Key Points
NrasG12D/+ induces proliferation and increases self-renewal and myeloid differentiation bias in HSCs.
ERK1/2 is constitutively hyperactivated in NrasG12D/+ HSCs and downregulation of the MEK/ERK signaling attenuates NrasG12D/+ HSC phenotypes.
Abstract
Oncogenic NRAS mutations are frequently identified in human myeloid leukemias. In mice, expression of endogenous oncogenic Nras (NrasG12D/+) in hematopoietic cells leads to expansion of myeloid progenitors, increased long-term reconstitution of bone marrow cells, and a chronic myeloproliferative neoplasm (MPN). However, acute expression of NrasG12D/+ in a pure C57BL/6 background does not induce hyperactivated granulocyte macrophage colony-stimulating factor signaling or increased proliferation in myeloid progenitors. It is thus unclear how NrasG12D/+ signaling promotes leukemogenesis. Here, we show that hematopoietic stem cells (HSCs) expressing NrasG12D/+ serve as MPN-initiating cells. They undergo moderate hyperproliferation with increased self-renewal. The aberrant NrasG12D/+ HSC function is associated with hyperactivation of ERK1/2 in HSCs. Conversely, downregulation of MEK/ERK by pharmacologic and genetic approaches attenuates the cycling of NrasG12D/+ HSCs and prevents the expansion of NrasG12D/+ HSCs and myeloid progenitors. Our data delineate critical mechanisms of oncogenic Nras signaling in HSC function and leukemogenesis.
Introduction
Hematopoietic stem cells (HSCs) undergo self-renewing divisions to sustain life-long hematopoiesis. HSC self-renewal is regulated by networks of proto-oncogenes and tumor suppressor genes.1 During leukemogenesis, genetically altered HSCs play an important role in initiating and/or maintaining leukemia phenotypes.2,3 These mutant HSCs often gain a competitive advantage over wild-type HSCs through increased cell division. In most cases, increased HSC proliferation tends to be associated with reduced HSC self-renewal and subsequently HSC depletion.4 In contrast, increased HSC cycling is associated with normal or increased self-renewal in HSCs deficient for p535,6 or p18INK4c.7,8 Thus, it remains unclear how the proliferation and self-renewal of HSCs are differentially regulated during leukemogenesis.
Ras proteins are master signaling switches that couple extracellular stimuli to the intracellular response machinery. Oncogenic RAS mutations are prevalent in essentially all human cancers.9 In human leukemias, although both NRAS and KRAS are mutated at significant frequencies, oncogenic NRAS mutations are significantly more prevalent than oncogenic KRAS mutations.10 In mice, expression of one copy of oncogenic Nras (NrasG12D/+) from its endogenous locus in hematopoietic cells leads to expansion of myeloid progenitors, increased long-term reconstitution of bone marrow (BM) cells, and a chronic myeloproliferative neoplasm (MPN) after a prolonged latency.11-13 However, acute expression of NrasG12D/+ in a pure C57BL/6 background does not induce hyperactivated granulocyte macrophage colony-stimulating factor signaling or increased proliferation in myeloid progenitors.13 In contrast to NrasG12D/+, stronger activation of oncogenic Ras signaling, such as KrasG12D/+ and two copies of oncogenic Nras (NrasG12D/G12D), leads to hyperactivated cytokine signaling, hyperproliferation, and expansion of myeloid progenitors as well as an acute MPN.2,3,13,14 It is thus unclear how NrasG12D/+ induces expansion of myeloid progenitors and promotes MPNs. We hypothesized that NrasG12D/+ promotes leukemogenesis by aberrantly regulating HSC function.
Here, we show that endogenous NrasG12D/+ induced proliferation and increased self-renewal and myeloid differentiation bias in HSCs. ERK1/2 is constitutively hyperactivated in NrasG12D/+ HSCs, and downregulation of MEK/ERK signaling attenuates NrasG12D/+ HSC phenotypes.
Materials and methods
Mice
Cre expression was induced through intraperitoneal injection of 250 μg (Sigma-Aldrich) or 7.5 μg/g body weight (GE Healthcare) polyinosinic-polycytidylic acid (pI-pC) every other day for two times. All experiments were conducted with the ethical approval of the International Association for Assessment and Accreditation of Laboratory Animal Care at the University of Wisconsin-Madison. Additional materials and methods are provided in the supplemental Data.
