Abstract
DNA probes for the studies of damaged strand excision during the nucleotide excision repair (NER) have been designed using the novel non-nucleosidic phosphoramidite reagents that contain N-[6-(9-antracenylcarbamoyl)hexanoyl]-3-amino-1,2-propandiol (nAnt) and N-[6-(5(6)-fluoresceinylcarbamoyl)hexanoyl]-3-amino-1,2-propandiol (nFlu) moieties. New lesion-imitating adducts being inserted into DNA show good substrate properties in NER process. Modified extended linear nFlu– and nAntr–DNA are suitable for estimation of specific excision activity catalysed with mammalian whole-cell extracts. The following substrate activity range was revealed for the model 137-bp linear double-stranded DNA: nAnt–DNA ≈ nFlu–DNA > Chol–DNA (Chol–DNA—legitimate NER substrate that contains non-nucleoside fragment bearing cholesterol residue). In vitro assay shows that modified DNA can be a useful tool to study NER activity in whole-cell extracts. The developed approach should be of general use for the incorporation of NER-sensitive distortions into model DNAs. The new synthetic extended linear DNA containing bulky non-nucleoside modifications will be useful for NER mechanism study and for applications.
INTRODUCTION
The DNA nucleotide excision repair (NER) system recognizes and removes a wide variety of structurally diverse helix-distorting bulky adducts from DNA, mostly modified nitrogen bases (Pu or Py) or covalently linked neighbouring bases, and results in the release of short oligonucleotides (24–32mer) containing the lesion (1). One of the advanced and upcoming approaches to DNA repair investigation is based on the application of synthetic DNA molecules, containing lesion-simulating adducts. The characterization of specific excision activities in cell extract and reconstituted systems is a significant problem and could represent a major concern for fundamental research and for medical applications. The availability of model DNA, which imitates NER substrate well, is the significant point of NER research progress (2–8). Methods for design of NER substrate analogues have been developed. A variety of purine nucleotide derivatives, such as acetylaminofluorene adducts, products of reactions between nucleobases and benzo[a]pyrene- or benzo[c]anthracene-diol-epoxides, products of intra-strand DNA cross-links by cis-dichlorodiamine platinum (II) and others, have been used for this purpose (9–11). Py–Py dimers (usually T–T dimers) were used as analogues of damaged pyrimidine bases (12). A number of investigations were carried out using model DNA containing the thymine mono-adduct 4′-hydroxymethyl-4,5′,8-trimethylpsoralen, or thymidine in which a fluorescein residue was introduced into C5 position via a linker fragment (9,13–15). However, synthesis of the model DNAs, containing damage-mimicking modification of the defined structure at the desirable position of DNA molecule, remains up to now actual problem. Studies are in progress to improve the synthesis and to extend the variety of model DNA lesions. The elaboration of ‘ultra-mild’ method of single dG-AAF adduct introduction in DNA by solid-phase DNA synthesis substantially facilitated production of oligonucleotides, containing these modifications (9,16). Fluorescein residue and several arylazide groups introduced via different linkers at the nucleobase were successfully applied to imitate DNA damages produced by the action of aromatic hydrocarbon derivatives. To synthesize these DNA substrates, the different methods were used, including the chemo-enzymatic approach (2,5,16–19). The improved enzymatic approach for synthesis of bulky substituted DNA structures allowed to synthesize the variety of functional analogues of NER substrates, including the photoactivable ones (20). There is also at least one type of legitimate NER substrate containing bulky non-nucleoside lesions, namely, Chol fragment, introduced by solid-phase DNA synthesis (21). This type of model DNA has been used to investigate both UVrABC and eukaryotic NER systems (22). Unfortunately, most of the NER substrates demonstrate low specific excision efficiency in vitro; therefore, usage of model DNA for evaluation of NER system activity is restricted, especially in cell or tissue extracts (23).
An effective and easily accessible standard substrate is necessary to enable the studies of a key stage of NER, the elimination of the damaged DNA fragment. This will permit to bring a further insight into the mechanisms of NER, its role in the development of degenerative diseases and to investigate the effects of gene–environment interactions on DNA repair.
The main goal of the present investigation was to design artificial lesions, which can be introduced at any specified DNA position regardless of nucleotide sequence and efficiently recognized and processed by NER system. This required three main issues to be resolved: (i) the efficient preparation of the appropriate phosphoramidites; (ii) the development of straightforward solid-phase DNA synthesis protocol for the incorporation of the new amidites that does not compromise further enzymatic integration of the modified ONT in DNA structures; (iii) the characterization of the substrate properties of the model DNA in the reaction of specific NER excision. Furthermore, it may serve as an example for the development of additional functional DNA repair tests, which are needed to gain further insight into the role of DNA repair in cancer risk and pathology.
MATERIALS AND METHODS
Reagents and equipment
Reagents obtained from commercial suppliers were used without further purification. Anhydrous solvents were prepared by distillation under an argon atmosphere in the presence of appropriate drying agents. DMF was distilled under reduced pressure and stored over 4 Å molecular sieves under argon. The following reagents were used: 4,4′-dimethoxytrityl chloride (Chem-IMPEX, USA), N,N-ethyldiisopropylamine and pivalic anhydride (DIEA, Fluka Chemie AG, Switzerland). The 5(6)-carboxy-3′,6′-O-dipivaloylfluorescein (24) was prepared as described. All other reagents were from Aldrich (USA). 1H and 31P nuclear magnetic resonance (NMR) spectra were recorded on a Bruker 400 and 300 MHz NMR spectrometers in deuterated solvents. Chemical shifts are reported in parts per million (ppm) relative to the tetramethylsilane as the internal standard. Matrix-assisted laser desorption/ionization-time of flight (MALDI–TOF) mass spectra were run on Autoflexspeed or Macroflex spectrometers (Bruker Daltonics, Germany) in positive-ion mode with dihydroxybenzoic acid as a matrix. Ion trap XTC ultra (Agilent Technologies, USA) mass spectrometer with an electrospray ionization (ESI) interface was also used for molecular mass measurements. Analytical thin-layer chromatography (TLC) was performed on Kieselgel 60 F254 pre-coated aluminium plates (Merck); spots were visualized under ultraviolet light (254 nm). Silica gel column chromatography was performed using Merck Kieselgel 60 0.040–0.063 mm. Cholesteryl–TEG–phosphoramidite (TEG - Dimethoxytrityloxy-3-O-(N-cholesteryl-3-aminopropyl)-triethyleneglycol-glyceryl-2-O-(2-cyanoethyl)-(N,N,-diisopropyl)-phosphoramidite) was from Glen Research. The oligonucleotide synthesis was performed on a DNA–RNA synthesizer ASM-800 (Biosset) using standard manufacturer’s protocols. The regular ONT for creation of model DNA duplexes and the cholesterol-containing ONT were produced at the laboratory of medical chemistry of ICBFM SB RAS (all the sequences are presented in Supplementary Table S1).
