Abstract
The development of mass spectrometry imaging technologies is of significant current research interest. Mass spectrometry potentially is capable of providing highly specific information about the distribution of chemical compounds on tissues at highly sensitive levels. The required in-situ analysis for the tissue imaging forced MS analysis being performed off the traditional conditions optimized in pharmaceutical applications with intense sample preparation. This critical review seeks to present an overview of the current status of the MS imaging with different sampling ionization methods and to discuss the 3D imaging and quantitative imaging capabilities needed to be further developed, the importance of the multi-modal imaging, and a balance between the pursuit of the high imaging resolution and the practical application of MS imaging in biomedicine.
Keywords: Mass spectrometry imaging, Secondary ion mass spectrometry, Matrix-assisted laser desorption ionization, Desorption electrospray ionization
INTRODUCTION
Biological imaging allows a correlation between the morphological features inside an organ with the pathological symptoms, which is important in finding the biomarkers and subsequently use them for the disease diagnosis. The distributions of the chemical and biological compounds in organs obviously could play an important role in the process, but the information had not been easily obtained until some of the advanced analytical technologies were used for the tissue analysis. With the visual observation of H&E (hematoxylin and eosin) stained tissues being practiced for pathology over a century, modern imaging tools are being developed to provide information about chemical distributions in tissue at molecule-specific levels. Among the various technologies used for the tissue imaging, mass spectrometry (MS) is capable of providing molecular information an incomparable specificity without using fluorescent or radioactive labels [1]. The high specificity in identifying a molecule is based on its molecular weight measured through mass analysis and its molecular structure elucidated through tandem mass spectrometry or MS/MS analysis. It provides a fragmentation pattern specific to the chemical structure a molecule that is obtained through energetic excitation of the molecular ions and subsequent mass analysis of the fragment ions.
The mass spectrometry imaging (MSI) is performed by recording MS spectra point by point over a large area on the sample surface and reconstructing artificial images correlating the relative intensities of one or a set of molecules with the original positions of the sampling. A wide range of mass analyzers can be selected for MS imaging of different types of the analytes or biomarkers including metal elements, organic metabolites, peptides and proteins. Though mass spectrometry measurement can be very sensitive, the analyte molecules need to be transformed into charged ions and physically sent into a mass spectrometer during the imaging process. This makes MS imaging significantly different from other imaging technologies, such as spectroscopic imaging or MRI (magnetic resonance imaging). The efficiency of the sampling ionization methods is of great importance for the MS imaging and its optimization is limited by many practical factors, such as the convenience of implementing the sampling ionization methods and the resolution desirable for the imaging.
The transfer of any analytical technology for biological and medical applications would typically experience two phases, the discovery of the biomarkers and the routine diagnosis with the biomarkers. Though the fundamental principles in the instrumentation and the analytical methods stay the same, the actual requirements in the technical development are usually significantly different for the two phases. The ability of observing wide ranges of chemical and biological compounds is very important for the discovery of the biomarkers, while the convenience of performing the biomarker monitoring and compatibility to the regulations are extremely important for the diagnosis. In this review, we briefly overview the current MS imaging capabilities enabled with the most commonly used sampling ionization methods and discuss their potentials as well as further development needed for finding biomarkers and performing diagnosis.
Sampling Methods
In the history of mass spectrometry, the marching of the MS analysis into a new field of application has always been enabled with a breakthrough in the ionization method. The sensitivity of the MS analysis of biological samples is always susceptible to the matrix effect and also relies on sophisticated sample cleaning up including the extraction and the chromatographic separation. Unfortunately, these procedures would not be an option for the MS imaging of a tissue section, where an analysis of the raw material is necessary to preserve the information on the spatial distribution of the constituents. The desorption ionization methods naturally became the first candidates for the MS imaging.