Results and discussion
NrasG12D/+ HSCs serve as MPN-initiating cells
We previously reported that more than 90% of recipient mice transplanted with NrasG12D/+ BM cells develop a chronic MPN, closely resembling human chronic myelomonocytic leukemia (CMML).11 To determine whether NrasG12D/+ HSCs are required for the initiation and/or maintenance of chronic MPNs, we transplanted different populations of cells from primary NrasG12D/+ mice or recipients that had developed chronic MPNs. NrasG12D/+ HSCs (Lin− CD41− CD48− c-Kit+ Sca-1+ CD150+)15, but not more committed myeloid progenitors, efficiently initiated CMML-like phenotypes in primary recipients, and only CMML cells containing HSCs could re-establish the disease in secondary recipients (supplemental Table 1). These data indicate that NrasG12D/+ HSCs are required to initiate and maintain CMML-like phenotypes in recipient mice and thus serve as MPN-initiating cells. Similar observations are reported in human chronic lymphocytic leukemia16 and in a mouse model of BCR-ABL–induced chronic myelogenous leukemia,17 suggesting a common role of genetically altered HSCs in the chronic phase of leukemias. Noticeably, CMML-like phenotypes that developed in the secondary recipients required a long latency similar to that observed in the primary recipients, suggesting that just as in KrasG12D/+-initiated MPNs,2 additional mutations promoting CMML development did not occur in NrasG12D/+ HSCs. With the second hits, we believe that myeloid progenitors gain short-term self-renewal capability but are not fully transformed to CMML-initiating cells, which explains why these cells fail to maintain disease phenotypes in secondary recipient mice.
NrasG12D/+ increases HSC proliferation and self-renewal capability
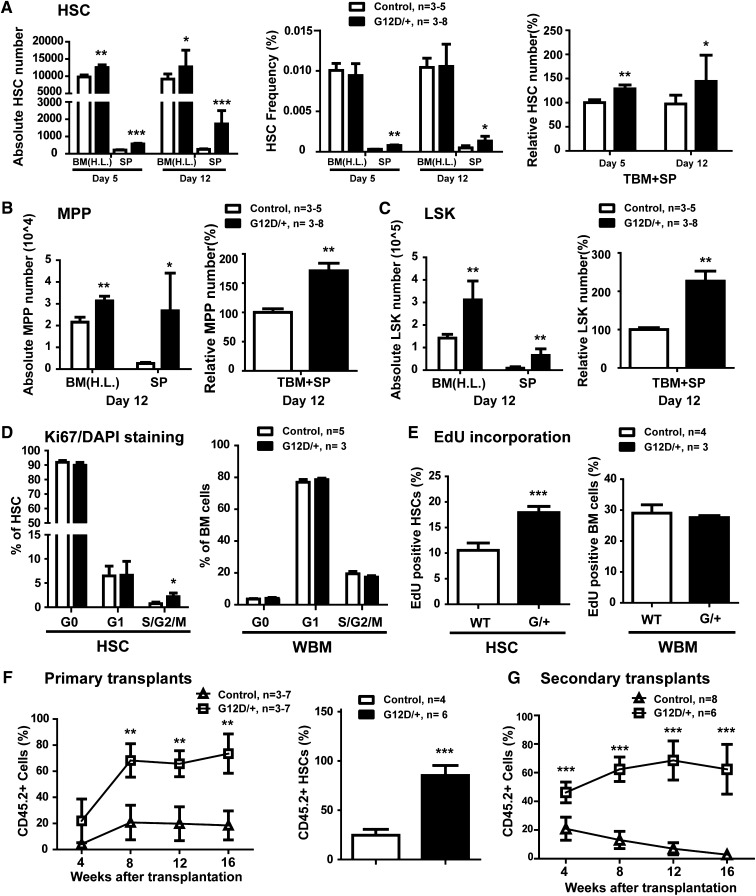
We further investigated how endogenous oncogenic Nras signaling regulates HSC function to promote leukemogenesis. We first examined the HSC compartment in Mx1-Cre; NrasG12D/+ mice (described in Wang et al11) at different time points after pI-pC injections. The conditional NrasG12D allele was recombined in almost 100% of HSCs in these mice, although the recombination frequency was 0% to 35% in Mx1-Cre; NrasG12D/+ mice without pI-pC treatment (supplemental Figure 1). The day of the first pI-pC injection is referred as day 1. On days 5 and 12 (Figure 1A),18 compared with control HSCs, NrasG12D/+ HSCs displayed a moderate but consistent expansion (∼25% increase). We did not observe pI-pC–induced Sca1 expression in total BM cells or defined HSCs (supplemental Figure 2). The numbers of multipotent progenitors (MPPs) and LSK cells in G12D/+ mice were also significantly elevated compared with controls (Figure 1B-C).
Figure 1.