α-[32P]-dCTP and (3000 Ci/mmol) were produced at ICBFM SB RAS. Taq DNA polymerase, T4 DNA ligase and T4 polynucleotide kinase were from Biosan (Novosibirsk), and exonuclease λ was kindly provided by A. I. Zakabunin (ICBFM SB RAS). Proteinase K was from Sigma. Dulbecco’s modified Eagle’s medium (DMEM) and DMEM/F12 mediums for HeLa and Chinese hamster ovarian (CHO) cells cultivation were produced by Invitrogen. SiHa and C33A cells biomasses were generous gift of Natalia and Fedor Kisselev (Institute of Carcinogenesis, Cancer Research Center, Russian Academy of Medical Sciences, Moscow).
The components for polyacrylamide gel preparation and the main components of buffer systems were produced by Sigma.
Synthesis of phosphoramidites
The organic synthesis initial stages
The organic synthesis initial stages are described in detail in Supplementary Material (see Supplementary Figures S1–S4 and attached description).
General procedure
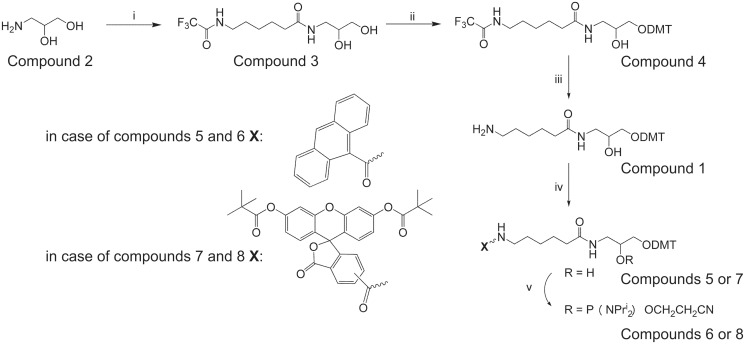
Compound 5 (7) (Figure 1) (1.05 mmol) was dissolved in a freshly distilled CH2Cl2 (10 ml), diisopropylammonium tetrazolide (90 mg, 0.525 mmol) was added, followed by addition of N,N,N′,N′-tetraisopropyl-(2-cyano)ethyl phosphodiamidite (0.50 ml, 1.575 mmol). After 2 h, the reaction mixture was evaporated, the residue was treated with hexane (30 ml) and the mixture was kept overnight at −20°C. Hexane was then decanted, and the residue was purified by chromatography on silica gel column with 1% Et3N in hexane:CH2Cl2 (1:1). Elution with 1% Et3N in CH2Cl2 gave the target fractions. Fractions containing the product were combined and evaporated in vacuo. The residue was then dissolved in dichloromethane (2 ml), and the product was precipitated with a 10-fold volume of hexane.
Figure 1.
The general scheme of the organic synthesis procedure. Reagents and conditions: (i) N-hydroxysuccinimide ester of TFA-NH-(CH2)5-COOH, Et3N, DMF; (ii) DMTCl, pyridine; (iii) aqueous NH3 (25%), pyridine; (iv) (a) 9-anthracenecarboxylic acid, N-hydroxybenzotriazole, DCC, CH2Cl2, Et3N, (b) 5(6)-carboxyfluorescein-NHS ester, Et3N, CH2Cl2; and (v) 2-cyanoethyl-N,N,N′,N′-tetraisopropylphosphodiamidite, diisopropylammonium tetrazolide, CH2Cl2.
Compound 6
Yield 6 74.9%, TLC (CH2Cl2/(CH3)2CO 4:1) Rf 0.74. 1H NMR (CDCl3, δ) 8.48 [1H, s, Ant-H10], 8.08 [2H, d, J 8.3, Ant-H1,8], 8.02 [2H, d, J 8.3, Ant-H1,8], 7.57–7.42 [4H, m, Ant-H2,3,6,7], 7.31–7.19 [9H, m, ArH, DMT], 6.82 [4H, m, J 8.8, ArH, DMT]; 4.04 [1 H, m, CHOH], 3.77 [s, 6 H, OCH3], 3.76–3.04 [10H, m, NCH, POCH2, NCH2(CH2)4 C(O), NHCH2CH(OH)CH2O], 2.58 (1 H, t, J 6.1, CH2CN), 2.43 (1 H, t, J 6.3, CH2CN), 2.15 [2H, m, N(CH2)4CH2C(O)], 1.49 [2H, m, NCH2CH2(CH2)3 C(O)], 1.29 [4H, m, N(CH2)2(CH2)2CH2C(O)], 1.23–1.10 [12H, m, CH(CH3)2]; 31P NMR (CDCl3, δ) 149.38 (s), 148.93 (s); ESI MS (M + Et3N): 1012.5 (calcd 1011.6).
Compound 8
Yield 8 64.5%, TLC (CH2Cl2/(CH3)2CO 4:1) Rf 0.81. 1H NMR (CD3)2CO, δ) 8.50 [0.6 H, s, 5-isomer Flu-H4], 8.34 [0.6 H, d, J 8.1, 5-isomer Flu-H6], 8.27 [0.4 H, d, J 8.0, 6-isomer Flu-H5], 8.14 [0.6 H, bt, NH], 8.08 [0.4 H, d, J 8.0, 6-isomer Flu-H4], 7.98 [0.4 H, bt, NH], 7.82 [0.4 H, s, 6-isomer Flu-H7], 7.52–7.18 [12H, m, 5-isomer Flu-H7, 4′,5′, ArH, DMT], 7.00–6.83 [8H m, ArH, DMT, Flu-H1′,2′,7′,8′], 4.11 [1 H, m, CHOH], 3.76 [s, 6 H, OCH3], 3.69–3.05 [10H, m, NCH, POCH2, NCH2(CH2)4 C(O), NHCH2CH(OH)CH2O], 2.72 (1 H, bt, CH2CN), 2.62 (1 H, bt, CH2CN), 2.15 [2H, m, N(CH2)4CH2C(O)], 1.60 [2H, m, NCH2CH2(CH2)3 C(O)], 1.34 [18H, s, C(CH3)3], 1.20 [4H, m, N(CH2)2(CH2)2CH2C(O)], 1.11 [12H, m, CH(CH3)2]; 31P NMR [(CD3)2CO, δ] 150.20 (s), 149.82 (s); ESI MS (M + Et3N): 1334.1 (calcd 1333.7).
