kume K, et al. Cell. 1999;98:193. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 6.Bunger MK, et al. Cell. 2000;103:1009. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Cell. 2001;107:855. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 8.Griffin EA, Jr, Staknis D, Weitz CJ. Science. 1999;286:768. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 9.Shirogane T, Jin J, Ang XL, Harper JW. J Biol Chem. 2005;280:26863. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 10.Reischl S, et al. J Biol Rhythms. 2007;22:375. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 11.Busino L, et al. Science. 2007;316:900. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 12.Siepka SM, et al. Cell. 2007;129:1011. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etchegaray JP, Lee C, Wade PA, Reppert SM. Nature. 2003;421:177. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 14.Ripperger JA, Schibler U. Nat Genet. 2006;38:369. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 15.Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Hosoda H, et al. Mol Brain. 2009;2:34. doi: 10.1186/1756-6606-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, et al. J Cell Sci. 2010;123:3547. doi: 10.1242/jcs.070300. [DOI] [PubMed] [Google Scholar]
- 18.Katada S, Sassone-Corsi P. Nat Struct Mol Biol. 2010;17:1414. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratmann M, Stadler F, Tamanini F, van der Horst GT, Ripperger JA. Genes De. 2010;24:1317. doi: 10.1101/gad.578810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TH, et al. Nature. 2005;436:876. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heintzman ND, et al. Nat Genet. 2007;39:311. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 22.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. Cell. 2007;130:77. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Carey M, Workman JL. Cell. 2007;128:707. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Hogenesch JB, Ueda HR. Nat Rev Genet. 2011;12:407. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 25.Masri S, Sassone-Corsi P. Nat Neurosci. 2010;13:1324. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng D, Lazar MA. Mol Cell. 2012;47:158. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatanaka F, et al. Mol Cell Biol. 2010;30:5636. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rey G, et al. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng D, et al. Science. 2011;331:1315. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho H, et al. Nature. 2012;485:123. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bugge A, et al. Genes Dev. 2012;26:657. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent WJ, et al. Genome Res. 2002;12:996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. BMC Bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panda S, et al. Cell. 2002;109:307. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhang EE, et al. Cell. 2009;139:199. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. Genes Dev. 2010;24:345. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamia KA, et al. Nature. 2011;480:552. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK. Nature. 2003;421:948. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 40.Ameur A, et al. Nat Struct Mol Biol. 2011;18:1435. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 41.Storch KF, et al. Nature. 2002;417:78. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 42.Ueda HR, et al. Nature. 2002;418:534. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 43.Hughes ME, et al. PLoS Genetics. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wuarin J, et al. J Cell Sci Suppl. 1992;16:123. doi: 10.1242/jcs.1992.supplement_16.15. [DOI] [PubMed] [Google Scholar]
- 45.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. Cell Metab. 2006;4:25. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Staiger D, Koster T. Cell Mol Life Sci. 2011;68:71. doi: 10.1007/s00018-010-0513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kojima S, Shingle DL, Green CB. J Cell Sci. 2011;124:311. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Genes Dev. 2004;18:2437. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 49.Fuda NJ, Ardehali MB, Lis JT. Nature. 2009;461:186. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones JC, et al. J Biol Chem. 2004;279:24957. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapman RD, et al. Science. 2007;318:1780. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 52.Barski A, et al. Cell. 2007;129:823. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Ong CT, Corces VG. Nat Rev Genet. 2011;12:283. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Creyghton MP, et al. Proc Natl Acad Sci U S A. 2010;107:21931. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rada-Iglesias A, et al. Nature. 2011;470:279. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Core LJ, Waterfall JJ, Lis JT. Science. 2008;322:1845. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahl PB, et al. Cell. 2010;141:432. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. J Biol Chem. 2011;286:25891. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Nat Rev Genet. 2010;11:109. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 60.Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Genes Dev. 2010;24:358. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duong HA, Robles MS, Knutti D, Weitz CJ. Science. 2011;332:1436. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Science. 2012 doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 63.Lee C, Weaver DR, Reppert SM. Mol Cell Biol. 2004;24:584. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reick M, Garcia JA, Dudley C, McKnight SL. Science. 2001;293:506. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 65.Eckner R, et al. Mol Cell Biol. 1996;16:3454. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nowak DE, Tian B, Brasier AR. Biotechniques. 2005;39:715. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 67.Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. Biotechniques. 2006;41:694. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- 68.Kim TK, et al. Nature. 2010;465:182. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinz S, et al. Mol Cell. 2010;38:576. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chow S, Ruskey F. Electronic Notes Theor Computer Sci. 2005;134:3. [Google Scholar]
- 71.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, et al. Bioinformatics. 2009;25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen R, et al. Mol Cell. 2009;36:417. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hughes ME, Hogenesch JB, Kornacker K. Journal of biological rhythms. 2010;25:372. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang R, Su Z. Bioinformatics. 2010;26:i168. doi: 10.1093/bioinformatics/btq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang B, Kirov S, Snoddy J. Nucleic acids research. 2005;33:W741. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duncan D, Prodduturi N, Zhang B. BMC Bioinformatics. 2010;11:P10. [Google Scholar]
- 79.Yoo SH, et al. Proc Natl Acad Sci U S A. 2004;101:5339. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.