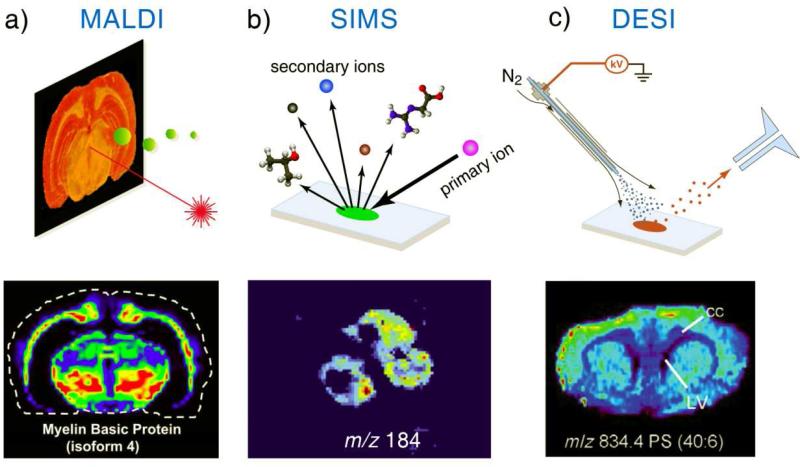
Though SIMS (secondary ion mass spectrometry) was the first widely used method for MS imaging of surface, MALDI (matrix-assisted laser desorption ionization) imaging was the first popular method applied for biological imaging of tissue (Figure 1a) [2]. MALDI has the advantage of ionizing biomolecules over a wide mass range, including DNA, peptides and proteins. The MS analysis with MALDI of extracted proteins relies on the mixing of organic matrices with the purified samples, which was transferred to MALDI imaging by spraying organic acids as the desorption matrix over the tissue. This adds an additional step of sample preparation and also raise the concerns of modification of the original chemical distribution on the tissue surface, but nevertheless enabled a powerful MS imaging tool. Though atmospheric pressure MALDI has been applied for imaging recently [3], MALDI imaging is mainly performed in a vacuum environment.
Figure 1.
Schematic illustrations of MSI with (a) MALDI, (b) SIMS, and (c) DESI and ion maps of (a) myelin basic protein (isoform 4) on sectioned rat brain by MALDI imaging [9], (b) phosphocholine head group on three PC12 cells by SIMS imaging [10], and (c) phosphatidylserine (PC 40:6) on sectioned rat brain by DESI imaging [8].
SIMS uses a highly energetic primary ion beam impinging the sample surface to generate the secondary ions from the analytes. It is used for imaging of inorganic and organic surfaces for many years but had not been applied for tissue imaging until the recent success in desorption ionization of large biomolecules using “softer” primary beams such as fullerene ions [5] or other cluster ions of high particle weights (Figure 1b). High kinetic energies of the primary ions in the range of 5-25 keV are used for the SIMS imaging, which requires a high vacuum environment to avoid the collisions before they reach the tissue surface. SIMS is highly efficient for mapping metal elements and small molecules (up to m/z 1200), while routine imaging of large peptides and proteins using SIMS remains challenging [4]. For both MALDI and SIMS, the enhancement in the sampling ionization efficiency is also explored with the modification of the sample substrates and the chemical coatings.
Besides SIMS and MALDI, many other ionization methods have been developed in the past for mass spectrometry analysis, following a main trend of ionizing larger and larger molecules, which led the MS applications evolved from elemental to organic and eventually biological analysis. Starting 2004, a new category of ionization methods called ambient ionization emerged, with a clear aim at the direct sampling ionization of raw samples so MS analysis can be performed without any sample preparation [6]. This philosophy of applying MS analysis suits well the imaging of raw tissue and many ambient ionization methods have been explored for the MS imaging [7]. The desorption electrospray ionization (DESI) is one of the first ambient ionization methods that has been intensively applied for the tissue imaging (Figure 1c) [8]. It utilizes the electrosprayed droplets to sample the tissue surface and generate the secondary ions for MS analysis. The distributions of the drug metabolites, fatty acids and lipids on the tissues samples could well be characterized with the DESI MS imaging.
Though many other features, such as the lateral resolution and optimized sensitivity for a particular set of biomarkers, would be considered for selection of an “ideal” sampling ionization method for the MS imaging, a primary concern could simply be if peptides and proteins are expected to be the biomarkers for the diagnostics. Ambient MS imaging using DESI provides the simplest solution without worrying about fixing tissue with special chemical coatings or substrates nor about transferring them into the vacuum; however, the desorption ionization of the peptides and proteins with DESI has not been shown to be as efficient as that for MALDI imaging.
While obtaining high resolutions for the MS images and observing wide range of chemical and biological compounds at high sensitivity will continue to be two main focuses for the technology development in the MS imaging, some other capabilities are being recognized to be essential for the MS imaging to play a real role in the biomedical applications.