Oligonucleotide synthesis
Oligonucleotide synthesis was performed using ASM-800 DNA/RNA synthesizer (Biosset) on a 0.1 μM scale. Standard phosphoramidite chemistry was applied for synthesis (25,26). The procedure consists of cycles of four chemical reactions: (i) removal of the DMTr-group (4,4'- Dimethoxytrityl) with 2% dichloroacetic acid in dichloromethane; (ii) coupling of phosphoramidite; (iii) capping with acetic anhydride and 1,4-dimethylaminopyridine; and (iv) oxidation of P(III) phosphite triester with 0.01 M iodine in tetrahydrofurane/pyridine/water to stable P(V) phosphate. Purification was accomplished using RP-Cartridge (ChemGenes Inc.) according to recommended methods. Oligonucleotide derivatives were analysed by polyacrylamide gel electrophoresis (PAGE) under denaturing conditions and in every case demonstrated single band purity.
The cholesterol residue attached to linker backbone instead of a nucleoside was introduced using TEG–phosphoramidite into the desired sequence by standard phosphoramidite chemistry at position of the top strand.
Synthesis of long DNA probes
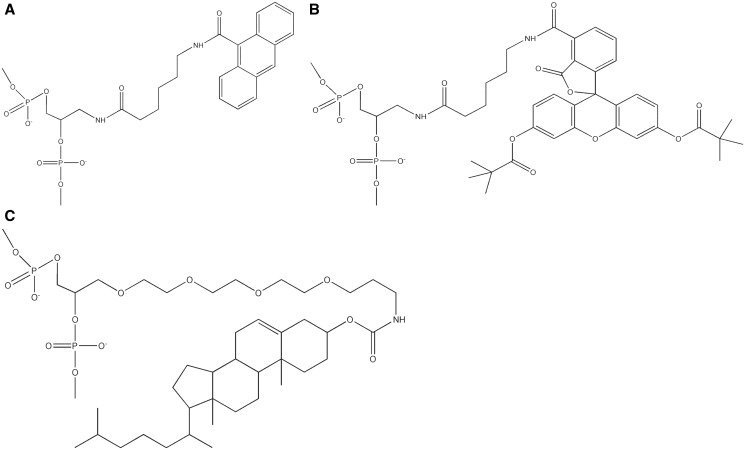
nAnt- and nFlu-modified oligonucleotides (Figure 2) of length corresponding to 16mer unmodified ONT were enzymatically phosphorylated at the 5′-ends using T4 polynucleotide kinase and adenosine triphosphate (ATP). They were ligated with flanking oligonucleotides (Supplementary Table S1) 1, 2 to yield single-stranded 137mer DNA. Resulting fragments were purified using PAGE under denaturing conditions. After spectrophotometric quantification, the 137mer ssDNA fragments were annealed with complementary unmodified strand and purified using non-denaturing PAGE. In the obtained duplexes, the internal modification was placed at the 68th position from the 5′-ends of the modified 137mer strand. The cholesterol-containing ONT was used to synthesize 137mer ssDNA for construction of DNA duplex, which served as positive control of NER activity in mammalian cells extracts. As a negative control, 137mer DNA strands synthesized with unmodified 16mer ONT were used for duplex formation. The ligation procedure was described in detail elsewhere (20).
Figure 2.
The non-nucleoside fragments of the modified DNA strand, containing N-[6-(9-antracenylcarbomoyl)hexanoyl]-3-amino-1,2-propandiol [nAnt, (A)], N-[6-(5(6)-fluoresceinylcarbomoyl)hexanoyl]-3-amino-1,2-propandiol [nFlu, (B)] or cholesterol residue [Chol, (C)].
Complementary unmodified 137mer ssDNA was produced using polymerase chain reaction (PCR) and subsequent treatment of PCR products with exonuclease λ. The template was pBR322 DNA (positions from 56 to 193 of the upper strand) and primers—5′-aattgctaacgcagtcagg-3′ and 5′-P-tggacgatatcccgcaagaggc-3′ (one of the primers was 5′-phosphorylated). Unmodified 137 nt ssDNA was produced via treatment of hemiphosphorylated PCR fragments with exonuclease λ. The reaction was performed under conditions and in the buffer supplied by New England Biolabs. To produce model dsDNA, the ssDNA was annealed with a complementary modified strand. The duplexes were purified in 6% polyacrylamide gel, 1 × TBE under non-denaturing conditions to remove remaining ssDNA.
Whole-cell extracts preparation
Human cervical cancer cell lines (SiHa, C33A and HeLa) and CHO cell line cells were used to prepare whole-cell extracts as described previously (27). Extracts were aliquoted and stored at −70°C.