Endogenous NrasG12D/+ induces increased proliferation and self-renewal in HSCs. (A-F) Control and NrasG12D/+ mice were treated with pI-pC and euthanized at various time points for analysis. We refer to the day of the first pI-pC injection as day 1. (A) Lin− CD41− CD48− c-Kit+ Sca-1+ CD150+ HSCs, (B) Lin− CD41− CD48− c-Kit+ Sca-1+ CD150− MPPs, and (C) LSK cells were quantified using flow cytometry. SP, spleen; H.L., hind limb, including tibia and femur; TBM, total bone marrow, ∼fourfold H.L. BM.18 (D) Cell cycle analysis of HSCs and WBM from control and NrasG12D/+ mice using Ki67/DAPI [4,6 diamidino-2-phenylindole] staining. (E) A 16-hour pulse of 5-ethynyl-2′-deoxyuridine (EdU) to quantify proliferating HSCs and WBM. (F) Twenty HSCs purified from control or NrasG12D/+ mice were transplanted with 2 × 105 congeneic BM cells into lethally irradiated mice. Donor-derived blood cells were regularly analyzed in the peripheral blood of recipients. Donor-derived HSCs were quantified in recipients 16 weeks after transplantation. (G) 2 × 106 BM cells isolated from primary recipients were transplanted into lethally irradiated secondary recipient mice. Donor-derived blood cells were regularly analyzed in the peripheral blood of secondary recipients. Data are presented as mean + standard deviation. *P < .05; **P < .01; ***P < .001.
Expression of endogenous NrasG12D/+ increased the division of HSCs, but not whole BM (WBM) cells. On day 12, consistent with a previous report,19 only ∼0.5% of control HSCs were in S/G2/M. In contrast, a significant increase of NrasG12D/+ HSCs was detected in S/G2/M, suggesting an increased proliferation in these HSCs (Figure 1D and supplemental Figure 3A). This result was further confirmed by using 5-ethynyl-2′-deoxyuridine (EdU) in vivo incorporation (Figure 1E and supplemental Figure 3B). We noticed that the increased proliferation in NrasG12D/+ HSCs is moderate compared with the strong hyperproliferation of KrasG12D/+ HSCs.20
To determine whether the proliferation of NrasG12D/+ HSCs was correlated with increased HSC self-renewal, we transplanted highly purified control or NrasG12D/+ HSCs along with wild-type WBM cells into lethally irradiated mice (Figure 1F). Compared with control cells, NrasG12D/+-derived blood cells were greatly overrepresented in peripheral blood of recipients in all lineages (supplemental Figure 4A). We euthanized some of the recipients 16 weeks posttransplant. Control donor HSCs represented ∼25% of the recipients’ BM HSC compartment, and NrasG12D/+ HSCs comprised >90% (Figure 1F). On the basis of our quantitative analysis, a single NrasG12D/+ HSC generated ∼10-fold as many HSCs as a single control HSC. Secondary transplantation further supported the increased self-renewal potential in NrasG12D/+ HSCs (Figure 1G and supplemental Figure 4B). These results indicate that NrasG12D/+ signaling enhances HSC self-renewal capability. The increased self-renewal potential in NrasG12D/+ HSCs is in sharp contrast to reduced self-renewal and depletion in HSCs expressing stronger oncogenic Ras, such as KrasG12D/+ or NrasG12D/+G12D 21 (G.K., Y.-I.C., and Jing Z., unpublished data), suggesting a distinct regulation of HSC function in an oncogenic Ras activity-dependent manner.
NrasG12D/+ hyperactivates ERK1/2 in HSCs
To investigate the signaling mechanism(s) underlying aberrant NrasG12D/+ HSC function, we studied the MEK/ERK and AKT signaling in HSCs and their downstream MPPs22 (Figure 2A) by using a recently developed HSC phospho-flow method.23 In the absence of serum and cytokines, ERK1/2 was constitutively hyperactivated in NrasG12D/+ HSCs (twofold over control HSCs) but not in MPPs (Figure 2A). The constitutive hyperactivation of ERK1/2 was inhibited by AZD6244, a potent MEK inhibitor (supplemental Figure 5). In contrast, AKT activation in NrasG12D/+ HSCs and MPPs was comparable to that in controls (Figure 2A). Upon stimulation with interleukin-3, ERK1/2 was further hyperactivated in NrasG12D/+ HSCs but not in MPPs (Figure 2B). The hyperactivation of ERK1/2 in NrasG12D/+ HSCs is in sharp contrast to the normal pERK1/2 level in NrasG12D/+ MPPs and Lin− cKit+ progenitor cells,13 indicating that HSC is the most sensitive cell type to oncogenic Nras mutation.