3D MS Imaging
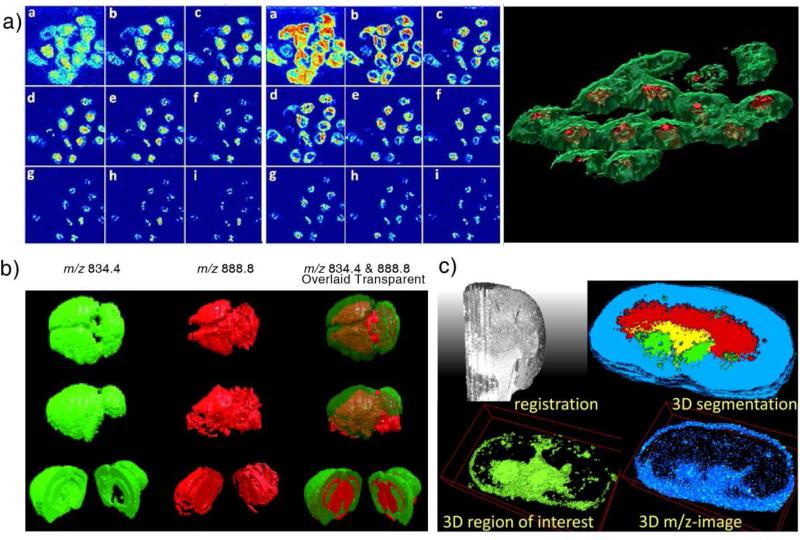
As shown by commercially available MRI, X-ray computed tomography (CT), and single-photon emission computed tomography (SPECT), the 3D imaging is powerful in providing a full set of imaging data allowing studies and diagnostics with the real spatial distributions of the features of interest. Performing 3D imaging using MS is particularly challenging due to the need to access the molecules of the internal parts of an organ for acquiring the spectra. Nevertheless, 3D MS imaging has been explored in various ways so highly molecule-specific distributions of chemicals could be obtained for 3D tissue volumes. Dynamic desorption for depth profiling uses the probing ion or laser beam to eliminate the tissue layers while performing 2D imaging (Figure 2a). A more promising method is to perform 2D imaging of a series of selected sections from an organ and reconstructing 3D images with extrapolation of the 2D imaging data (Figure 2b and c). Spatial distributions of the metabolites, lipids and proteins have be extracted using this method[11-13].
Figure 2.
(a) SIMS depth profiling of the distribution of adenine (m/z 136.1, left panel) and phosphocholine head group (m/z 184.1, middle panel) in nine layers at different depth. Right panel: 3D visualization of the membrane (green, phosphocholine head group) and nucleus (red, adenine) [13]. (b) 3D images of mouse brain obtained with DESI. 3D reconstructions from different view angles show the distributions of PS 18:0/22:6 (m/z 834.4) in green, ST 24:1 (m/z 888.8) in red, and the overlaid distribution [12]. (c) 3D imaging of mouse kidney with MALDI [17].
The practical difficulties in the 3D MS imaging are caused by the intensive work for getting large numbers of tissue sections imaged for a sizable organ and the lack of suitable software for the data processing [14]. A mouse brain can be sectioned to 560 pieces of 20 μm thick tissue sections [12] and 2D imaging of all of them would take a very long time from a month to more than a year, depending on the resolution. Only a representative set of sections could be imaged and data extrapolation would be needed for filling the gaps in data processing. Currently there is few commercially available means for data processing of 3D MS imaging. 3D images were produced with limited number of chemical compounds and reconstructed without original MS information retained, which makes the 3D images completely unsuitable for further data analysis. The size of the raw data is larger than 150 MB for 2D imaging of one section of a mouse brain (about 1 cm2) at a low spectral resolution (about 1000) and a relatively low image resolution (about 200 μm). Data reduction [15] is necessary for processing data in 3D MS imaging with high spectral and spatial resolutions. The reconstruction of the 3D data space is also challenged by practical issues such as the deformation of tissue sections, misalignment between the sections, and the need for the intersection intensity normalization. Some quantitation measure is needed in 3D imaging to overcome the artificial differences between the 2D images due to the section-to-section inconsistencies. A proper constructed 3D MS data would enable powerful analysis of the features in spatial distribution of chemicals [11,14,16] and allow the correlation and comparison of images acquired using different methods in multi-modal studies. However, much more effort is required for the development of proper protocols and capable software tools for the data processing in 3D MS imaging.
Quantitative MS Imaging
The success in application of mass spectrometry to the drug discovery, environmental and food regulations heavily relies on the quantitation capability of MS analysis at trace levels. Mass analysis suffers severely the matrix effects but has been overcome with proper mixing of the internal standards into the sample. For imaging of intact tissue, a direct transfer of this method is somewhat difficult. However, quantitation of the absolute amounts of the chemicals in the tissue, besides the relative distributions, would allow MS images better correlated to information acquired using other methods [18,19].
Though the quantitation in the MS imaging could be performed with external calibrations, relatively good accuracy might not be achieved due to the tissue heterogeneity and the scan-to-scan variations. For MALDI imaging, quantitation was explored by spraying internal standards on the sample substrate prior to applying tissue section or mixed the internal standards into the MALDI matrices to be coated on the tissue section. Accuracy better than 5% (RSD) has been achieved for quantitation of acetyl-L-carnitine in piglet brain [20]. Quantitative DESI imaging has also been explored by micro pipetting internal standards onto selected spots on the tissue sections and a relative standard deviation better than 17% was obtained for a quantitation of the clozapine in the brain tissue of dosed rats [21].
