Abstract
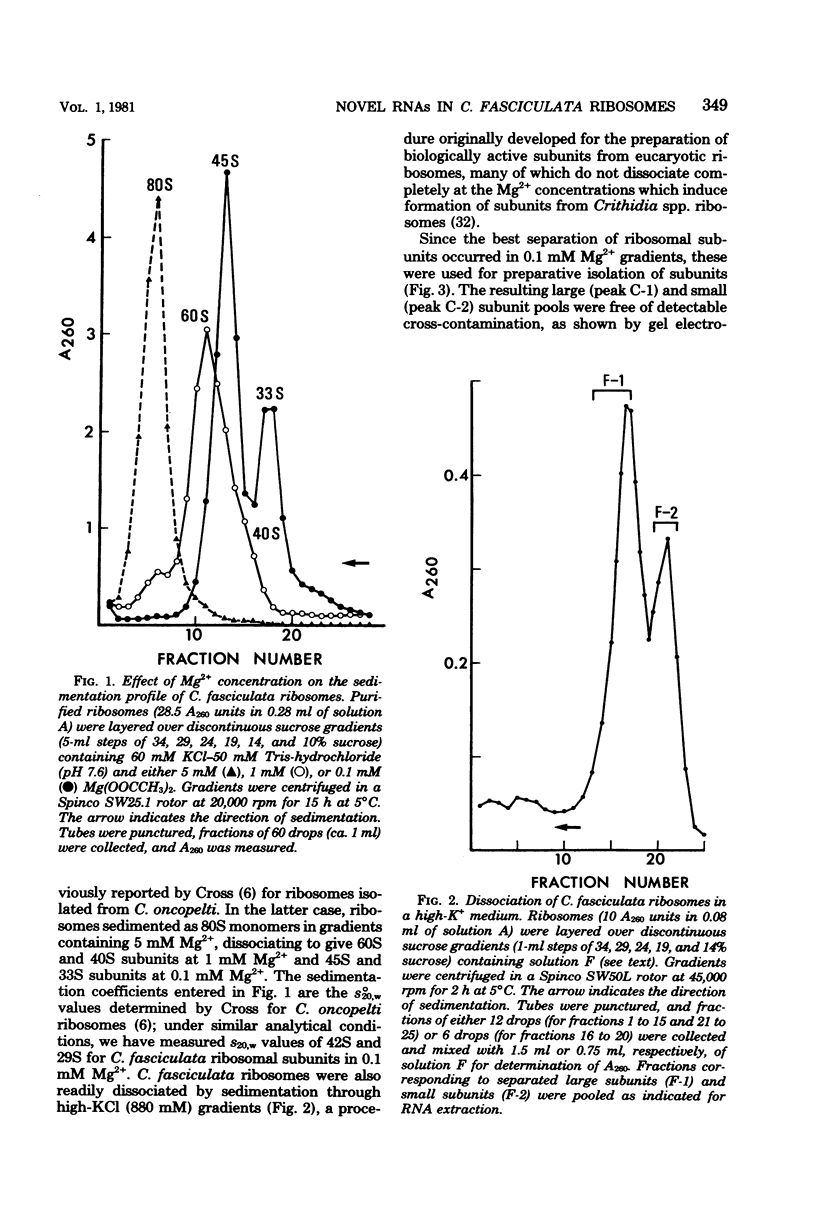
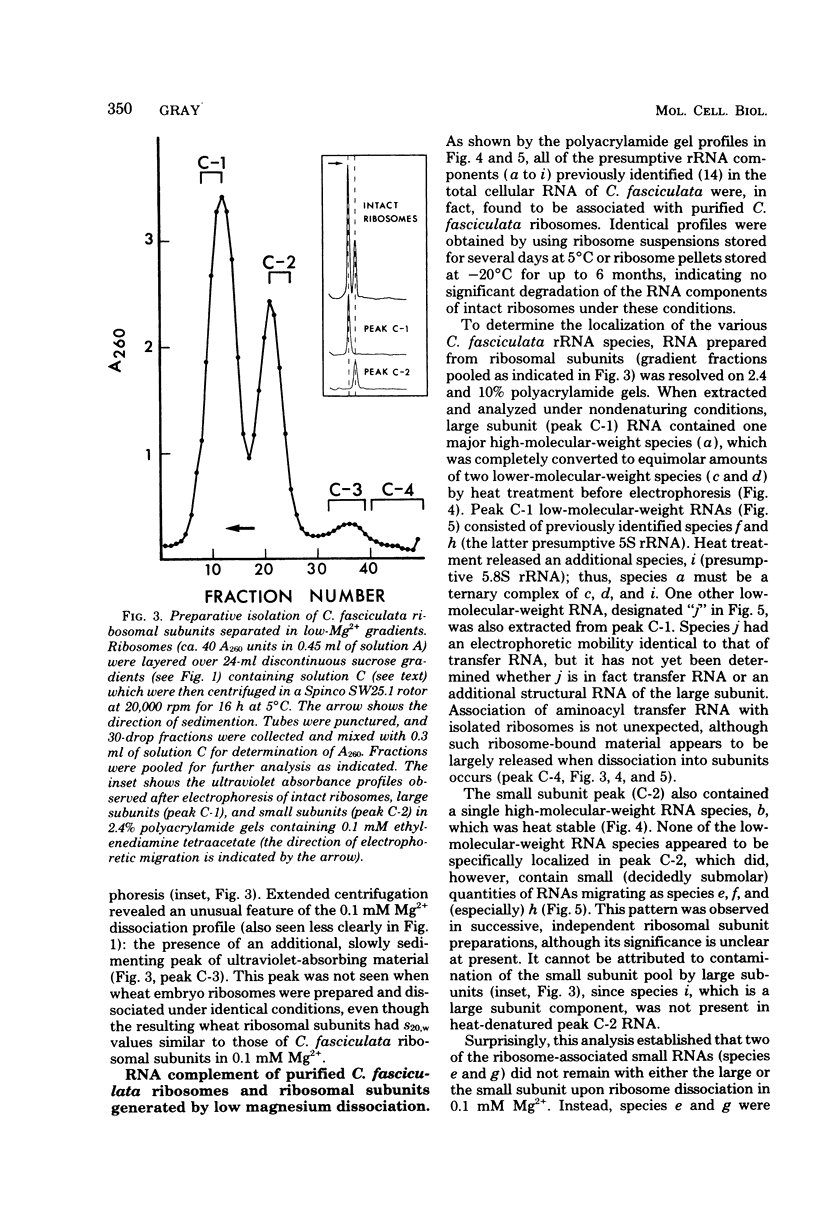
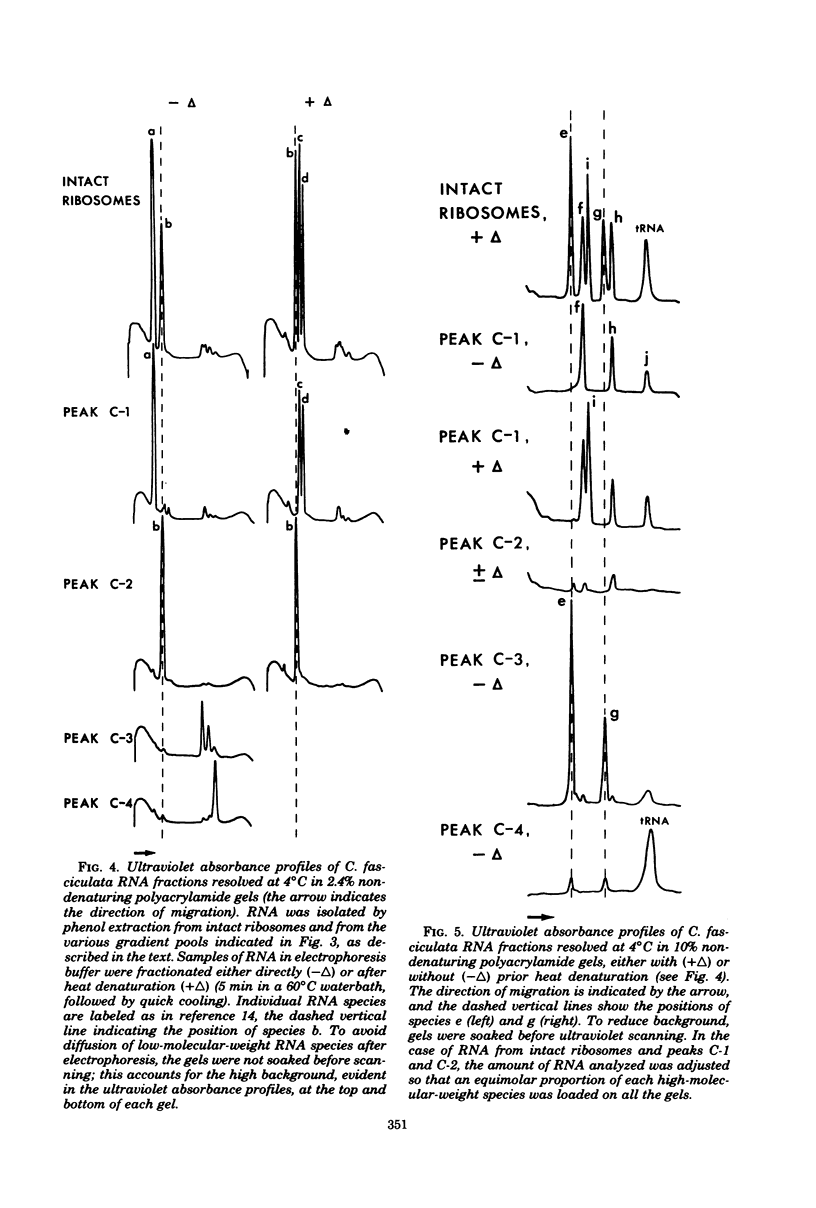
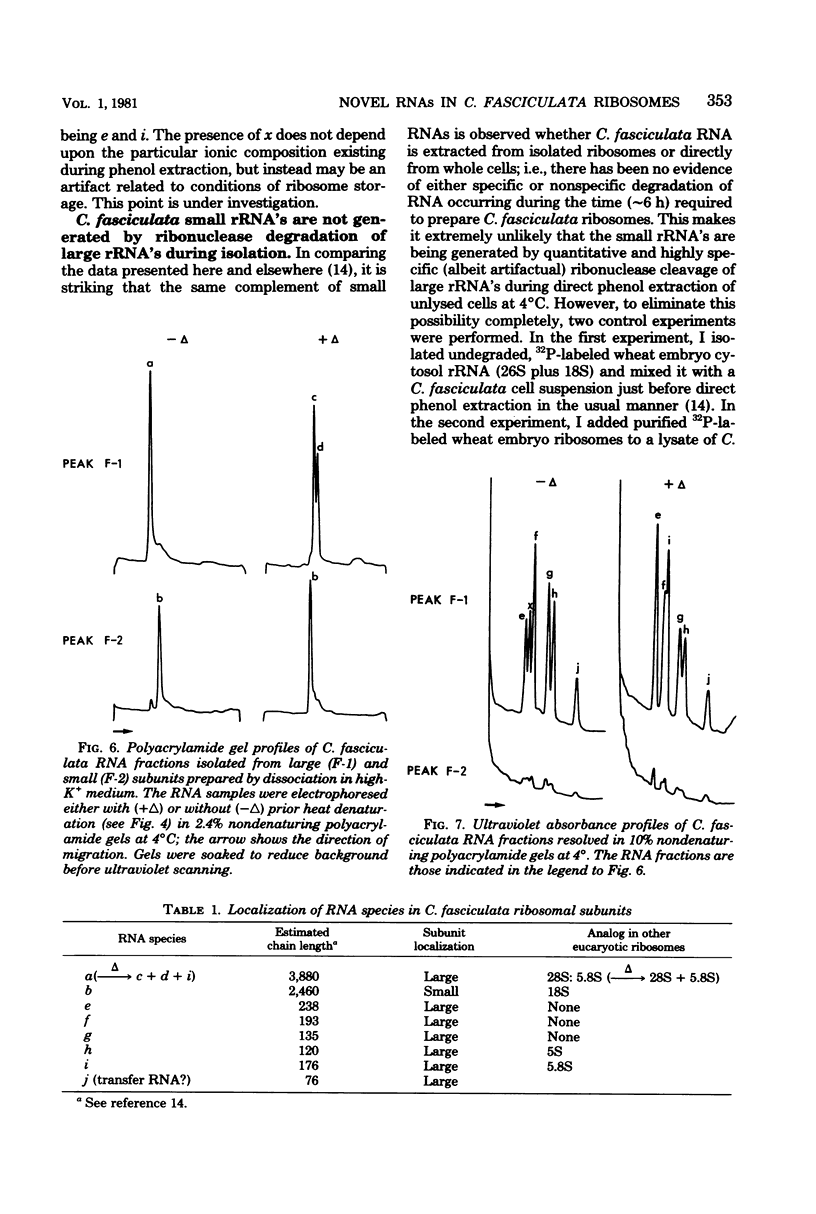
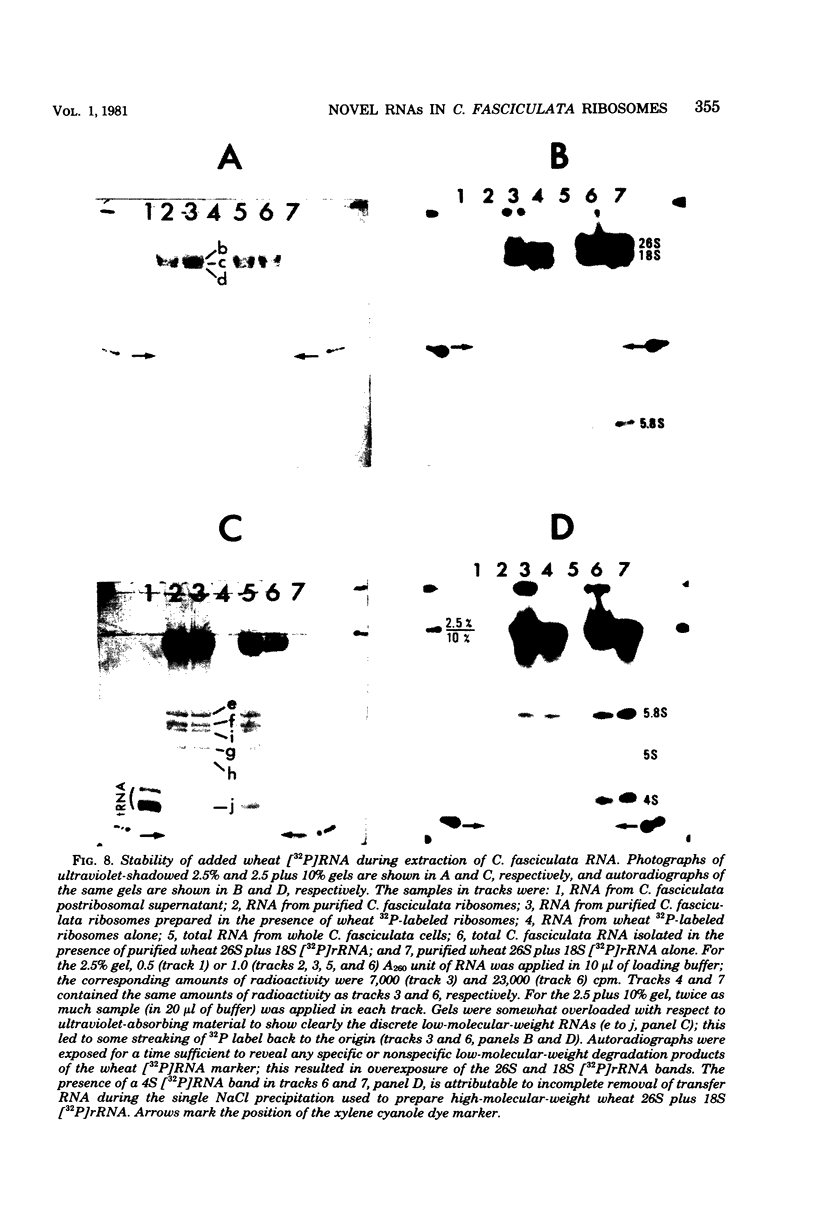
In a previous study from this laboratory, presumptive ribosomal ribonucleic acid (RNA) species were identified in the total cellular RNA directly extracted from intact cells of the trypanosomatid protozoan Crithidia fasciculata (M. W. Gray, Can. J. Biochem. 57:914-926, 1979). The results suggested that the C. fasciculata ribosome might be unusual in containing three novel, low-molecular-weight ribosomal RNA components, designated e, f, and g (apparent chain lengths 240, 195, and 135 nucleotides, respectively), in addition to analogs of eucaryotic 5S (species h) and 5.8S (species i) ribosomal RNAs. In the present study, all of the presumptive ribosomal RNAs were indeed found to be associated with purified C. fasciculata ribosomes, and their localization was investigated in subunits produced under different conditions of ribosome dissociation. When ribosomes were dissociated in a high-potassium (880 mM K+, 12.5 mM Mg2+) medium, species e to i were all