Abstract
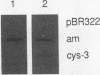
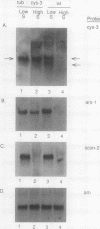
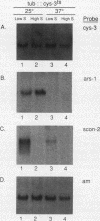
The cys-3+ gene of Neurospora crassa encodes a bZIP (basic region-leucine zipper) regulatory protein that is essential for sulfur structural gene expression (e.g., ars-1+). Nuclear transcription assays confirmed that cys-3+ was under sulfur-regulated transcriptional control and that cys-3+ transcription was constitutive in sulfur controller (scon)-negative regulator mutants. Given these results, I have tested whether expression of cys-3+ under high-sulfur (repressing) conditions was sufficient to induce sulfur gene expression. The N. crassa beta-tubulin (tub) promoter was fused to the cys-3+ coding segment and used to transform a cys-3 deletion mutant. Function of the tub::cys-3 fusion in homokaryotic transformants grown under high-sulfur conditions was confirmed by Northern (RNA) and Western immunoblot analysis. The tub::cys-3 transformants showed arylsulfatase gene expression under normally repressing high-sulfur conditions. A tub::cys-3ts fusion encoding a temperature-sensitive CYS3 protein was used to confirm that the induced structural gene expression was due to CYS3 protein function. Constitutive CYS3 production did not induce scon-2+ expression under repressing conditions. In addition, a cys-3 promoter fusion to lacZ showed that CYS3 production was sufficient to induce its own expression and provides in vivo evidence for autoregulation. Finally, an apparent inhibitory effect observed with a strain carrying a point mutation at the cys-3 locus was examined by in vitro heterodimerization studies. These results support an interpretation of CYS3 as a transcriptional activator whose regulation is a crucial control point in the signal response pathway triggered by sulfur limitation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burton E. G., Metzenberg R. L. Novel mutation causing derepression of several enzymes of sulfur metabolism in Neurospora crassa. J Bacteriol. 1972 Jan;109(1):140–151. doi: 10.1128/jb.109.1.140-151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri E. B., Jensen B. C., Schabtach E., Selker E. U. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989 Jun 30;244(4912):1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Fields S. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 1990 Apr;4(4):492–502. doi: 10.1101/gad.4.4.492. [DOI] [PubMed] [Google Scholar]
- Dwarki V. J., Montminy M., Verma I. M. Both the basic region and the 'leucine zipper' domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 1990 Jan;9(1):225–232. doi: 10.1002/j.1460-2075.1990.tb08099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D. J., Paluh J. L., Plamann M., Sachs M. S., Yanofsky C. cpc-1, the general regulatory gene for genes of amino acid biosynthesis in Neurospora crassa, is differentially expressed during the asexual life cycle. Mol Cell Biol. 1991 Feb;11(2):928–934. doi: 10.1128/mcb.11.2.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. J Biol Chem. 1990 Jul 15;265(20):11942–11947. [PubMed] [Google Scholar]
- Fu Y. H., Paietta J. V., Mannix D. G., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA-binding element. Mol Cell Biol. 1989 Mar;9(3):1120–1127. doi: 10.1128/mcb.9.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R., Rauscher F. J., 3rd, Abate C., Curran T. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science. 1989 Mar 31;243(4899):1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988 Aug 25;334(6184):721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Hanson M. A., Marzluf G. A. Regulation of a sulfur-controlled protease in Neurospora crassa. J Bacteriol. 1973 Nov;116(2):785–789. doi: 10.1128/jb.116.2.785-789.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan M. N., Marzluf G. A. Mutational analysis of the DNA-binding domain of the CYS3 regulatory protein of Neurospora crassa. Mol Cell Biol. 1991 Sep;11(9):4356–4362. doi: 10.1128/mcb.11.9.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketter J. S., Marzluf G. A. Molecular cloning and analysis of the regulation of cys-14+, a structural gene of the sulfur regulatory circuit of Neurospora crassa. Mol Cell Biol. 1988 Apr;8(4):1504–1508. doi: 10.1128/mcb.8.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird J. H., Keighren M. A., Kinsey J. A., Eaton M., Fincham J. R. Cloning of the am (glutamate dehydrogenase) gene of Neurospora crassa through the use of a synthetic DNA probe. Gene. 1982 Dec;20(3):387–396. doi: 10.1016/0378-1119(82)90207-4. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature. 1989 Aug 17;340(6234):568–571. doi: 10.1038/340568a0. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Loros J. J., Dunlap J. C. Neurospora crassa clock-controlled genes are regulated at the level of transcription. Mol Cell Biol. 1991 Jan;11(1):558–563. doi: 10.1128/mcb.11.1.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G. A. Genetic and metabolic controls for sulfate metabolism in Neurospora crassa: isolation and study of chromate-resistant and sulfate transport-negative mutants. J Bacteriol. 1970 Jun;102(3):716–721. doi: 10.1128/jb.102.3.716-721.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg R. L. Implications of some genetic control mechanisms in Neurospora. Microbiol Rev. 1979 Sep;43(3):361–383. doi: 10.1128/mr.43.3.361-383.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg R. L., Parson J. W. Altered repression of some enzymes of sulfur utilization in a temperature-conditional lethal mutant of Neurospora. Proc Natl Acad Sci U S A. 1966 Mar;55(3):629–635. doi: 10.1073/pnas.55.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V., Akins R. A., Lambowitz A. M., Marzluf G. A. Molecular cloning and characterization of the cys-3 regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 Jul;7(7):2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V., Marzluf G. A. Gene disruption by transformation in Neurospora crassa. Mol Cell Biol. 1985 Jul;5(7):1554–1559. doi: 10.1128/mcb.5.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Molecular cloning and analysis of the scon-2 negative regulatory gene of Neurospora crassa. Mol Cell Biol. 1990 Oct;10(10):5207–5214. doi: 10.1128/mcb.10.10.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Molecular cloning and regulatory analysis of the arylsulfatase structural gene of Neurospora crassa. Mol Cell Biol. 1989 Sep;9(9):3630–3637. doi: 10.1128/mcb.9.9.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall M. L. Amino acid transport in Neurospora crassa. IV. Properties and regulation of a methionine transport system. Biochim Biophys Acta. 1971 Mar 9;233(1):201–214. doi: 10.1016/0005-2736(71)90372-5. [DOI] [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. Characterization of Neurospora CPC1, a bZIP DNA-binding protein that does not require aligned heptad leucines for dimerization. Mol Cell Biol. 1991 Feb;11(2):935–944. doi: 10.1128/mcb.11.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert W. R., Patel V. B., Giles N. H. Genetic regulation of the qa gene cluster of Neurospora crassa: induction of qa messenger ribonucleic acid and dependency on qa-1 function. Mol Cell Biol. 1981 Sep;1(9):829–835. doi: 10.1128/mcb.1.9.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Ransone L. J., Lamph W. W., Verma I. M. Direct interaction between fos and jun nuclear oncoproteins: role of the 'leucine zipper' domain. Nature. 1988 Dec 15;336(6200):692–695. doi: 10.1038/336692a0. [DOI] [PubMed] [Google Scholar]
- Smeal T., Angel P., Meek J., Karin M. Different requirements for formation of Jun: Jun and Jun: Fos complexes. Genes Dev. 1989 Dec;3(12B):2091–2100. doi: 10.1101/gad.3.12b.2091. [DOI] [PubMed] [Google Scholar]
- Smith D. E., Fisher P. A. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984 Jul;99(1 Pt 1):20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Maniatis T. Simian virus 40 enhancer increases number of RNA polymerase II molecules on linked DNA. Nature. 1985 May 2;315(6014):73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- Turner R., Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989 Mar 31;243(4899):1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]