The quantitative MS imaging is still in its infancy stage. The development in this field will rely on the development of practically useful operation protocols for introducing the calibration standards for tissue imaging and corresponding treatments in the data processing. Putting internal standard spots in the selected areas on sample substrates might be broadly practiced for calibration purposes in the future MS imaging. The significant impacts with the improvements in the quantitation will include the convenience of inter-section normalization for 3D MS imaging, providing absolute quantitation of the chemical distribution, and quantitatively correlating of results by MS with other imaging technologies in multimodal studies or diagnosis.
Multimodal Imaging
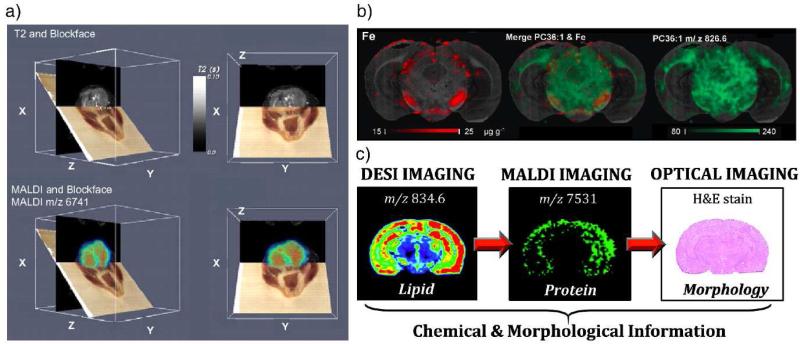
Development, validation, and application of a MS imaging method is a process constantly involving correlations of the molecular distributions observed with MS to the morphological and cellular features obtained through other methods, at least but certainly not the last, with the optical images by H&E staining [22] or immunostaining. Multimodal imaging involving MS and other techniques, such as radiography, ultrasound, optical spectroscopy, or MRI, would be very powerful for establishing the correlations between the histology and pathology by providing the most complementary and comprehensive information about the object of study or diagnosis [23]. Correlation of MALDI images with optical images and MRI data has been demonstrated characterization of the tumor in a mouse brain[9,24].
Biological samples are very complex. A single tissue section contains a large variety chemical species ranging from salts, amino acid, lipids, to peptides and proteins. The MS imaging itself could benefit from multi-modal sampling ionization methods due to the difference in the sampling efficiency for different types of the molecules in tissue. Use of multiple types of sampling ionization methods, each optimized for different types of molecules, could enhance the overall quality of the MS images. The laser ablation inductively coupled plasma mass spectrometry (LA-ICPMS) and MALDI (Figure 3b) has been used for imaging elements and biomolecules, respectively, on the adjacent tissue sections from a mouse brain [25]. The elemental and the lipid images were co-registered with the corresponding optical images. Multimodal imaging of the tissue section has been demonstrated with a series methods starting with DESI, followed with MALDI, prior to applying H&E staining on the same tissue section (Figure 3c) [26]. Chemical information on the distributions of both lipids and proteins was obtained for comparison with the histological features. The serial multimodal MS imaging could be applied as a general approach, but the sample treatments for optimizing the individual imaging steps need to be compatible with each other and a special sequence of the imaging would be critical.
Figure 3.
(a) Co-registered block face optical images (brown plane), MRI data (black plane), and protein images (colored plane) by MALDI from a mouse head with a tumor [9]. (b) Multimodal imaging on mouse brain section. Elemental concentration map (Fe in red) obtained by laser ablation inductively coupled plasma mass spectrometry and lipid map (PC 36:1 in green) obtained by MALDI could be combined to show the chemical distribution of different species [25]. (c) DESI ion image of a mouse brain coronal section showing the distribution of lipid PS(18:0/22:6) m/z 834.6, followed by MALDI imaging to obtain the distribution of neurogranin (m/z 7531) and finally tested by H&E stain to show the morphology of the same tissue [26].
Imaging Resolution
As for all the imaging techniques, a huge emphasis has been put onto the resolution improvement in MS imaging. A desirable resolution by default is better than a sub-cellular or sub-micrometer level. This would allow a distinction between scattered malignant cells from its surrounding healthy cells and allow a direct cell-to-cell comparison between the MS images and the optical images of the stained tissue section. The lateral resolution in the 2D imaging is largely dependent on the sampling probe size, but is also limited by the sampling efficiency and the time consumed for imaging a sizable sample.
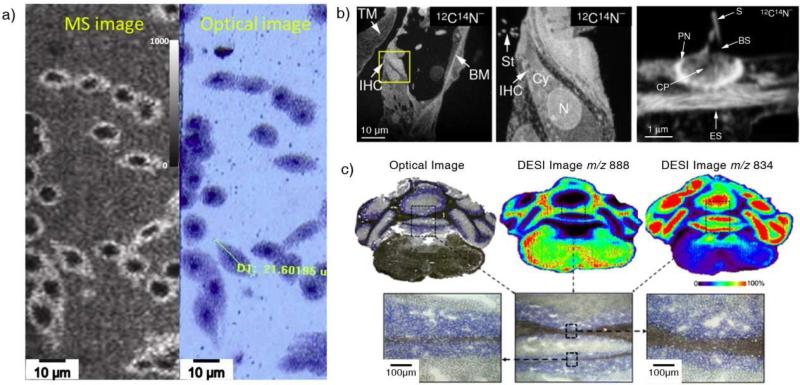
A highest resolution at 10 nm level has been speculated for SIMS based on the impinging area by a single ion; however, practically it is impossible to have an ion beam at an intensity high enough consistently focused into such a small area. A resolution below 100 nm has been reported for imaging cyanide (12C14N) in a mouse cochlea using SIMS (Figure 4b), which is hard to achieve for imaging of proteins. The resolution of MALDI imaging is generally limited to the laser spot size (50-150 μm) but can be limited by the size of the matrix droplets, which cause the diffusion of the analytes on the tissue [27]. MALDI imaging at a spatial resolution of sub-micron was reported for cell analysis (Figure 4a) with a transmission mode, for which the laser beam irradiates from the back side of the samples [28]. For DESI, the single droplet can be as small as 1 μm in diameter, but the size of the overall sampling plume generated by spray is significantly larger. A highest lateral resolution of 35 μm has been recently reported (Figure 4c) [29]. Based on the improved spatial resolution, lipid profiling on mouse ovaries was carried out and the temporal changes during the ovulatory cycle were observed and characterized. Instead of making the probe size smaller, an interesting approach has been explored by Sweedler's group, with which the tissue sections were stretched before the imaging, instead of shrinking the sampling spots. This practically resulted in an improved image resolution [30].
Figure 4.
(a) High-resolution MALDI imaging on HEK-293 cells. MS image of m/z 782 (left) and optical image (right) after MALDI imaging [28]. (b) High resolution SIMS imaging on a section of a mouse cochlea. From left to right: SIMS imaging on the tissue section at mass 12C14N; SIMS imaging on the boxed area at a higher resolution; an even higher resolution mass image of stereocilia [35]. (c) Optical image and DESI ion images of a mouse brain section. The high resolution DESI image was obtained with a modified DESI emitter and optimized working conditions [29].
Performing high resolution MS imaging inevitably results in reduced signal intensity for each sample spot due to a relatively lower intensity of the probing photons, ions or charged droplets reaching the spot. The total amount of analytes on sample surface is proportional to the imaging resolution while the time required for the imaging is reversely proportional to the imaging resolution. It takes 20 hours to image a tissue section of about 1 cm2 size at an image resolution of 50 μm [31]. Further improvement of the resolution requires much longer time for both covering larger number of pixels as well as to spend longer sampling time at each spot. Different from spectroscopic imaging, the sampling by MS probes does consume the material on the sample surface, which makes a super long sampling impractical. When performing 3D imaging, the ultimate spatial resolution is limited by the thickness of the tissue section for 2D imaging, which is 20 μm. However, the current limitation is the gap between the tissue sections selected for 3D imaging, which can be chosen to be relatively large to save time for imaging a 3D tissue volume. For examples, inter-section resolutions of 130 to 300 μm have been used [12,32].
An interesting alternative approach that might lead to a significant improvement to the speed of MS imaging is the method so called microscope MS imaging. Instead of using fine laser beam to analyze the sample surface pixel by pixel, a laser beam of relatively large size (hundreds of microns [33]) is used to desorb and ionize the analytes from the surface. A mass analyzer retaining the lateral positions of the ions, such as a time-of-flight, and a position-sensitive detector are needed to obtain the images for the chemical distributions of the entire area sampled. An image resolution in micron range was claimed for microscope MALDI imaging of whole-rat cross sections and rat brain sections[34]. The speed of MS imaging could be improved by 2-4 orders of magnitude in microscope mode, while the image resolution would be highly dependent on the retention of the relative positions of the chemical species during the ionization and MS analysis.
Diagnostic Tissue Profiling
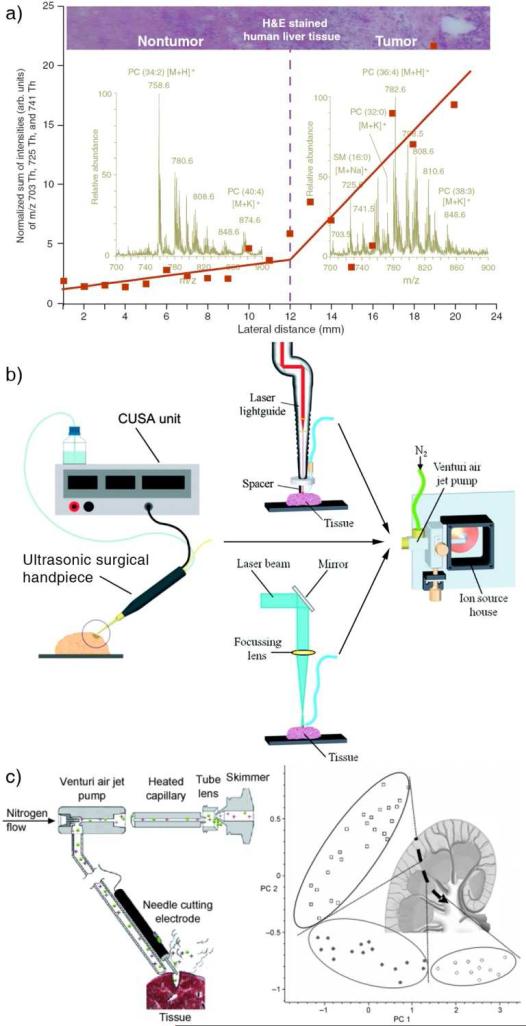
With the molecule specific information provided by MS imaging, we would hope the pathologists soon be able to use them in identifying the tumor margin for surgery purpose, instead of the examination of the stained tissue, which is a process highly dependent on the pathologist's experience. The high spatial resolution MS imaging taking tens of hours may not be practical or necessary for the tumor surgery. A fast profiling of the tissue [36] might serve as a solution for real-time tissue examination (Figure 5a). While the profiling could be performed with a tissue section taken from the patient during a surgery, real-time chemical profiling on organ has also been explored. The challenges of doing so include the sampling ionization of the chemicals on the organ in a fashion compatible and safe with the surgical protocol and the efficient transfer of the ions to a mass spectrometer for the analysis. It has been demonstrated that aerosol containing tissue analytes can be directly generated from the organ during the organ excision in the electrosurgery [37] and laser surgery [38]. Using a Cavitron ultrasonic surgical aspirator combined with a Venturi device, the gaseous particles were transferred over several meters to a mass spectrometer, where they were sampled and ionized by a sonic-spray. (Figure 5b and 5c) [39].
Figure 5.
(a) A DESI profiling on human liver adenocarcinoma. The red plots show the trend of signal intensity of three ions (m/z 703, 725, 741) from the non-tumor tissue to tumor tissue [6]. (b) Real time analysis of tissue with different ionization methods: sonic spray mass spectrometry combined with ultrasonic surgical aspiration, ultraviolet and infrared laser desorption ionization, respectively [38,39]. (c) Setup of rapid evaporative ionization based on electrosurgical unit for real time analysis of tissue. A porcine kidney was tested with rapid evaporative ionization in a profiling manner to distinguish the three parts of kidney: □ cortex, ● medulla, ○ pelvis [37].
OUTLOOK
What mass spectrometry brings to the tissue imaging is the potential capability in quantitation of a wide range of chemical and biological compounds in tissues with high specificity. Analysis of the analytes directly from tissues forces MS being performed at conditions significantly different from those for pharmaceutical applications, which involves intensive sample preparation for highly sensitive and highly quantitative results. The tandem mass spectrometry, which is the key to the ultimate specificity, is not routinely applied for MS imaging yet due to the time and sample it might take for compounds in a relatively wide mass range. Fast survey over a broad area at low mass resolution and low image resolution combined with zoom-in scans of target areas with high resolutions and MS/MS at selected masses might be necessary to make the MS imaging practically useful for the biomedical research in the future. The data processing capability for the MS imaging certainly needs to be developed much further to handle large size data files, quantify the abundances, perform the statistical analysis, and to compare the images acquired by MS and other technologies. A proper combination of the ambient ionization methods with miniature mass spectrometers [40] might also produce small benchtop tissue imagers or profilers for the biologists or physicians in the future.
REFERENCES
- 1.Chughtai K, Heeren RMA. Mass Spectrometric Imaging for Biomedical Tissue Analysis. Chem Rev. 2010;110(5):3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7(4):493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 3.Guenther S, Rompp A, Kummer W, Spengler B. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 μm spatial resolution and high mass accuracy. Int J Mass Spectrom. 2011;305(2-3):228–237. [Google Scholar]
- 4.Fletcher JS, Vickerman JC. Secondary Ion Mass Spectrometry: Characterizing Complex Samples in Two and Three Dimensions. Anal Chem. 2013;85:610–639. doi: 10.1021/ac303088m. [DOI] [PubMed] [Google Scholar]
- 5.Piehowski PD, Carado AJ, Kurczy ME, Ostrowski SG, Heien ML, Winograd N, Ewing AG. MS/MS Methodology To Improve Subcellular Mapping of Cholesterol Using TOF-SIMS. Anal Chem. 2008;80(22):8662–8667. doi: 10.1021/ac801591r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Ambient mass spectrometry. Science. 2006;311(5767):1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 7.Sampson JS, Hawkridge AM, Muddiman DC. Construction of a Versatile High Precision Ambient Ionization Source for Direct Analysis and Imaging. J Am Soc Mass Spectrom. 2008;19(10):1527–1534. doi: 10.1016/j.jasms.2008.06.013. doi:DOI 10.1016/j.jasms.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiseman JM, Ifa DR, Song QY, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Ed. 2006;45(43):7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 9.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci USA. 2008;105(47):18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, Sjovall P, Ewing AG. Time of Flight Mass Spectrometry Imaging of Samples Fractured In Situ with a Spring-Loaded Trap System. Anal Chem. 2010;82(15):6652–6659. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J Am Soc Mass Spectrom. 2005;16(7):1093–1099. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-Dimensional Vizualization of Mouse Brain by Lipid Analysis Using Ambient Ionization Mass Spectrometry. Angew Chem Int Ed. 2010;49(5):873–876. doi: 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher JS, Rabbani S, Henderson A, Lockyer NP, Vickerman JC. Three-dimensional mass spectral imaging of HeLa-M cells - sample preparation, data interpretation and visualisation. Rapid Commun Mass Spectrom. 2011;25(7):925–932. doi: 10.1002/rcm.4944. [DOI] [PubMed] [Google Scholar]
- 14.Xiong XC, Xu W, Eberlin LS, Wiseman JM, Fang X, Jiang Y, Huang ZJ, Zhang YK, Cooks RG, Ouyang Z. Data Processing for 3D Mass Spectrometry Imaging. J Am Soc Mass Spectrom. 2012;23(6):1147–1156. doi: 10.1007/s13361-012-0361-7. [DOI] [PubMed] [Google Scholar]
- 15.McDonnell LA, van Remoortere A, de Velde N, van Zeijl RJM, Deelder AM. Imaging Mass Spectrometry Data Reduction: Automated Feature Identification and Extraction. J Am Soc Mass Spectrom. 2010;21(12):1969–1978. doi: 10.1016/j.jasms.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008;5(1):101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 17.Trede D, Schiffler S, Becker M, Wirtz S, Steinhorst K, Strehlow J, Aichler M, Kobarg JH, Oetjen J, Dyatloy A, Heldmann S, Walch A, Thiele H, Maass P, Alexandrov T. Exploring Three-Dimensional Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry Data: Three-Dimensional Spatial Segmentation of Mouse Kidney. Anal Chem. 2012;84(14):6079–6087. doi: 10.1021/ac300673y. [DOI] [PubMed] [Google Scholar]
- 18.Clemis EJ, Smith DS, Camenzind AG, Danell RM, Parker CE, Borchers CH. Quantitation of Spatially-Localized Proteins in Tissue Samples using MALDI-MRM Imaging. Anal Chem. 2012;84(8):3514–3522. doi: 10.1021/ac202875d. [DOI] [PubMed] [Google Scholar]
- 19.Lanekoff I, Sjovall P, Ewing AG. Relative Quantification of Phospholipid Accumulation in the PC12 Cell Plasma Membrane Following Phospholipid Incubation Using TOF-SIMS Imaging. Anal Chem. 2011;83(13):5337–5343. doi: 10.1021/ac200771g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirman DA, Yost RA. Quantitative Tandem Mass Spectrometric Imaging of Endogenous Acetyl-L-carnitine from Piglet Brain Tissue Using an Internal Standard. Anal Chem. 2011;83(22):8575–8581. doi: 10.1021/ac201949b. [DOI] [PubMed] [Google Scholar]
- 21.Vismeh R, Waldon DJ, Teffera Y, Zhao ZY. Localization and Quantification of Drugs in Animal Tissues by Use of Desorption Electrospray Ionization Mass Spectrometry Imaging. Anal Chem. 2012;84(12):5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- 22.Gerbig S, Golf O, Balog J, Denes J, Baranyai Z, Zarand A, Raso E, Timar J, Takats Z. Analysis of colorectal adenocarcinoma tissue by desorption electrospray ionization mass spectrometric imaging. Anal Bioanal Chem. 2012;403(8):2315–2325. doi: 10.1007/s00216-012-5841-x. [DOI] [PubMed] [Google Scholar]
- 23.Rompp A, Guenther S, Schober Y, Schulz O, Takats Z, Kummer W, Spengler B. Histology by Mass Spectrometry: Label-Free Tissue Characterization Obtained from High-Accuracy Bioanalytical Imaging. Angew Chem Int Ed. 2010;49(22):3834–3838. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- 24.Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, Dawant BM, Caprioli RM, Gore JC. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat Methods. 2008;5(1):57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matusch A, Fenn LS, Depboylu C, Klietz M, Strohmer S, McLean JA, Becker JS. Combined Elemental and Biomolecular Mass Spectrometry Imaging for Probing the Inventory of Tissue at a Micrometer Scale. Anal Chem. 2012;84(7):3170–3178. doi: 10.1021/ac203112c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eberlin LS, Liu XH, Ferreira CR, Santagata S, Agar NYR, Cooks RG. Desorption Electrospray Ionization then MALDI Mass Spectrometry Imaging of Lipid and Protein Distributions in Single Tissue Sections. Anal Chem. 2011;83(22):8366–8371. doi: 10.1021/ac202016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaletas BK, van der Wiel IM, Stauber J, Dekker LJ, Guzel C, Kros JM, Luider TM, Heeren RMA. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9(10):2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 28.Zavalin A, Todd EM, Rawhouser PD, Yang JH, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J Mass Spectrom. 2012;47(11):1473–1481. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 29.Campbell DI, Ferreira CR, Eberlin LS, Cooks RG. Improved spatial resolution in the imaging of biological tissue using desorption electrospray ionization. Anal Bioanal Chem. 2012;404(2):389–398. doi: 10.1007/s00216-012-6173-6. [DOI] [PubMed] [Google Scholar]
- 30.Tucker KR, Lanni EJ, Serebryannyy LA, Rubakhin SS, Sweedler JV. Stretched Tissue Mounting for MALDI Mass Spectrometry Imaging. Anal Chem. 2011;83(23):9181–9185. doi: 10.1021/ac201857k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaurand P, Schwartz SA, Caprioli RM. Profiling and imaging proteins in tissue sections by MS. Anal Chem. 2004;76(5):86a–93a. [PubMed] [Google Scholar]
- 32.Chen RB, Hui LM, Sturm RM, Li LJ. Three Dimensional Mapping of Neuropeptides and Lipids in Crustacean Brain by Mass Spectral Imaging. J Am Soc Mass Spectrom. 2009;20(6):1068–1077. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jungmann JH, MacAleese L, Buijs R, Giskes F, de Snaijer A, Visser J, Visschers J, Vrakking MJJ, Heeren RMA. Fast, High Resolution Mass Spectrometry Imaging Using a Medipix Pixelated Detector. J Am Soc Mass Spectrom. 2010;21(12):2023–2030. doi: 10.1016/j.jasms.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Klerk LA, Altelaar AFM, Froesch M, McDonnell LA, Heeren RMA. Fast and automated large-area imaging MALDI mass spectrometry in microprobe and microscope mode. Int J Mass Spectrom. 2009;285(1-2):19–25. [Google Scholar]
- 35.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, Park KM, Ito S, Schwartz M, Benichou G, Slodzian G. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Biol. 2006;5(6):20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiseman JM, Puolitaival SM, Takats Z, Cooks RG, Caprioli RM. Mass spectrometric profiling of intact biological tissue by using desorption electrospray ionization. Angew Chem Int Ed. 2005;44(43):7094–7097. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 37.Schafer KC, Denes J, Albrecht K, Szaniszlo T, Balog J, Skoumal R, Katona M, Toth M, Balogh L, Takats Z. In Vivo, In Situ Tissue Analysis Using Rapid Evaporative Ionization Mass Spectrometry. Angew Chem Int Ed. 2009;48(44):8240–8242. doi: 10.1002/anie.200902546. [DOI] [PubMed] [Google Scholar]
- 38.Schafer KC, Szaniszlo T, Gunther S, Balog J, Denes J, Keseru M, Dezso B, Toth M, Spengler B, Takats Z. In Situ, Real-Time Identification of Biological Tissues by Ultraviolet and Infrared Laser Desorption Ionization Mass Spectrometry. Anal Chem. 2011;83(5):1632–1640. doi: 10.1021/ac102613m. [DOI] [PubMed] [Google Scholar]
- 39.Schafer KC, Balog J, Szaniszlo T, Szalay D, Mezey G, Denes J, Bognar L, Oertel M, Takats Z. Real Time Analysis of Brain Tissue by Direct Combination of Ultrasonic Surgical Aspiration and Sonic Spray Mass Spectrometry. Anal Chem. 2011;83(20):7729–7735. doi: 10.1021/ac201251s. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang Z, Cooks RG. Miniature Mass Spectrometers. Annu Rev Anal Chem. 2009;2:187–214. doi: 10.1146/annurev-anchem-060908-155229. [DOI] [PubMed] [Google Scholar]