found in the large ribosomal subunit, which also contained an additional, transfer RNA-sized component (species j). However, when subunits were prepared in a low-magnesium (60 mM K+, 0.1 mM Mg2+) medium, two of the novel species (e and g) did not remain with the large subunit, but were released, apparently as free RNAs. Control experiments have eliminated the possibility that the small RNAs are generated by quantitative and highly specific (albeit artifactual) ribonuclease cleavage of large ribosomal RNAs during isolation. In terms of RNA composition and dissociation properties, therefore, the ribosome of C. fasciculata is the most "atypical" eucaryotic ribosome yet described. These observations raise interesting questions about the function and evolutionary origin of C. fasciculata ribosomes and about the organization and expression of ribosomal RNA genes in this organism.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chesters J. K. Protein synthesis by cell-free extracts of Crithidia oncopelti. Biochim Biophys Acta. 1966 Feb 21;114(2):385–397. doi: 10.1016/0005-2787(66)90318-2. [DOI] [PubMed] [Google Scholar]
- Cox R. A. Structure and function of prokaryotic and eukaryotic ribosomes. Prog Biophys Mol Biol. 1977;32(3):193–231. [PubMed] [Google Scholar]
- Cox R. A., Thompson R. D. Distribution of sequences common to the 25--28S-ribonucleic acid genes of Xenopus laevis and Neurospora crassa. Biochem J. 1980 Apr 1;187(1):75–90. doi: 10.1042/bj1870075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross G. A. Sedimentation roperties of polyribosomes, ribosomes and ribosomal subunits from Crithidia oncopelti. Biochim Biophys Acta. 1970 Apr 15;204(2):470–477. doi: 10.1016/0005-2787(70)90167-x. [DOI] [PubMed] [Google Scholar]
- Curgy J. J., Ledoigt G., Stevens B. J., André J. Mitochondrial and cytoplasmic ribosomes from Tetrahymena pyriformis. Correlative analysis by gel electrophoresis and electron microscopy. J Cell Biol. 1974 Mar;60(3):628–640. doi: 10.1083/jcb.60.3.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F. Postmaturational cleavage of 23s ribosomal ribonucleic acid and its metabolic control in the blue-green alga Anacystis nidulans. J Bacteriol. 1973 Mar;113(3):1256–1263. doi: 10.1128/jb.113.3.1256-1263.