In vitro NER assay
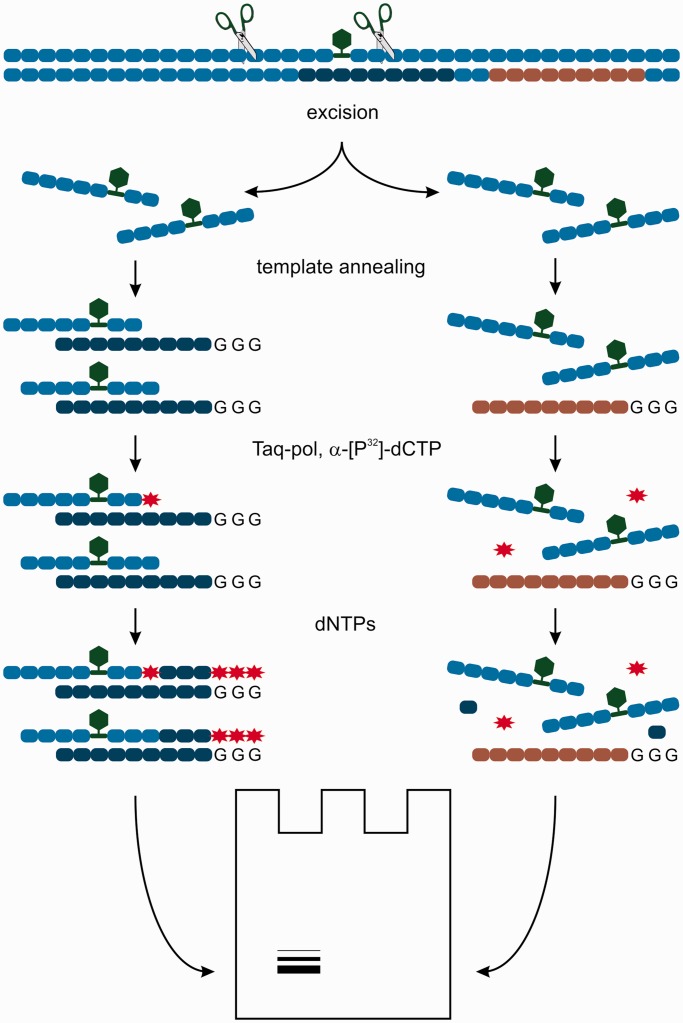
The efficiency of removal of the DNA region containing model lesions was determined using 3′-end-labelling (or ‘fill-in’ synthesis) method as described previously (28), with several modifications. The reaction mixtures containing 20 nM model DNA duplex, 500 nM template 5′-gggggctcggcaccgtcaccctggatgctgtagg-p-3′ (template 1) and NER-competent cell extract (the amounts of proteins and extract type specified in the legends for figures) in 1 × NER buffer (25 mM Tris–HCl, pH 7.8, 45 mM NaCl, 4.4 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid and 4 mM ATP) were incubated for 15–60 min at 30°C. Unlike the original method (28), in vitro NER reaction was performed in the presence of the excess of template 1 to decrease degradation of the excised oligonucleotide by extract nucleases. After incubation, the reaction mixture was heated to 95°C and slowly cooled down to room temperature for annealing of excised oligonucleotide to specific template. The 10 × Taq-polymerase buffer (1/10 of reaction mixture volume), Taq polymerase (5–10 activity units) and α-[32P] dCTP (500–600 Bq) were added, and the probes were incubated for 5 min at 37°C, then 1.2 µl of dNTP mixture containing 100 µM dATP, dGTP, dTTP and 50 µM dCTP was added, and incubation was continued further for 15 min. The reaction was stopped by adding proteinase K solution (0.5 µl, 4 mg/ml). After 60 min of proteinase treatment at 37°C, reaction mixtures were ethanol precipitated and analysed by PAGE under denaturing conditions followed by quantitative autoradiography (Molecular Imager FX Pro+, Quantity One software, BioRad). Quantitative analysis was performed relatively to total radioactivity in each lane, using control lane signal as baseline. The influence of CHO extract protein concentration on the intensity of the excision products was analysed at the initial step, and the optimal concentration of the extract proteins (1.6 mg/ml) was selected for the following analysis.
The control reaction mixtures were processed and analysed according to the procedure described earlier in the text. The first type of control mixture contained non-modified DNA (nm DNA) of the same sequence instead of modified duplex. The second control contained the modified DNAs, but the non-specific template 5′-ggggtcaggcaccgtgtatgaaatctaacaat-p-3′ (template 2), which is complementary to undamaged segment of DNA (see Figure 3 for details). DNA incubated in 1 × NER buffer in the absence of extract and processed as described earlier in the text was used as additional control.
Figure 3.
The scheme of NER in vitro assay. Green hexagon represents a bulky modification; red asterisk, radioactive label; dark blue and grey ovals, the sequence of specific and non-specific templates, respectively.
RESULTS AND DISCUSSION
Many previous investigations with artificial NER substrates have been performed using circular structures (phage or plasmid DNA) bearing lesions in the target position or statistically distributed in the DNA molecule (8,15,29,30).The primary evaluation of the substrate properties of recently suggested photoreactive lesions Fap-dC (-dU) introduced in pBSK II DNA was described (16). Another type of model substrates for the eukaryotic NER system is represented by a long (>120 bp length) linear double-stranded modified DNA (ldsmDNA) bearing bulky modification in the internal position of the DNA strand at approximately equal distances from the ends of the duplex. Despite reported problems in the usage of linear NER substrates [e.g. NER inhibition with several cell extracts proteins, non-specific nuclease degradation and so forth (23,31,32)], modified dsDNA is universal and is a widely used model for in vitro NER investigations. Linear NER substrate analogues of the different length were successfully used to estimate correlation among the damage d DNA structure and repair efficiency, as well as for evaluation of kinetic and affinity parameters of NER proteins interaction with DNA substrates (33–35). Linear NER substrates were used in affinity cross-linking experiments (2,20,36–38) and in modern proteomics (36).
The aim of the present study was to design the new effective DNA probes for detection of damaged strand excision in the NER. The optimal strategy of synthesis and good substrate properties of ldsmDNA demonstrate their usability for NER activity measurement. The solid-phase synthesis using bulky modified non-nucleoside phosphoamidites was suggested as most straightforward approach for creation of model DNA independently of the length and nucleotide sequence. Taking into account the knowledge of the NER recognizable DNA lesions (1,3,4,30), the fluorescein and antracene derivatives have been chosen as bulky substituents.
The method of new non-nucleosidic phosphoramidite reagents synthesis has been elaborated
The synthesis of new non-nucleoside phosphoramidites and their incorporation into oligodeoxyribonucleotides are reported herein.
Building block 1 was synthesized from commercially available 3-amino-1,2-propanediol, as depicted in Figure 1. Initial N-hydroxysuccinimide ester of N-TFA-hexanoic acid (TFA—trifluoroacetic acid) was condensed with 2 to give diol/3, which was dimetoxytritylated without separation and purification to yield the dimetoxytrityl protected diol 4. Universal intermediate 1 was obtained from compound 4 by removal the TFA-protecting group with NH3.
On the basis of the non-nucleosidic reagent 1 (Figure 1), phosphoramidites with fluorophores and other modifications can be obtained. The synthesis of the compound 1 is convenient and involves only three steps. The main difference from the earlier described phosphoramidites containing 1,2-diol structural type is the distance between the amino-group and secondary hydroxyl. An amino group separated from the backbone by the extended linker should allow easy introduction of the phosphoramidites with bulky groups into oligonucleotides. Universal intermediate 1 was converted to derivatives 5 and 7 by its reaction with 9-anthracenecarboxylic acid in the presence of HOBt (hydroxybenzotriazole) and DCC (N,N′-dicycloxeyl-carbodiimid) or with 5(6)-carboxyfluorescein-NHS ester, respectively (Figure 1). Activated esters were synthesized in situ without additional purification. Compounds 5 and 7 were converted to phosphoramidites with N,N,N′,N′-tetraisopropyl-(2-cyano)ethyl phosphodiamidite. The structures of the compounds were confirmed by 1H, 31P NMR and by high-resolution mass spectra.
Synthesized phosphoramidites (6 and 8) are readily soluble in dry acetonitrile and were used for the internal functionalization of the oligonucleotide chain during DNA synthesis. The average coupling efficiency of the modified phosphoramidites at 0.1 M concentration in dry acetonitrile and 5 min reaction time was found to be 95–97%. Oligonucleotides (ONTs) were assembled, removed from their supports, de-protected by concentrated aqueous ammonia treatment at 60°C overnight and analysed by reverse-phase high-performance liquid chromatography and MALDI–TOF MS. Sequences and properties of the oligodeoxyribonucleotides bearing various modifications are summarized in Table 1. The obtained yields were good, and all the molecular masses corresponded to the expected values within the accuracy of their measurement.
Table 1.
The sequences and masses of oligodeoxyribonucleotides with non-nucleosidic modifications (positions in bold indicate insertion of non-nucleosidic phosphoramidites: M1 – 6, M2 – 8)
| No | Sequence 5′→3′ | Calc. masses (M + H)+ | Observed masses (M + H)+ |
|---|---|---|---|
| I-Ant | ATCCAGGG-M1-GACGGTG | 5129.0 | 5129.8 |
| I-Flu | ATCCAGGG-M2-GACGGTG | 5281.0 | 5283.7 |
| II-Ant | CACCGTCG-M1-CCTGGAT | 5000.0 | 5002.2 |
| II-Flu | CACCGTCG-M2-CCTGGAT | 5152.0 | 5153.3 |
The synthesized ONTs (Table 1) contain 15 standard nucleotides and 1 non-nucleoside fragment (Figure 2) and have to imitate 16mer ONT.
We have analysed properties of all modified oligonucleotides (Table 1) as substrates of template-dependent enzyme—T4 polynucleotide ligase. 5′-phosphorylated ONT, containing n-Ant and n-Flu were efficiently incorporated in long DNA structures. Good substrate properties revealed by modified oligonucleotides in the reaction of enzymatic ligation allowed synthesis of a set of 137-nt DNA, containing bulky non-nucleoside groups. After purification (PAGE, 7 M urea) and quantification, the modified 137mer ssDNA was used for annealing with complementary DNA to create 137-bp double-stranded model DNA.
Testing of the model DNA as substrates for in vitro NER assay. Analysis of the NER reaction in the different cell extracts
The 137-bp linear dsDNAs containing bulky modification in the internal position of the molecule (position 68 from 5′-end) have to meet the DNA size and damage-position requirements to allow assembly of the functional NER machinery on artificial NER substrates (3). Substrate properties of nAnt– and nFlu–DNA have been evaluated using NER-competent extracts of eukaryotic cells. The excision reaction (1) results in the release of short oligonucleotides (24–32mer) containing the lesion. It should be noted that the levels of substrate conversion are usually low for in vitro NER assays (10,11,30). Also, the specific excision of the damaged fragments from linear DNA–substrate might be additionally decreased because of the action of several proteins presented in cell extracts (31,32).
Both of most widely used methods of dual incision products detection are based on 32[P]-label use.
The method of direct detection of NER cleavage was developed in 1992 (39) and was used to perform an impressive number of investigations (3,40–43). However, this approach has some disadvantages. First of all, high amount of 32[P] should be used (27) to produce the substrate DNA of high-specific radioactivity. Moreover, the process of DNA–substrate synthesis consists of complicated procedures, which have to be performed with high-radioactive substances, especially at the initial stages. Also, the half-life of radioactive substrates is relatively short, and application of synthesized radioactive DNA for full-scale research is time-limited.
These problems were mainly solved by the development of indirect detection of dual incision, catalysed by NER system. This method was suggested by Wood and colleagues (28). 32[P]-label was introduced into 3′-ends of the specific excision products using DNA polymerase, α-[32P]-dNTP and complementary template ONT. It was clearly demonstrated that the bands obtained by this method correspond to the products of specific excision catalysed by NER system. For now, this method represents the accepted technique to analyse activity of NER system and is widely used for a variety of model DNA substrates (2,16,44,45). Indirect analysis has several advantages in comparison with direct method of cleavage detection. First of all, indirect method requires substantially less amount of radioactivity but shows the superior sensitivity. In addition, the non-radioactive model DNA substrates are stable when stored. The scheme of the approach used in this study is presented in Figure 3.
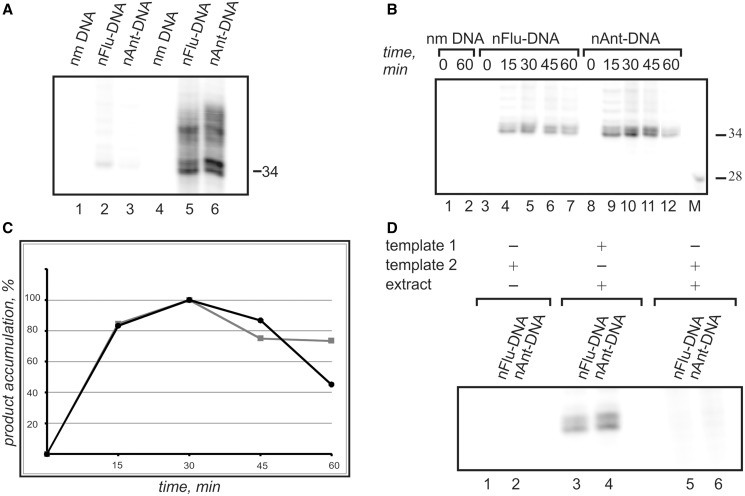
It was known from our previous study that the whole-cell extracts may contain non-specific nucleases. Such nucleases effectively digest single-stranded DNA, which is the main product of the excision reaction. To improve the original procedure (28), the subsequent modification was developed here. The excess of template 1 was introduced into reaction mixture before incubation of the model NER substrates with cell extracts to protect single-stranded excision products from their digestion by nucleases of cell extracts. It permits to increase significantly the resulting signal of the excision products (Figure 4A).
Figure 4.
The NER dual incision activity of CHO cells extract. The excision products were detected by annealing to the template followed by end-labelling using α-[32P]-dCTP and Taq DNA polymerase. The reaction products were resolved on a 10% denaturing polyacrylamide gel. (A) The comparison between original (28) (lanes 1–3) and modified (lanes 4–6) in vitro NER assays. In contrast to the original method the presence of template 1 in reaction mixture during the incubation of the model DNA substrate with cell extract increases the observed signal intensity. Model DNAs were incubated for 30 min at 30°C with cell extracts prepared from CHO cells (20 nМ DNA, 1.6 mg/ml of extract proteins with or without 500 nM template 1). (B) Specific product accumulation. The 20 nМ model DNAs was incubated for 15–60 min at 30°C with 1.6 mg/ml of extract proteins and 500 nM template 1. (C) Time dependence of the specific excision products accumulation. Grey squares correspond to nFlu–DNA; black circles, nAnt–DNA. The maximal products accumulation was taken as 100% and was detected for samples incubated for 30 min. The subsequent incubation leads to non-specific products degradation. (D) Comparison of the results obtained with template 1 and template 2.
Wild-type CHO cells were used to prepare ‘NER-competent’ extracts (27). Incubation of nFlu– and nAnt–DNA with proteins of NER-competent extract derived of CHO cells followed by ‘fill-in’ synthesis resulted in the appearance of the major bands of the radioactive DNA fragments of specific length and sequence (Figure 4B). The maximal accumulation of the excision products was detected after incubation of samples for 30 min (Figure 4C). The appearance of the multiple bands reflects the existence of different sites of excision.
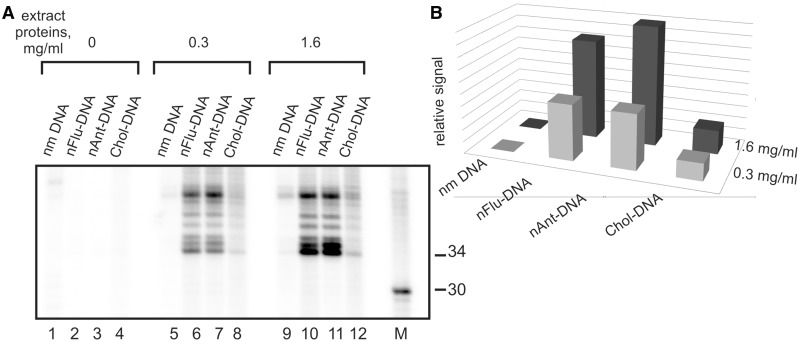
As irrelevant nuclease activities are present in the whole-cell extracts, the specificity of excision reaction of bulky lesion from ldsmDNA catalysed by proteins of the extracts was the subject under particular investigation. The products of specific excision were not observed when the control non-modified DNA was incubated with the extract (Figure 4B, lines 1, 2), or in the case when ‘non-specific’ template 2 (see Figure 2 for details) was used in the experiments (Figure 4D). The nFlu– and nAnt–DNA were evaluated as three to five times more efficient substrates in comparison with Chol–DNA (Figure 5). To the best of our knowledge, this is the first direct comparison of the excision efficiency for Chol-containing DNA with other non-nucleoside substrates.
Figure 5.
(A) The NER dual incision activity of CHO cells extract. Model DNAs were incubated for 30 min at 30°C with cell extracts prepared from CHO cells (20 nM model DNA, 0.3 or 1.6 mg/ml of extract proteins and 500 nM template 1).The excision products, containing nFlu, nAnt or Chol, were detected by annealing to a template followed by end-labelling using α-[32P]-dCTP and Taq DNA polymerase. The reaction products were resolved on a 10% denaturing polyacrylamide gel. Non-modified 137-bp DNA was used as a negative control; 32P-labelled oligonucleotides were used as size marker (lane M). (B) Relative signal of the target products for non-modified, nFlu– and nAnt–DNA after incubation with different concentrations of the extract proteins.
Recently the series of 137mer DNAs bearing bulky modifications as substitutions of nucleobase (including Flu-dU and Ant-dC) were analysed in vitro NER assay. The affinity of the model DNA bearing this type of lesion to NER proteins (46–48) and their capability to be efficient competitors for DNA substrate, containing benz[a]pyrene residue attached to dG, was found. However, the excision reaction of the relatively low level was found only for linear 137mer Flu-dU–DNA (20). The products of NER-specific excision in HeLa cell extract were also not observed using linear 120mer DNA containing well-defined damage, namely, AAF-dG (9). AAF-dG was efficiently repaired by NER system in vitro only when it was introduced into circular DNA (9,15). As it was aforementioned, the other activities in the cell extracts, such as a non-specific nucleases and DNA-binding proteins of high affinity to DNA, particularly Ku 70–80 antigen, the protein, high content of which is so typiсal for cancer cells extracts, can impede detection of the NER reaction for ldsmDNA (23,31,32).
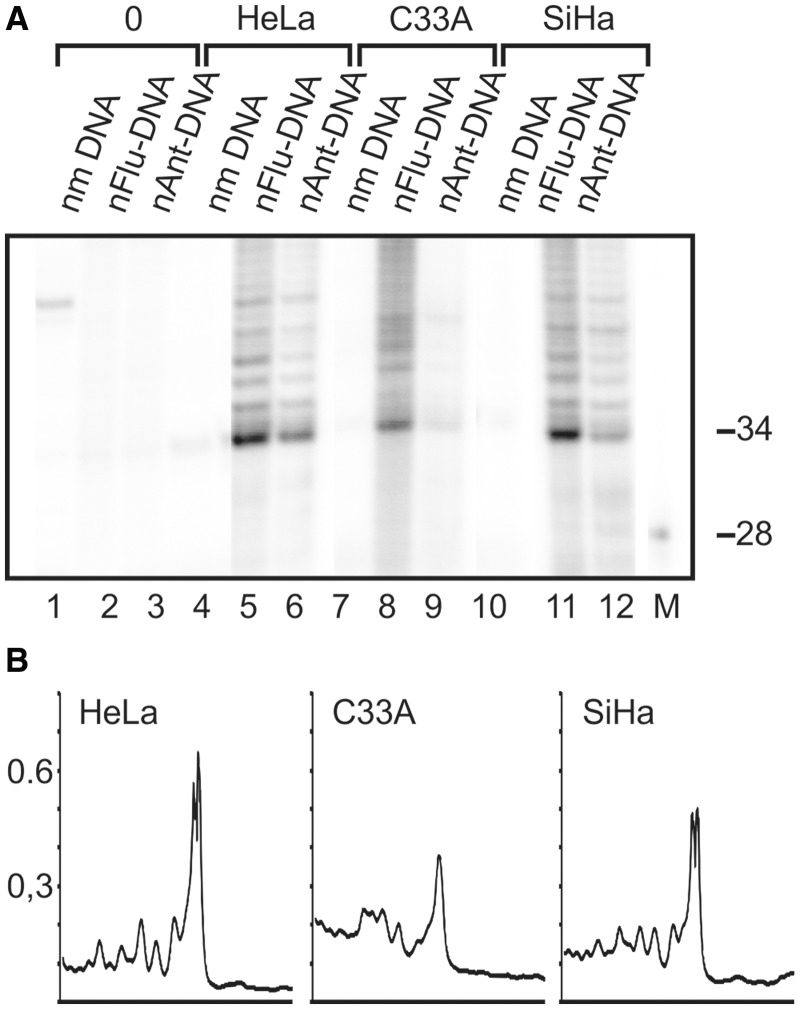
In the present investigation, the new model DNAs were used to test NER dual incision activity of cancer cells extracts produced using HeLa, SiHa and C33A human cancer cell lines. It was observed that NER system of these cells also efficiently removed the damaged DNA fragments from nFlu– and nAnt–DNAs (Figure 6).
Figure 6.
(A) The NER dual incision activity of cancer cells extracts. Model DNAs were incubated for 30 min at 30°C with cell extracts prepared from HeLa, C33A or SiHa cells (20 nМ DNA, 1.6 mg/ml of extract proteins and 500 nM template 1). The excision products were detected by annealing to template followed by end-labelling using α-[32P]-dCTP and Taq DNA polymerase. The reaction products were resolved on a 10% denaturing polyacrylamide gel. Non-modified 137-bp DNA was used as a negative control; 32P-labelled oligonucleotides were used as size marker. (B) Densitometric analysis of the lanes 5 (HeLa, left), 8 (C33A, middle) and 11 (SiHa, right).
The results obtained indicate that the new model lesions imitate efficient NER substrates.
SUMMARY AND CONCLUSIONS
We have reported the synthesis of the novel non-nucleoside phosphoamidites for the solid-phase synthesis of deoxyoligoribonucleotides containing bulky modifications. The method of their incorporation into DNA and analysis of the obtained model DNAs as substrates of mammalian NER system were developed. The designed phosphoamidites can be used in the standard solid-phase synthesis protocol to synthesize similar oligonucleotides of any length and sequence. Mammalian NER system processes long double-stranded model DNA containing nFlu- and nAnt-lesions with high efficiency in different cell extracts. The possibility to generate modified DNA containing NER-recognizable lesions using solid-phase synthesis is a significant advance. It provides unimpaired access to the well-defined efficient substrates for NER studies. The additional advantages of the new DNA derivatives are good substrate properties of the relatively short nAnt- and nFlu-oligonucleotides in the reactions of enzymatic ligation using T4 polynucleotide ligase and elongation using DNA polymerases). This opens new perspectives for design of the efficient DNA probes for affinity cross-linking and for modern proteomic research. New artificial DNA, containing bulky non-nucleoside modifications will be useful for medical research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Russian Fund for Basic Research [RFBR-12-04-00487а], [RFBR-12-04-33162]; Russian Ministry of Education and Science [projects 14.B37.21.0188 and 02.740.11.0079]; Program of Russian Academy of Science on Molecular and Cellular Biology. Funding for open access charge: Russian Fund for Basic Research [RFBR-12-04-00487а]; Program of Russian Academy of Science on Molecular and Cellular Biology.
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Professor Alexandre Boutorine (Museum National d’Histoire Naturelle, Paris) for careful reading of the manuscript and his advices.
REFERENCES
- 1.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 2.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang JC, Sancar A. Determination of minimum substrate size for human excinuclease. J. Biol. Chem. 1994;269:19034–19040. [PubMed] [Google Scholar]
- 4.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 5.DellaVecchia MJ, Croteau DL, Skorvaga M, Dezhurov SV, Lavrik OI, Van Houten B. Analyzing the handoff of DNA from UvrA to UvrB utilizing DNA-protein photoaffinity labeling. J. Biol. Chem. 2004;279:45245–45256. doi: 10.1074/jbc.M408659200. [DOI] [PubMed] [Google Scholar]
- 6.Hermanson-Miller IL, Turchi JJ. Strand-specific binding of RPA and XPA to damaged duplex DNA. Biochemistry. 2002;41:2402–2408. doi: 10.1021/bi0112863. [DOI] [PubMed] [Google Scholar]
- 7.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair (Amst) 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillet LC, Alzeer J, Scharer OD. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified ‘ultra-mild' DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, et al. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell. Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan QA, Kohlhagen G, Marshall R, Austin CA, Kalena GP, Kroth H, Sayer JM, Jerina DM, Pommier Y. Position-specific trapping of topoisomerase II by benzo[a]pyrene diol epoxide adducts: implications for interactions with intercalating anticancer agents. Proc. Natl Acad. Sci. USA. 2003;100:12498–12503. doi: 10.1073/pnas.2032456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alzeer J, Scharer OD. A modified thymine for the synthesis of site-specific thymine-guanine DNA interstrand crosslinks. Nucleic Acids Res. 2006;34:4458–4466. doi: 10.1093/nar/gkl587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maltseva EA, Rechkunova NI, Gillet LC, Petruseva IO, Scharer OD, Lavrik OI. Crosslinking of the NER damage recognition proteins XPC-HR23B, XPA and RPA to photoreactive probes that mimic DNA damages. Biochim. Biophys. Acta. 2007;1770:781–789. doi: 10.1016/j.bbagen.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Khodyreva SN, Lavrik OI. Photoaffinity labeling technique for studying DNA replication and DNA repair. Curr. Med. Chem. 2005;12:641–655. doi: 10.2174/0929867053202179. [DOI] [PubMed] [Google Scholar]
- 18.Maltseva EA, Rechkunova NI, Petruseva IO, Silnikov VN, Vermeulen W, Lavrik OI. Interaction of nucleotide excision repair factors RPA and XPA with DNA containing bulky photoreactive groups imitating damages. Biochemistry (Mosc) 2006;71:270–278. doi: 10.1134/s0006297906030060. [DOI] [PubMed] [Google Scholar]
- 19.Petruseva IO, Tikhanovich IS, Maltseva EA, Safronov IV, Lavrik OI. Photoactivated DNA analogs of substrates of the nucleotide excision repair system and their interaction with proteins of NER-competent HeLa cell extract. Biochemistry (Mosc) 2009;74:491–501. doi: 10.1134/s0006297909050034. [DOI] [PubMed] [Google Scholar]
- 20.Evdokimov AN, Petruseva IO, Pestryakov PE, Lavrik OI. Photoactivated DNA analogs of substrates of the nucleotide excision repair system and their interaction with proteins of NER-competent extract of HeLa cells. Synthesis and application of long model DNA. Biochemistry (Mosc) 2011;76:157–166. doi: 10.1134/s0006297911010159. [DOI] [PubMed] [Google Scholar]
- 21.Matsunaga T, Mu D, Park CH, Reardon JT, Sancar A. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- 22.Verhoeven EE, Wyman C, Moolenaar GF, Hoeijmakers JH, Goosen N. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J. 2001;20:601–611. doi: 10.1093/emboj/20.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langie SA, Cameron KM, Waldron KJ, Fletcher KP, von Zglinicki T, Mathers JC. Measuring DNA repair incision activity of mouse tissue extracts towards singlet oxygen-induced DNA damage: a comet-based in vitro repair assay. Mutagenesis. 2011;26:461–471. doi: 10.1093/mutage/ger005. [DOI] [PubMed] [Google Scholar]
- 24.Adamczyk M, Chan CM, Fino JR, Mattingly PG. Synthesis of 5- and 6-hydroxymethylfluorescein phosphoramidites. J. Org. Chem. 2000;65:596–601. doi: 10.1021/jo991449h. [DOI] [PubMed] [Google Scholar]
- 25.Agrawal S. Protocols for Oligonucleotides and Analogs: Synthesis and Properties in Methods in Molecular Biology. Totowa, NJ: Humana Press; 1993. [Google Scholar]
- 26.Gait M, editor. Oligonucleotide Synthesis. A Practical Approach. IRL Press Lim: Oxford; 1984. [Google Scholar]
- 27.Reardon JT, Sancar A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- 28.Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 29.Hess MT, Gunz D, Naegeli H. A repair competition assay to assess recognition by human nucleotide excision repair. Nucleic Acids Res. 1996;24:824–828. doi: 10.1093/nar/24.5.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buterin T, Hess MT, Gunz D, Geacintov NE, Mullenders LH, Naegeli H. Trapping of DNA nucleotide excision repair factors by nonrepairable carcinogen adducts. Cancer Res. 2002;62:4229–4235. [PubMed] [Google Scholar]
- 31.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl Acad. Sci. USA. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calsou P, Frit P, Salles B. Double strand breaks in DNA inhibit nucleotide excision repair in vitro. J. Biol. Chem. 1996;271:27601–27607. doi: 10.1074/jbc.271.44.27601. [DOI] [PubMed] [Google Scholar]
- 33.Trego KS, Turchi JJ. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45:1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002;41:6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]
- 35.Hey T, Lipps G, Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry. 2001;40:2901–2910. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 36.Guggenheim ER, Xu D, Zhang CX, Chang PV, Lippard SJ. Photoaffinity isolation and identification of proteins in cancer cell extracts that bind to platinum-modified DNA. Chembiochem. 2009;10:141–157. doi: 10.1002/cbic.200800471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khodyreva SN, Prasad R, Ilina ES, Sukhanova MV, Kutuzov MM, Liu Y, Hou EW, Wilson SH, Lavrik OI. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1) Proc. Natl Acad. Sci. USA. 107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukhanova MV, D'Herin C, van der Kemp PA, Koval VV, Boiteux S, Lavrik OI. Ddc1 checkpoint protein and DNA polymerase varepsilon interact with nick-containing DNA repair intermediate in cell free extracts of Saccharomyces cerevisiae. DNA Repair (Amst) 2011;10:815–825. doi: 10.1016/j.dnarep.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 39.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansen J, Olsen AK, Wiger R, Naegeli H, de Boer P, van Der Hoeven F, Holme JA, Brunborg G, Mullenders L. Nucleotide excision repair in rat male germ cells: low level of repair in intact cells contrasts with high dual incision activity in vitro. Nucleic Acids Res. 2001;29:1791–1800. doi: 10.1093/nar/29.8.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol. Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 42.Ding S, Kropachev K, Cai Y, Kolbanovskiy M, Durandina SA, Liu Z, Shafirovich V, Broyde S, Geacintov NE. Structural, energetic and dynamic properties of guanine(C8)-thymine(N3) cross-links in DNA provide insights on susceptibility to nucleotide excision repair. Nucleic Acids Res. 40:2506–2517. doi: 10.1093/nar/gkr1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2012;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petruseva IO, Tikhanovich IS, Chelobanov BP, Lavrik OI. RPA repair recognition of DNA containing pyrimidines bearing bulky adducts. J. Mol. Recognit. 2008;21:154–162. doi: 10.1002/jmr.877. [DOI] [PubMed] [Google Scholar]
- 47.Krasikova YS, Rechkunova NI, Maltseva EA, Petruseva IO, Lavrik OI. Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair. Nucleic Acids Res. 2011;38:8083–8094. doi: 10.1093/nar/gkq649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krasikova YS, Rechkunova NI, Maltseva EA, Petruseva IO, Silnikov VN, Zatsepin TS, Oretskaya TS, Scharer OD, Lavrik OI. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry (Mosc) 2008;73:886–896. doi: 10.1134/s0006297908080063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.