Figure 2.
NrasG12D/+ hyperactivates ERK1/2 in HSCs and downregulation of the MEK/ERK signaling attenuates NrasG12D/+ HSC phenotypes. (A-B) Total BM cells from control or NrasG12D/+ mice were enriched for Sca1+ cells. CD150+ CD41− cells (enriched for HSCs) and CD150− CD41− cells (enriched for MPPs) were subsequently sorted from Sca1+-enriched cells. (A) Sorted cells were serum- and cytokine-starved for 30 minutes at 37°C. (B) Interleukin-3 (IL-3) stimulation was performed for 10 minutes at 37°C after starvation. Levels of phosphorylated ERK1/2 and AKT were measured using phospho-specific flow cytometry. HSCs (defined as [Lin CD48]−/low cKit+ cells from sorted CD150+ CD41− cells) and MPPs (defined as [Lin CD48]−/low cKit+ cells from sorted CD150− CD41− cells) were gated for data analysis. To quantify the activation of ERK1/2, median intensities of pERK1/2 at different IL-3 concentrations are compared with their respective control cells at 0 ng/mL, which is arbitrarily set at 1. (C) Total BM cells were serum- and cytokine-starved for 1 hour and stimulated with or without 10 ng/mL of granulocyte macrophage colony-stimulating factor (GM-CSF) at 37°C for 10 minutes. AZD6244 or vehicle was mixed with cells for 30 minutes before GM-CSF stimulation. Levels of pERK1/2 or pSTAT5 were measured using phospho-specific flow cytometry. Non-neutrophil Lin− c-Kit+ cells were gated for data analysis. Results obtained from one representative experiment are shown. (D) Quantification of BM and SP HSCs from control and NrasG12D/+ (G/+) mice treated with vehicle or AZD6244 for 7 days. (E) Quantification of BM and SP common myeloid progenitors (CMPs) from control and NrasG12D/+ (G/+) mice treated with vehicle or AZD6244 for 7 days. (F) EdU incorporation to quantify proliferating HSCs and WBM in control, NrasG12D/+, Erk2+/−, and NrasG12D/+; Erk2+/− mice. Data are presented as mean + standard deviation. WT, wild-type. *P < .05; **P < .01; ***P < .001.
Gene set enrichment analysis (GSEA) identifies a gene signature associated with myeloid differentiation in NrasG12D/+ HSCs
To better understand the molecular mechanisms underlying oncogenic Nras signaling-induced aberrant HSC function, we performed a microarray analysis (accession number: GSE46948) using highly purified HSCs. Compared with control HSCs, NrasG12D/+ HSCs did not show significant differential expression in genes or pathways that are associated with HSC self-renewal4 (supplemental Figure 6A-B).
Because NrasG12D/+ HSCs support leukemia development predominantly in myeloid cells (supplemental Table 1 and Wang et al11), we sought to determine whether these HSCs are associated with a myeloid differentiation bias. In support of this idea, GSEA of our HSC microarray results identified a gene signature associated with myeloid differentiation in NrasG12D/+ HSCs (supplemental Figure 6C). Consistent with the GSEA results, the common myeloid progenitor (CMP) compartment was greatly expanded in NrasG12D/+ mice (Figure 2E, Li et al,12 and Wang et al13), although common lymphoid progenitor (CLP) compartment was comparable to that in control mice (supplemental Figure 6D). Our current result of myeloid differentiation bias in NrasG12D/+ HSCs explains the selective expansion of myeloid progenitor compartment in NrasG12D/+ mice, in which granulocyte macrophage colony-stimulating factor signaling and cell proliferation are indistinguishable from those in control cells.11,13
Downregulation of the MEK/ERK signaling attenuates the HSC phenotypes in NrasG12D/+ mice
To test whether hyperactivation of MEK/ERK signaling contributes to the effects of NrasG12D/+ on HSCs, we treated control and NrasG12D/+ mice with vehicle or AZD6244,24 an efficacious and specific MEK inhibitor (Figure 2C). AZD6244 prevented the expansion of NrasG12D/+ HSC and CMP compartment (Figure 2D-E) and restored slightly enlarged spleen to a normal size in NrasG12D/+ mice (supplemental Figure 7). In addition, we generated NrasG12D/+; Erk2+/− mice to downregulate the MEK/ERK signaling using a genetic approach. Loss of one copy of Erk2 partially downregulated ERK signing (supplemental Figure 8) and significantly decreased the cycling of NrasG12D/+ HSCs without affecting the cycling of WBM cells (Figure 2F). The slightly enlarged spleen remained a similar size (supplemental Figure 9).
In summary, our data suggest that moderate activation of oncogenic Ras/MEK/ERK signaling contributes to the increased long-term competitiveness in mutant HSCs during leukemogenesis.
Supplementary Material
Acknowledgments
The authors thank Dr Kevin Haigis and Dr Tyler E. Jacks for generously providing conditional oncogenic Nras mice. The authors are grateful to Drs Emery Bresnick and Qiang Chang for helpful discussion and critical comments on the manuscript and are appreciative of Dr Ya-Huei Kuo for sharing her EdU labeling protocol. The authors thank the University of Wisconsin Carbone Comprehensive Cancer Center for use of its Shared Services to complete this research.
This work was supported by the National Cancer Institute (NCI) (1R01CA152108), a Shaw Scientist Award from the Greater Milwaukee Foundation, an ASH Scholar Award from the American Society of Hematology (ASH), a V Scholar Award from the V Foundation for Cancer Research, and an Investigator Initiated Grant from the University of Wisconsin Carbone Comprehensive Cancer Center (J.Z.), a grant from the National Natural Science Foundation of China (31271457; J.W.), and a grant from the Canadian Institutes of Health Research (MOP-119327; S.M.). S.M. holds the Canada Research Chair in Cellular Signaling. This work was also supported in part by a grant from the NCI/National Institutes of Health Comprehensive Cancer Center (P30 CA014520-UW).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.W. and G.K. provided experimental design and execution; Y.L., J.D., Y.-I.C., S.R.T., A.D., and Jingfang Z. provided experimental execution; X.Z. performed microarray analysis; E.A.R. provided histopathologic analysis and edited the manuscript; M.K.S.-E.-L. and S.M. generated the Erk2 conditional knockout allele; and Jing Z. provided experimental design and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jinyong Wang, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 190 Kaiyuan Rd, Guangzhou Science Park, Guangzhou, China 510530; e-mail: wang_jinyong@gibh.ac.cn; and Jing Zhang, McArdle Laboratory for Cancer Research, 1400 University Ave, University of Wisconsin, Madison, WI; e-mail: zhang@oncology.wisc.edu.
References
- 1.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, Fleming MD. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009;113(6):1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabnis AJ, Cheung LS, Dail M, et al. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009;7(3):e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9(2):115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Elf SE, Miyata Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4(1):37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milyavsky M, Gan OI, Trottier M, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7(2):186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y, Shen H, Franklin DS, Scadden DT, Cheng T. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat Cell Biol. 2004;6(5):436–442. doi: 10.1038/ncb1126. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Yuan Y, Shen H, Cheng T. Hematopoietic stem cell exhaustion impacted by p18 INK4C and p21 Cip1/Waf1 in opposite manners. Blood. 2006;107(3):1200–1206. doi: 10.1182/blood-2005-02-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 10.Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120(17):3397–3406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JY, Liu YG, Li ZY, et al. Endogenous oncogenic Nras mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood. 2010;116(26):5991–6002. doi: 10.1182/blood-2010-04-281527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Haigis KM, McDaniel A, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117(6):2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JY, Liu YG, Li ZY, et al. Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood. 2011;118(2):368–379. doi: 10.1182/blood-2010-12-326058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan IT, Kutok JL, Williams IR, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113(4):528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiel MJ, He S, Ashkenazi R, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449(7159):238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikushige Y, Ishikawa F, Miyamoto T, et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20(2):246–259. doi: 10.1016/j.ccr.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 17.Reynaud D, Pietras E, Barry-Holson K, et al. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20(5):661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boggs DR. The total marrow mass of the mouse: a simplified method of measurement. Am J Hematol. 1984;16(3):277–286. doi: 10.1002/ajh.2830160309. [DOI] [PubMed] [Google Scholar]
- 19.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441(7092):475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 20.Du J, Liu Y, Meline B, et al. Loss of CD44 attenuates aberrant GM-CSF signaling in Kras G12D hematopoietic progenitor/precursor cells and prolongs the survival of diseased animals. Leukemia. 2013;27(3):754–757. doi: 10.1038/leu.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Liu Y, Tan LX, et al. Distinct requirements of hematopoietic stem cell activity and Nras G12D signaling in different cell types during leukemogenesis. Cell Cycle. 2011;10(17):2836–2839. doi: 10.4161/cc.10.17.17195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Du J, Wang J, Kong G, et al. Signaling profiling at the single-cell level identifies a distinct signaling signature in murine hematopoietic stem cells. Stem Cells. 2012;30(7):1447–1454. doi: 10.1002/stem.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh TC, Marsh V, Bernat BA, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13(5):1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.