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Garvin R. T., Hill R. C., Weber M. M. The atypical RNA components of cytoplasmic ribosomes from Crithidia fasciculata. Arch Biochem Biophys. 1978 Dec;191(2):774–781. doi: 10.1016/0003-9861(78)90419-8. [DOI] [PubMed] [Google Scholar]
- Gerbi S. A. Fine structure of ribosomal RNA. I. Conservation of homologous regions within ribosomal RNA of eukaryotes. J Mol Biol. 1976 Sep 25;106(3):791–816. doi: 10.1016/0022-2836(76)90265-5. [DOI] [PubMed] [Google Scholar]
- Glover C. V., Gorovsky M. A. Amino-acid sequence of Tetrahymena histone H4 differs from that of higher eukaryotes. Proc Natl Acad Sci U S A. 1979 Feb;76(2):585–589. doi: 10.1073/pnas.76.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. The ribosomal RNA of the trypanosomatid protozoan Crithidia fasciculata: physical characteristics and methylated sequences. Can J Biochem. 1979 Jun;57(6):914–926. doi: 10.1139/o79-111. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol. 1976;31(2):95–144. doi: 10.1016/0079-6107(78)90006-8. [DOI] [PubMed] [Google Scholar]
- Hassur S. M., Whitlock H. W., Jr UV shadowing--a new and convenient method for the location of ultraviolet-absorbing species in polyacrylamide gels. Anal Biochem. 1974 May;59(1):162–164. doi: 10.1016/0003-2697(74)90020-7. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Evolution of ribosomal RNA. Comp Biochem Physiol B. 1977;58(1):1–7. doi: 10.1016/0305-0491(77)90116-x. [DOI] [PubMed] [Google Scholar]
- Jordan B. R. Demonstration of intact 26 S ribosomal RNA molecules in Drosophila cells. J Mol Biol. 1975 Oct 15;98(1):277–280. doi: 10.1016/s0022-2836(75)80117-3. [DOI] [PubMed] [Google Scholar]
- Kahan D., Zahalsky A. C., Hutner S. H. Protein synthesis in cell-free preparation of Crithidia fasciculata. J Protozool. 1968 May;15(2):385–390. doi: 10.1111/j.1550-7408.1968.tb02142.x. [DOI] [PubMed] [Google Scholar]
- Kaulenas M. S. Rapid isolation of insect ribosomal subunits by ethanol-magnesium precipitation. Anal Biochem. 1971 May;41(1):126–131. doi: 10.1016/0003-2697(71)90197-7. [DOI] [PubMed] [Google Scholar]
- Kramer R. A., Philippsen P., Davis R. W. Divergent transcription in the yeast ribosomal RNA coding region as shown by hybridization to separated strands and sequence analysis of cloned DNA. J Mol Biol. 1978 Aug 15;123(3):405–416. doi: 10.1016/0022-2836(78)90087-6. [DOI] [PubMed] [Google Scholar]
- Lau R. Y., Kennedy T. D., