Abstract
The usability of many high-throughput lab-on-a-chip devices in point-of-care applications is currently limited by the manual data acquisition and analysis process, which are labor intensive and time consuming. Based on our original design in the biochemical reactions, we proposed here a universal approach to perform automatic, fast, and robust analysis for high-throughput array-based microfluidic immunoassays. Inspired by two-dimensional (2D) barcodes, we incorporated asymmetric function patterns into a microfluidic array. These function patterns provide quantitative information on the characteristic dimensions of the microfluidic array, as well as mark its orientation and origin of coordinates. We used a computer program to perform automatic analysis for a high-throughput antigen/antibody interaction experiment in 10 s, which was more than 500 times faster than conventional manual processing. Our method is broadly applicable to many other microchannel-based immunoassays.
INTRODUCTION
The usability of many high-throughput lab-on-a-chip devices in point-of-care applications is limited by the manual data acquisition and analysis process, which are labor intensive and time consuming.1 One such example is the DNA/protein microarray.2, 3 From the past development of microarray technologies, we could see that image analysis may even influence experimental conclusions and hence critical for a right implementation of high-throughput assays.4 Advanced array-based analytical devices further enabled massively parallel detection of proteins or cells from various kinds of biological samples,5, 6 which has the same biochemical principle of specific interaction between targets and receptors.7 The fluorescence image of the microarray could be obtained quickly, however, some portion of image processing and data analysis must be performed manually by an experienced technician,8 which determines the important parameters, such as the size of the reaction region of interest (ROI) and the location of each array grid. This problem is not so obvious in low-throughput bioassays,7 but is highly prominent for two-dimensional (2D) bioassays,9, 10, 11 which have greater information storage capacity than one dimensional (1D) bioassay structures.12, 13 Toward a high-throughput assay system, recent developments in technologies allow controlled surface modification and enable a single microfluidic device to contain dozens of microfluidic channels,14, 15, 16 and the analytes in each channel can be serially diluted, such that the concentration of targets can be quantified;17 however, the effort required for manual interventions increases quadratically with the number of microfluidic channels.
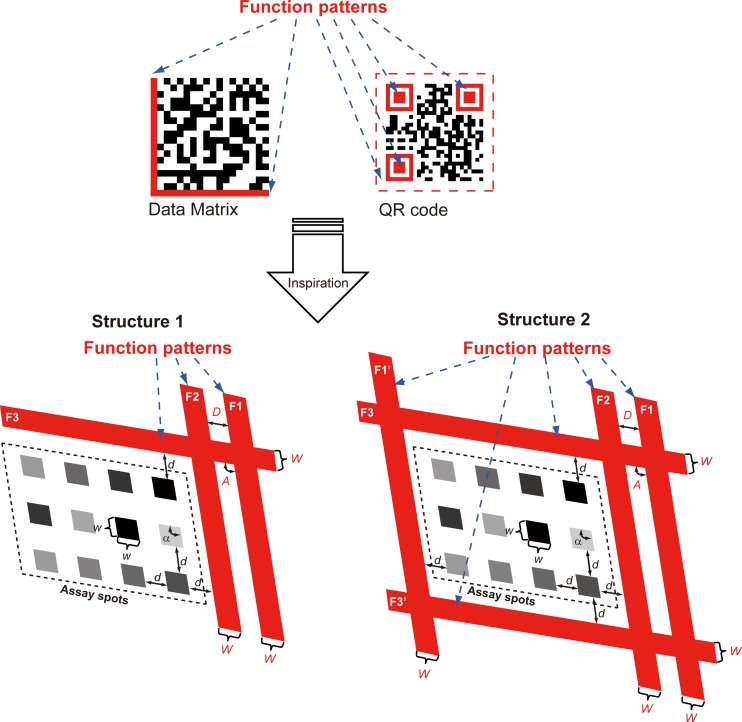
Inspired by 2D barcodes (or matrix barcodes), we presented here an interdisciplinary approach to facilitate automatic and accurate analysis of 2D bioassays. Most 2D barcodes, such as the widely used Data Matrix18 and QR Code19 illustrated in Fig. 1, contain “function patterns,” which are asymmetric features within the 2D barcode. The function patterns define the orientation and the area of the ROI, as well as the characteristic dimensions of the data elements in the 2D barcode. This allows hand-held scanners to automatically decode 2D barcodes, regardless of the orientation or size of the 2D image. Conventional high-throughput bioassays do not contain such function patterns. So far, in order to acquire data from the bioassays, the position of the ROI had to be manually defined. Current softwares used for microarray analysis commonly achieve this by predefining the shape mask of the ROI, and the shape mask is then manually adjusted in the software to fit the ROI. This is the initial step known as “gridding” for microarray image analysis, and the automation for gridding will open the door to efficient high-throughput analysis.20 State-of-the-art approaches discussed in recent papers for an automatic gridding in microarray were exclusively based on image processing techniques,21, 22, 23, 24, 25, 26 thus the corresponding algorithms are complex in order to tolerate with various possible situations, even so, some of them still have only limited applicabilities. For example, the power spectra of the image need to be calculated repeatedly in the rotate correcting algorithm,21 or both of the Radon transform and Bayesian image analysis take huge computational time,22, 24, 25 resulting in increased computational complexity even making them inefficient on large data sets. The other approaches such as hierarchical refinement algorithm23 are powerless in the presence of large experimental contaminations or reaction spots with too low intensities; the shock filter-based approach26 reduced computational complexity but does not deal with the rotation problem. In comparison to these limited studies in the field of microarray, researchers care much less than they do about the array-based immunoassay. Fewer groups focused on the bottleneck problem and disclosed a few details about data analysis for the construction of their automated immunoassay system.27, 28 However, these strategies may not completely satisfy the needs of data analysis, e.g., some are based on a pre-defined rectangular digital mask that may be restricted by the precision of perpendicular alignment of two pieces of microfluidic chips and the positioning of the planar substrate to the CCD imaging array, which can be mechanically complicated and thus not sufficiently robust. Others require sufficiently large (1 mm) spots in the array in order to improve the relative tolerances of the processing algorithm. Previous studies and reported approaches, therefore, were not convincingly demonstrated for their processing performance.
Figure 1.
Schematic illustration of the microfluidic framework with built-in function patterns (in red), inspired by 2D barcodes. Two structures are proposed. Structure 1 is a semi-open frame structure, similar to the Data Matrix scheme. Structure 2 is a closed-frame structure, similar to the QR code scheme. F1, F2, F3, F1′, F3′: function pattern. Α, D, W: characteristic angle, spacing, and width of the function patterns. α, d, w: characteristic angle, spacing, and width of the array spots.
Instead of complex algorithms or programming, we proposed a biochemical approach, and designed high-throughput 2D immunoassay arrays with built-in function patterns. Briefly, we introduced some auxiliary objects (i.e., the function patterns) into the original array, and these objects can serve as the orientation and the characteristic dimensions of the array. More importantly, the function patterns are realized by our specially designed biochemical reactions representing positive controls. We carried out antigen-antibody interaction experiment as a typical example, and automatically identified and characterized the incorporated function patterns from the immunoassay images, which determined the positions of the individual array spots, as well as read and analyzed the fluorescence intensity for each array spot. In this way, the entire analyzing steps are automated and can be done within 10 s, dramatically speeding up the high-throughput immunoassay as a whole.
MATERIALS AND METHODS
Materials
SU-8 2100 photoresist was from MicroChem. Polydimethylsiloxane (PDMS) (Sylgard 184) was from Dow Corning. Albumin bovine serum-fraction V (BSA) was from Merck. Rabbit IgG (affinity purified) and rabbit antiserum against BSA were from Bioss. Recombinant human immunodeficiency virus (HIV)-1 envelope gp41 antigen (6.85 μM) and rabbit antiserum against gp41 antigen (concentration unknown) were from ProSpec-Tany. Cy3-labeled goat anti-rabbit IgG was from Beyotime Institute of Biotechnology. Phosphate-buffered saline (PBS, pH 7.2) was from Beyotime Institute of Biotechnology, and we added 0.05% (v/v) Tween-20 (from Sigma) into the PBS to prepare PBST as the wash solution.
Microfluidic design
The automatic gridding for the array leads to an automated analysis, and automatic gridding can be achieved by extracting several characteristic dimensions from the function patterns. The incorporation of the function patterns into the microfluidic network is illustrated in Fig. 1.
We proposed two function pattern structures, as illustrated in Fig. 1. In structure 1, the function patterns consist of three solid bands (F1, F2, and F3). F1 and F2 are parallel to each other. The corner where F1/F2 intersects with F3 marks the origin of the array. These solid bands can be automatically detected with the aid of a pattern recognition algorithm. The beginning (and the ending) of the array can be determined by the recognized bands, indicating the start point of following recursive operation for the matrix. All necessary parameters for determining the grid can then be characterized by the function patterns. The asymmetric arrangement of the function patterns defines the orientation of the image. For both structures: the spacing between adjacent spots (d) is equal to the distance between F1 and F2 (D); the width of each spot (w) is equal to the width of F1 (W); the angle of the array (α) is equal to the intersectional angle between F1 and F3 (A), which is only determined after the recognition of function patterns. If the characteristic dimensions of the array are known (only for structure 1), the array grids can be determined without ambiguity from the predefined d and w. The signal intensity (i.e., gray value) of each spot can be automatically extracted.
In many applications, the characteristic dimensions of the array may be unknown prior to the image analysis. Thus, it would be helpful to build in a method to determine the array size online. There inevitably are cumulative errors during recursive operation in structure 1 caused by single pixel misrecognition for the edges of F1 ∼ F3 if we do not predefine d and w in the program. The determination of the unknown array size is achieved in structure 2, where F1′ (parallel to F1) and F3′ (parallel to F3) are added to enclose the ROI of the array. Structure 2 is also useful for the analysis of large arrays aiming at assays with higher throughput. In structure 2, the cumulative errors can be reduced through the subtraction of coordinates of the opposite sides followed by division by the number of channels.
Device fabrication
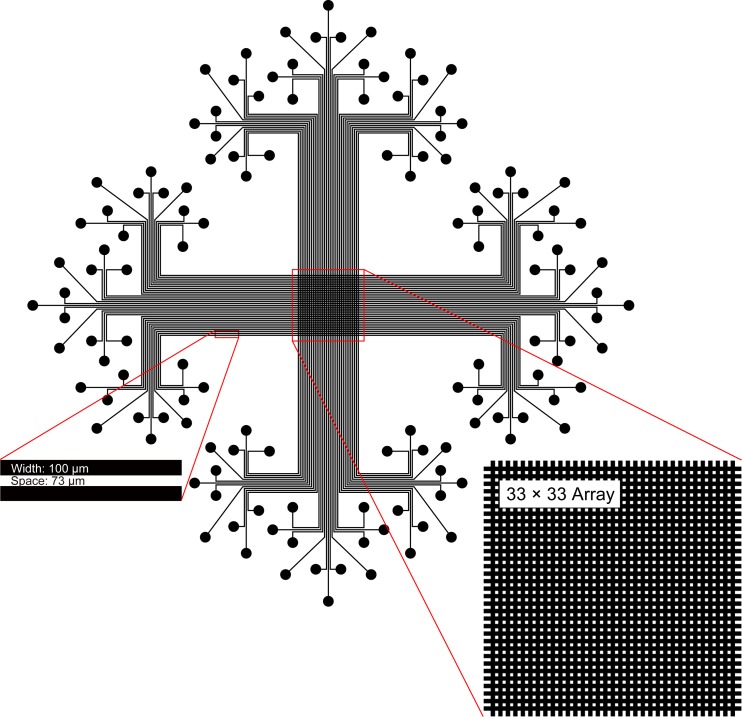
We designed the photomask by cad software and fabricated the master consisting of 33 channels by photolithography, in accordance with our previous report.29 We transferred the relief structure to the PDMS by replica molding. We assembled the PDMS channels with a PDMS slab to generate the sealed channels which guide the liquid flow. This operation is quite simple and only takes a couple of seconds. We generated a 33 × 33 matrix by peeling off the first piece of PDMS channels and then assembling the second piece of PDMS channels perpendicular to the previous one with the same PDMS slab (Fig. 2).
Figure 2.
Layout of the 2D microfluidic chip.
Arrayed immunoassays
We carried out immunoassays by a modified protocol, compared with conventional arrayed microfluidic immunoassays.9 This protocol can provide the function patterns with fluorescence (Figs. 4a, 5a).
Figure 4.
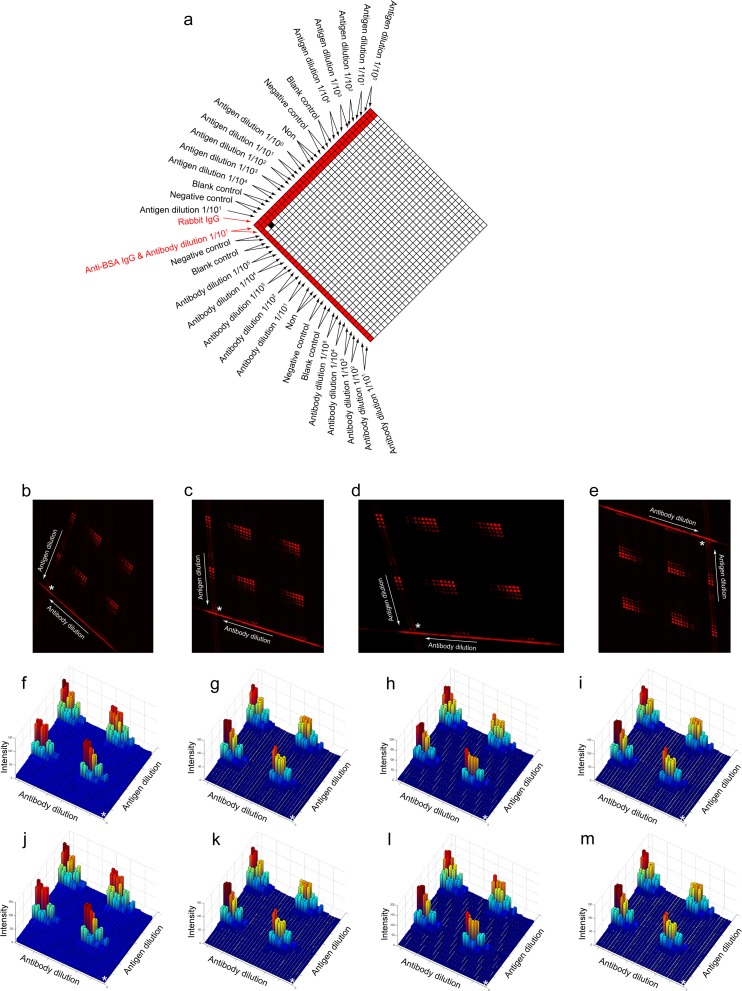
The high throughput immunoassay using the semi-opened function pattern-incorporated microfluidic network (structure 1). (a) The scheme for sample introduction. This is a concentration gradient experiment. We highlight the function patterns in red. (b)–(e) We captured the fluorescence images in different styles. (f)–(i) The 3D plots of the arithmetic mean for (b)–(e), respectively. (j)–(m) The 3D plots of the median for (b)–(e), respectively. The white asterisks in (f)–(m) indicate the origin of the 3D plots and correspond to the white asterisks in (b)–(e), respectively. This origin is shown in (a) in a black square.
Figure 5.
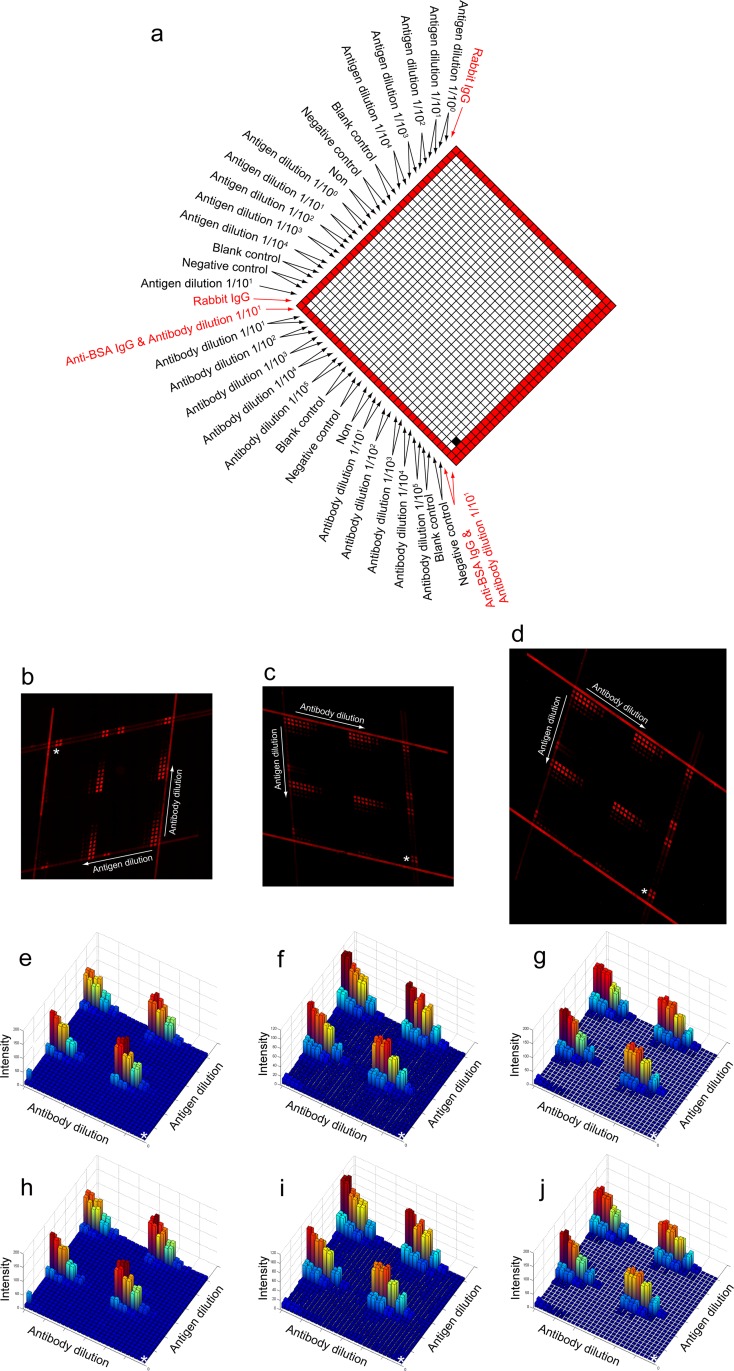
The high throughput immunoassay using the closed function pattern-incorporated microfluidic network (structure 2). (a) The scheme for sample introduction. This is a concentration gradient experiment. We highlight the function patterns in red. (b)–(d) We captured the fluorescence images in different styles. (e)–(g) The 3D plots of the arithmetic mean for (b)–(d), respectively. (h)–(j) The 3D plots of the median for (b)–(d), respectively. The white asterisks in (e)–(j) indicate the origin of the 3D plots and correspond to the white asterisks in (b)–(d), respectively. This origin is shown in (a) in a black square.
We introduced serially diluted gp41 antigen (including the blank control and negative control) in duplicate into each channel except for the last one (for structure 1) or for the first one and the last one (for structure 2) where we introduced rabbit IgG. After incubation, we removed the antigen solutions and the PDMS channels. The gp41 antigen and rabbit IgG were immobilized onto the surface in a form of strips. We blocked the other regions of the surface by 5% BSA solution. After washing and drying, we assembled a new piece of PDMS channels perpendicular to the strips of immobilized antigens with the PDMS slab. We introduced serially diluted rabbit antiserum against gp41 antigen (also including the blank control and negative control) into each channels except for the last two (for structure 1) or for the first one and the last two (for structure 2) where we introduced a mixture of rabbit antiserum against gp41 antigen and rabbit antiserum against BSA. After incubation, we removed the antibody solution and the PDMS channels. The antigen-antibody interaction (i.e., gp41 antigen and anti-gp41 antibody, BSA and anti-BSA antibody) occurred in the cross-areas. Finally, we incubated the whole region by anti-rabbit secondary antibody labeled with Cy3 fluorophore. The secondary antibody bound to the rabbit antibodies (i.e., rabbit anti-gp41 antibody and rabbit anti-BSA antibody) and rabbit IgG. We washed the surface of reaction substrate by PBST and dried it before taking fluorescence images.
Image acquisition
The fluorescence images were captured by a microscope (DMI 6000B, Leica) with a filter set (excitation 515-516 nm; dichromatic mirror: 580 nm; suppression: 590 nm), and the original microscope files were exported into the digital images which were used for the following automatic analyses.
Programming
We developed a matlab program as well as graphical user interface (GUI). The basic flow of the program is in Fig. 3. This program aims at the automatic acquisition of quantitative data.
Figure 3.

Flow chart of the program.
We used the function “edge” and the edge detector “Canny”30 provided by matlab to perform edge detection.31 There are several detectors for edge detection, among which Canny is the most powerful edge detector supported by the function edge. It uses two thresholds to detect strong and weak edges respectively, and includes the weak edges in the final output only if they are connected to strong edges. Other detectors (e.g., Sobel, Prewitt, Roberts, LoG, and Zerocross) are more sensitive to noises in an image, and nowadays may only be used in some hardware realization because of their simple calculation. Canny is therefore less likely than the others to be fooled by the noise, and more suitable for detecting true weak edges. The algorithm of Canny uses Gaussian filter to reduce the noise of the edge and performs edge linking by incorporating the weak pixels. Hence, we could obtain the exquisite edge that is favorable for realizing the Hough transform followed.
After the edge detection, the image turned to a binary image consisting of discontinuous pixels. We used Hough transform to perform the line detection, i.e., assemble edge pixels into meaningful edges.32 The Hough space contains peaks if the binary image contains lines. In our study, we should detect multiple lines (6 lines for structure 1, 10 lines for structure 2), that is to say we need to find out multiple peaks through the Hough transform. So we used the function “houghpeaks” provided by matlab to perform the peak detection. At this point in the program, we accomplished the most crucial part of the processing. We obtained a group of 4 parallel lines (F1, F2) and another group of 2 parallel lines (F3) for structure 1, and a group of 4 parallel lines (F3, F3′) and another group of 6 parallel lines (F1, F2, F1′) for structure 2, respectively. We thus determined the beginning of the array, as well as the orientation of the image.
The following work is the gridding of the image, based on the above parameters we obtained. Particularly for structure 1, we measured the characteristic width W (100 μm, 38.5 pixels in our microfluidic chip) and the characteristic distance D (73 μm, 28.1 pixels in our microfluidic chip) beforehand, and input these values into the program. We predefined that the central point of the image locates within the region of assay spots (because we normally capture images in the center of the visual field). So we know the positive direction of the axes of the gridding network. On the other hand for structure 2, the region surrounded by the 10 lines is the assay region, and the characteristic dimensions (i.e., the width W and the distance D) can be determined by the opposite groups of lines and the number of the channels between these lines. This strategy used in structure 2 could apparently reduce the cumulative errors. Because of the vague edges for some assay spots, we shrank the grids inward. We performed the gridding by simple recursion for the whole image, according to the width, spacing, angle, beginnings, and positive directions. Each spot will be surrounded by four grid lines, so the coordinate of each spot can be determined.33
After gridding, we could extract the gray value of the spots according to the coordinates determined above. In our study, the images captured by the microscope equipped with a monochrome CCD (Leica DFC350 FX) is pseudo-color. We used Cy3 as the fluorescent dye (emission max: 570 nm) and defined its color as red. So we extracted the gray value from the Red Channel of the image.
Finally, the program saves the data in tabular format into a text file and displays 3D-plots representing the intensities of every spots.33
RESULTS AND DISCUSSION
Efficiency evaluation
The clear advantage of our strategy is the automatic process. We just need to load the resulting image and wait for the results for a moment without any other manual intervention. We realized the “input to output” mode, which allows individuals with little training to handle the process. This feature will especially benefit the integration for rapid point-of-care applications.
We calculated how much time was saved when using the 2D barcode-inspired approach. Using a conventional personal computer, we analyzed an image consisting of 31-by-31 (=961) arrayed spots (for structure 2, that is 30 × 30 = 900 spots) with high resolution over ten megapixels.33 The entire process took 9 s. Previously, a standard method for the data extraction was using the commercial software packaged with the microscope (for Leica, the software is las af lite). We had to manually circle and quantify each ROI individually, and the time spent on each operation on average is about 5 s. The minimum time needed to carry out such an analysis is therefore about 80 min (5 s × 961). So we saved the time consumption over 500 (derived by 5 × 961 ÷ 9) times. We dramatically improved the efficiency for high throughput data processing, and the increase in efficiency will be even more dramatic if we analyze images consisting of more spots.
We should review the high-throughput immunoassay from a broader perspective, as the time consumption for analyzing results should also be included into a normal immunoassay. As the proposed principle, we have to add a few auxiliary channels into the microfluidic network to construct the function patterns. The additional labor for the additional fluid operation during entire immunoassay only takes a few minutes, but we saved the time consumption for following analysis 500 times (from ∼80 min to < 10 s) by our strategy. More importantly, the function patterns serve as quality control (i.e., positive control) that ensures the normal state of the fluorescent antibody, just as the incorporation of the control line in widely used lateral flow test strips.34, 35, 36, 37 The normal state of the fluorescent antibody guarantees the integrity of the 2D barcode-like patterns, otherwise the array cannot be analyzed automatically because of the disappearance of the function patterns. The high performance of the automatic analysis directly resulted from the simple algorithms and the corresponding code, which was realized by our interdisciplinary approach combining biochemistry, microfabrication and mathematics in the development of this analytical device.
Robustness evaluation
Our microfluidic design is very robust and flexible, because no matter how the substrate is rotated, or the intersection angle of two pieces of microfluidic channels is changed, or even the number of the channels is changed, the image can be analyzed automatically with high accuracy.
To prove the robustness of our program in different situations, we intentionally introduced factors that would make conventional analysis very difficult. There are two primary factors resulting in analytical difficulties in the arrayed immunoassay: one is the rotation problem of the entire reaction region caused by the relative position between substrate and image sensor; the other one is the registration problem of those two pieces of microfluidic chips caused by sequential assembly. Particularly, the latter case did not exist in previous studies, because previous studies only dealt with the round spots in conventional microarrays, while our microfluidic chip produces quadrangular spots and the quadrangular spots will be affected by the registration angle. We tested the rotation angle over 180° and also tested different intersection angles (Figs. 4b–4e and Figs. 5b–5d), i.e., obtuse angle or acute angle. Regardless of the orientation or the intersection angle, we can obtain the intensities consistently. The results from our fully automatic analyses showed that the variable trends of the extracted intensities are consistent with each other. For the typical cases shown in Figs. 45, we could get the same optimal concentration/dilution for antigen (6.85 μM) or antibody (103 times dilution) from any trials, based on respective dynamic range. At the above determined optimal concentrations, the linearity of each assay in row or column can then be calculated, for example (Fig. 4f), the quadruplicate assays for antibody detection using undiluted antigen have high coefficient of determination (i.e., R2) of 0.988, 0.985, 0.983 and 0.999 across the linear range (from 10−2 to 10−4) of the antibody and show the same sigmoid relationship across the entire dilution range (from 10−1 to 10−5). Our approach, therefore, is not only automatic but also robust.
The gridding based on recursion could give the precise position of the spots with low signal intensity. The spots with low intensity normally represent the important blank control or negative control. Their precise positioning is a prerequisite for the precise quantification, and the precise quantification of the control groups is fundamental to the definition of the threshold and other values. In conventional manual approaches, it is difficult to locate the coordinates of spots with too low intensity. Our automatic approach overcame this problem.
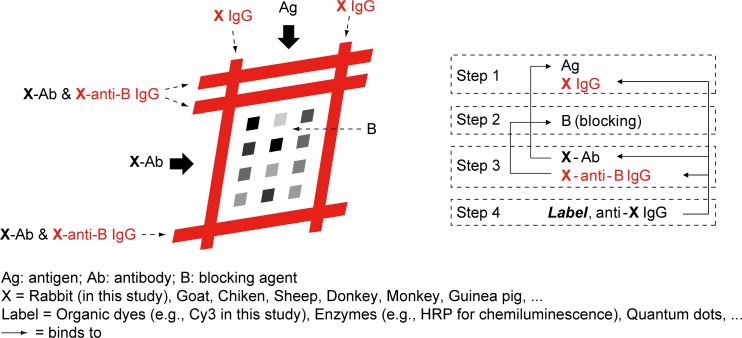
Constructing principle of function patterns
We specially designed the function patterns by a proper selection of the marker proteins (Figs. 4a, 5a), which results in a constant appearance of the function patterns regardless of the results of assays and can be used as positive control. Because the final fluorescent antibody (Cy3- labeled goat anti-rabbit IgG) was specific to rabbit, we utilized the rabbit IgG and the rabbit antiserum (which is specific to the blocking molecule) for the construction of the function patterns. Thus, the function patterns and the arrayed spots are functionally independent from each other, physically isolated, and chemically interrelated with each other. Given the final reporting antibody specific to a given species X (X could be rabbit, mouse, goat, etc.), in general we should utilize the X-IgG and the X-antiserum specific to the blocking molecule for the construction of the function patterns (Fig. 6). This is the generalized description about the construction of function patterns in the 2D barcode-inspired approach. Existing approaches solving automatic analyses only relied on mathematical algorithms and lengthy codes, while ours provided an alternative way to the goal by combining the biochemical design together, which greatly simplified the code and hence resulted in the superiority on the calculation as well as the efficiency of analysis.
Figure 6.
The generalized approach for the construction of function patterns. The function patterns and the corresponding molecules used in the construction are highlighted in red.
Further discussion
Some microarray studies based on spotting have already shown the “prototype” of asymmetric arrangement of some arrayed spots called references or markers,11, 38, 39, 40, 41 in order to facilitate the final interpretation of the result by the people (still a manual intervention). It is impossible to realize such asymmetric reference spots in the microchannel-based immunoassay, because the spotting is no longer suited to this system. Thereupon, we proposed the idea to construct the asymmetric structure, namely the function patterns, using microchannels and demonstrated the feasibility of the automatic analysis for the arrayed microfluidic immunoassay.
The function pattern-incorporated microfluidic design does not limit the dimension of the array, nor the throughput of the assay. It only requires the dimension of the microfluidic channels to be regular (a constant width and a constant spacing), which is convenient for interrogating the size information.29 This limitation does not affect the throughput of assays, and we usually use the chip with fixed channel width and fixed channel spacing. The advantage of our approach will be much more clear with the increase of the throughput. Besides, all program-based processing is limited by the hardware of the computer. This program also potentially has this limitation, but we demonstrated that the image with ten-megapixel resolution can be analyzed by a normal home computer. The smallest dimension of one pixel in conventional DNA microarray images is about 5 ∼ 10 μm (e.g., LuxScan 10 K, CapitalBio, or Typhoon Trio+, GE Healthcare), while ours is about 2.6 μm. This resolution is sufficient. We extracted the gray value from the Red Channel of the image, because we used Cy3 as the fluorescent dye and defined its color as red. While for other types of reporting probes (e.g., organic dyes or quantum dots for fluorescence, enzymes for chemiluminescence, etc.), obviously, the program could be modified to ensure the correct extraction of intensities, and matlab supports many different cases for the intensity extraction from an image. Toward an integrally miniaturized analytical system, we belive that our 2D barcode-based approach can be integrated with several miniaturized detection apparatus,42, 43 although our current proof-of-conceptual experiments utilized the standard microscope to readout the result.
As shown in Fig. 6, our strategy seamlessly connects with conventional arrayed immunoassays. Our own work and many works by other groups in the arrayed immunoassay have shown the wide applicability to disease biomarkers, small molecules, cell surface receptors, and so forth.29, 44, 45, 46 It is straightforward to combine our proposed 2D barcode principle with these systems to greatly enhance the overall efficiency. Because the function patterns and the arrayed spots are relatively independent of each other, the modification to the conventional immunoassay does not affect the analytical performance of such system.
CONCLUSIONS
Originally, the lack of “function patterns” leads to the inefficient analysis for arrayed microfluidic immunoassays. Inspired by the 2D barcodes, we designed a microfluidic network with built-in function patterns to automate the image analysis as well as the data acquisition for the high-throughput immunoassays and developed a program to realize this approach. We carried out the immunoassays by using arrayed microfluidic chips, and analyzed images consisting of near a thousand spots. We showed the feasibility, convenience and robustness of our approach. In contrast to many microfluidic assays where the time necessary for analysis increases with the throughput, our approach allows for better efficiency with increased throughput of assays. We strongly believe that this type of approach will advance the whole integration and the practical applications of microfluidic immunoassays, as well as provide inspiration for highly efficient analysis in the context of other regularly arrayed patterns.
ACKNOWLEDGMENTS
We gratefully acknowledge the National Natural Science Foundation of China (21025520), the Chinese Academy of Sciences (KJCX2-YW-M15), and the Ministry of Science and Technology of the People's Republic of China (2012AA030608) for financial support.
References
- Aguilera-Herrador E., Cruz-Vera M., and Valcarcel M., Analyst 135, 2220 (2010). 10.1039/c0an00307g [DOI] [PubMed] [Google Scholar]
- Heller M. J., Annu. Rev. Biomed. Eng. 4, 129 (2002). 10.1146/annurev.bioeng.4.020702.153438 [DOI] [PubMed] [Google Scholar]
- Allison D. B., Cui X., Page G. P., and Sabripour M., Nat. Rev. Genet. 7, 55 (2006). 10.1038/nrg1749 [DOI] [PubMed] [Google Scholar]
- Qin L.-X., Kerr K. F., and Consortium C. M. o. t. T. R., Nucleic Acids Res. 32, 5471 (2004). 10.1093/nar/gkh866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revzin A., Maverakis E., and Chang H. C., Biomicrofluidics 6, 021301 (2012). 10.1063/1.4706845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Tam V., Stybayeva G., Pan T., and Revzin A., Biomicrofluidics 7, 024105 (2013). 10.1063/1.4795423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhang L., Jing W., Liu S., Tachibana H., Cheng X., and Sui G., Biomicrofluidics 7, 011101 (2013). 10.1063/1.4793222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M. M., in Computational Genomics: Theory and Application, edited by Grant R. P. (Horizon Scientific Press, Norwich, 2004), p. 225. [Google Scholar]
- Bernard A., Michel B., and Delamarche E., Anal. Chem. 73, 8 (2001) 10.1021/ac0008845. [DOI] [PubMed] [Google Scholar]
- Rowe C. A., Scruggs S. B., Feldstein M. J., Golden J. P., and Ligler F. S., Anal. Chem. 71, 433 (1999). 10.1021/ac980798t [DOI] [PubMed] [Google Scholar]
- Mendoza L. G., McQuary P., Mongan A., Gangadharan R., Brignac S., and Eggers M., BioTechniques 27, 778 (1999). [DOI] [PubMed] [Google Scholar]
- Delamarche E., Bernard A., Schmid H., Michel B., and Biebuyck H., Science 276, 779 (1997). 10.1126/science.276.5313.779 [DOI] [PubMed] [Google Scholar]
- Delamarche E., Bernard A., Schmid H., Bietsch A., Michel B., and Biebuyck H., J. Am. Chem. Soc. 120, 500 (1998). 10.1021/ja973071f [DOI] [Google Scholar]
- Song L. S., Zhang Y., Wang W. J., Ma L. Y., Liu Y., Hao Y. L., Shao Y. M., Zhang W., and Jiang X. Y., Biomed. Microdevices 14, 631 (2012). 10.1007/s10544-012-9644-9 [DOI] [PubMed] [Google Scholar]
- Zheng C., Wang J., Pang Y., Wang J., Li W., Ge Z., and Huang Y., Lab Chip 12, 2487 (2012). 10.1039/c2lc40145b [DOI] [PubMed] [Google Scholar]
- Wang Y., Lowe R. D., Mejia Y. X., Feindt H., Steltenkamp S., and Burg T. P., Biomicrofluidics 7, 026503 (2013). 10.1063/1.4802270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. Y., Ng J. M. K., Stroock A. D., Dertinger S. K. W., and Whitesides G. M., J. Am. Chem. Soc. 125, 5294 (2003). 10.1021/ja034566+ [DOI] [PubMed] [Google Scholar]
- International Organization for Standardization, ISO/IEC 16022:2006 (International Organization for Standardization, 2006).
- International Organization for Standardization, ISO/IEC 18004:2006 (International Organization for Standardization, 2006).
- Yang Y. H. and Speed T., DNA Microarrays: A Molecular Cloning Manual, edited by Bowtell D. and Sambrook J. (Cold Spring Harbor Laboratory Press, 2003), p. 526. [Google Scholar]
- Deng N. and Duan H. L., in 27th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Shanghai, China, 1–4 September 2005 (IEEE, New York, USA, 2005), p. 898.
- Ceccarelli M. and Antoniol G., IEEE Trans. Image Process. 15, 3178 (2006). 10.1109/TIP.2006.877488 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ma M. Q., Zhang K., and Shih F. Y., Inf. Sci. 177, 1123 (2007). 10.1016/j.ins.2006.07.004 [DOI] [Google Scholar]
- Bariamis D., Iakovidis D. K., and Maroulis D., BMC Bioinformatics 11, 49 (2010). 10.1186/1471-2105-11-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda L. and Rezaeian I., BMC Bioinformatics 12, 113 (2011). 10.1186/1471-2105-12-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belean B., Borda M., Le Gal B., and Terebes R., Comput. Med. Imaging Graph. 36, 419 (2012). 10.1016/j.compmedimag.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Feldstein M., Golden J., Rowe C., MacCraith B., and Ligler F., Biomed. Microdevices 1, 139 (1999). 10.1023/A:1009900608757 [DOI] [PubMed] [Google Scholar]
- Rowe-Taitt C. A., Golden J. P., Feldstein M. J., Cras J. J., Hoffman K. E., and Ligler F. S., Biosens. Bioelectron. 14, 785 (2000). 10.1016/S0956-5663(99)00052-4 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang X. W., Song L. S., Xu C. L., Ma L. Y., Li Z. H., Xi J. Z., and Jiang X. Y., Anal. Methods 4, 3466 (2012). 10.1039/c2ay25485a [DOI] [Google Scholar]
- Canny J., IEEE Trans. Pattern Anal. Mach. Intell. 8, 679 (1986) 10.1109/TPAMI.1986.4767851. [DOI] [PubMed] [Google Scholar]
- Gonzalez R. C., Woods R. E., and Eddins S. L., Digital Image Processing Using MATLAB (Gatesmark, 2009), p. 541. [Google Scholar]
- Gonzalez R. C., Woods R. E., and Eddins S. L., Digital Image Processing Using MATLAB (Gatesmark, 2009), p. 549. [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4811278 for the entire process for analyzing an image.
- Yager P., Edwards T., Fu E., Helton K., Nelson K., Tam M. R., and Weigl B. H., Nature 442, 412 (2006). 10.1038/nature05064 [DOI] [PubMed] [Google Scholar]
- Ngom B., Guo Y. C., Wang X. L., and Bi D. R., Anal. Bioanal. Chem. 397, 1113 (2010). 10.1007/s00216-010-3661-4 [DOI] [PubMed] [Google Scholar]
- Parolo C. and Merkoci A., Chem. Soc. Rev. 42, 450 (2013). 10.1039/c2cs35255a [DOI] [PubMed] [Google Scholar]
- Tang D. P., Cui Y. L., and Chen G. A., Analyst 138, 981 (2013). 10.1039/c2an36500f [DOI] [PubMed] [Google Scholar]
- Marimon J. M., Monasterio A., Ercibengoa M., Pascual J., Prieto I., Simon L., and Perez-Trallero E., J. Microbiol. Methods 80, 274 (2010). 10.1016/j.mimet.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Le Goff G. C., Corgier B. P., Mandon C. A., De Crozals G., Chaix C., Blum L. J., and Marquette C. A., Biosens. Bioelectron. 35, 94 (2012). 10.1016/j.bios.2012.02.025 [DOI] [PubMed] [Google Scholar]
- Andresen H., Grotzinger C., Zarse K., Birringer M., Hessenius C., Kreuzer O. J., Ehrentreich-Forster E., and Bier F. F., Sens. Actuators B 113, 655 (2006). 10.1016/j.snb.2005.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Dhumpa R., Bang D. D., Hogberg J., Handberg K., and Wolff A., Lab Chip 11, 1457 (2011). 10.1039/c0lc00528b [DOI] [PubMed] [Google Scholar]
- Yao B., Luo G., Wang L., Gao Y., Lei G., Ren K., Chen L., Wang Y., Hu Y., and Qiu Y., Lab Chip 5, 1041 (2005). 10.1039/b504959h [DOI] [PubMed] [Google Scholar]
- Zhu H., Yaglidere O., Su T.-W., Tseng D., and Ozcan A., Lab Chip 11, 315 (2011). 10.1039/c0lc00358a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M., Juncker D., Michel B., Hunziker P., and Delamarche E., Biosens. Bioelectron. 19, 1193 (2004). 10.1016/j.bios.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Murphy B. M., He X. Y., Dandy D., and Henry C. S., Anal. Chem. 80, 444 (2008). 10.1021/ac7019046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M., Zimmermann M., Delamarche E., and Hunziker P., Biomed. Microdevices 9, 135 (2007). 10.1007/s10544-006-9006-6 [DOI] [PubMed] [Google Scholar]