Abstract
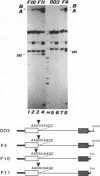
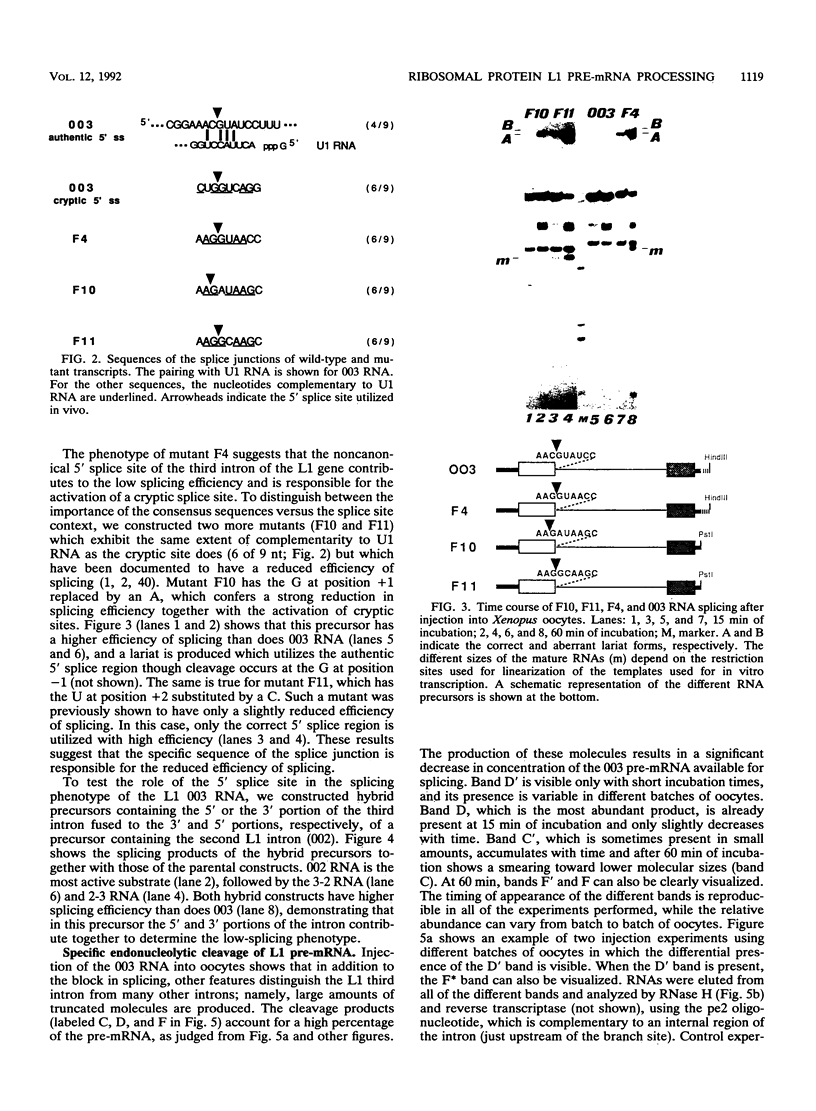
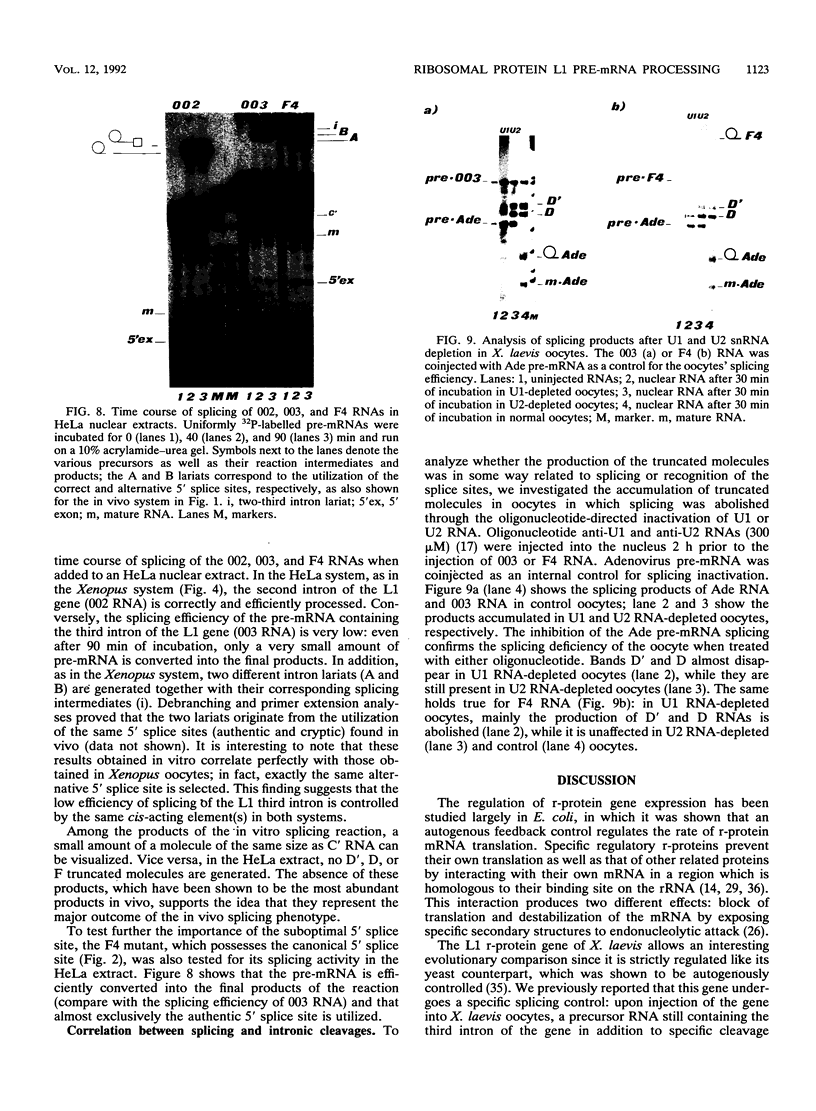
Splicing of the regulated third intron of the L1 ribosomal protein gene of Xenopus laevis has been studied in vivo by oocyte microinjection of wild-type and mutant SP6 precursor RNAs and in vitro in the heterologous HeLa nuclear extract. We show that two different phenomena combine to produce the peculiar splicing phenotype of this intron. One, which can be defined constitutive, shows the same features in the two systems and leads to the accumulation of spliced mRNA, but in very small amounts. The low efficiency of splicing is due to the presence of a noncanonical 5' splice site which acts in conjunction with sequences present in the 3' portion of the intron. The second leads to the massive conversion of the pre-mRNA into site specific truncated molecules. This has the effect of decreasing the concentration of the pre-mRNA available for splicing. We show that this aberrant cleavage activity occurs only in the in vivo oocyte system and depends on the presence of an intact U1 RNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Al-Atia G. R., Fruscoloni P., Jacobs-Lorena M. Translational regulation of mRNAs for ribosomal proteins during early Drosophila development. Biochemistry. 1985 Oct 8;24(21):5798–5803. doi: 10.1021/bi00342a017. [DOI] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bowman L. H. The synthesis of ribosomal proteins S16 and L32 is not autogenously regulated during mouse myoblast differentiation. Mol Cell Biol. 1987 Dec;7(12):4464–4471. doi: 10.1128/mcb.7.12.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzoni I., Fragapane P., Annesi F., Pierandrei-Amaldi P., Amaldi F., Beccari E. Expression of two Xenopus laevis ribosomal protein genes in injected frog oocytes. A specific splicing block interferes with the L1 RNA maturation. J Mol Biol. 1984 Dec 25;180(4):987–1005. doi: 10.1016/0022-2836(84)90267-5. [DOI] [PubMed] [Google Scholar]
- Caffarelli E., Fragapane P., Gehring C., Bozzoni I. The accumulation of mature RNA for the Xenopus laevis ribosomal protein L1 is controlled at the level of splicing and turnover of the precursor RNA. EMBO J. 1987 Nov;6(11):3493–3498. doi: 10.1002/j.1460-2075.1987.tb02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Dabeva M. D., Post-Beittenmiller M. A., Warner J. R. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5854–5857. doi: 10.1073/pnas.83.16.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J. N. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol. 1972 Feb;136(2):153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B. Ras C-terminal processing enzymes--new drug targets? Cell. 1991 Apr 5;65(1):1–4. doi: 10.1016/0092-8674(91)90352-y. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Mattaj I. W. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989 Oct 6;59(1):159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990 Oct 5;63(1):109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Kakizuka A., Ingi T., Murai T., Nakanishi S. A set of U1 snRNA-complementary sequences involved in governing alternative RNA splicing of the kininogen genes. J Biol Chem. 1990 Jun 15;265(17):10102–10108. [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990 Feb;10(2):696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Keller W. Purification of a protein required for the splicing of pre-mRNA and its separation from the lariat debranching enzyme. EMBO J. 1985 Dec 16;4(13A):3571–3581. doi: 10.1002/j.1460-2075.1985.tb04119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Loreni F., Ruberti I., Bozzoni I., Pierandrei-Amaldi P., Amaldi F. Nucleotide sequence of the L1 ribosomal protein gene of Xenopus laevis: remarkable sequence homology among introns. EMBO J. 1985 Dec 16;4(13A):3483–3488. doi: 10.1002/j.1460-2075.1985.tb04107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariottini P., Amaldi F. The 5' untranslated region of mRNA for ribosomal protein S19 is involved in its translational regulation during Xenopus development. Mol Cell Biol. 1990 Feb;10(2):816–822. doi: 10.1128/mcb.10.2.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis L., Vu L., Sor F., Nomura M. Retroregulation of the synthesis of ribosomal proteins L14 and L24 by feedback repressor S8 in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Jan;86(2):448–452. doi: 10.1073/pnas.86.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O., Thompson E. A., Jr, Perry R. P. Glucocorticoids selectively inhibit translation of ribosomal protein mRNAs in P1798 lymphosarcoma cells. Mol Cell Biol. 1987 Aug;7(8):2691–2699. doi: 10.1128/mcb.7.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi R. N., Baker B. S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990 Jan;4(1):89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989 Feb;9(2):726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierandrei-Amaldi P., Beccari E., Bozzoni I., Amaldi F. Ribosomal protein production in normal and anucleolate Xenopus embryos: regulation at the posttranscriptional and translational levels. Cell. 1985 Aug;42(1):317–323. doi: 10.1016/s0092-8674(85)80127-6. [DOI] [PubMed] [Google Scholar]
- Pierandrei-Amaldi P., Bozzoni I., Cardinali B. Expression of the gene for ribosomal protein L1 in Xenopus embryos: alteration of gene dosage by microinjection. Genes Dev. 1988 Jan;2(1):23–31. doi: 10.1101/gad.2.1.23. [DOI] [PubMed] [Google Scholar]
- Pierandrei-Amaldi P., Campioni N., Beccari E., Bozzoni I., Amaldi F. Expression of ribosomal-protein genes in Xenopus laevis development. Cell. 1982 Aug;30(1):163–171. doi: 10.1016/0092-8674(82)90022-8. [DOI] [PubMed] [Google Scholar]
- Presutti C., Ciafré S. A., Bozzoni I. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 1991 Aug;10(8):2215–2221. doi: 10.1002/j.1460-2075.1991.tb07757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said B., Cole J. R., Nomura M. Mutational analysis of the L1 binding site of 23S rRNA in Escherichia coli. Nucleic Acids Res. 1988 Nov 25;16(22):10529–10545. doi: 10.1093/nar/16.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Thomas M. S., Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: mutations that define the target site for repression by L1. Nucleic Acids Res. 1987 Apr 10;15(7):3085–3096. doi: 10.1093/nar/15.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Aebi M. In vitro splicing of mRNA precursors: 5' cleavage site can be predicted from the interaction between the 5' splice region and the 5' terminus of U1 snRNA. Nucleic Acids Res. 1988 Jan 25;16(2):471–486. doi: 10.1093/nar/16.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]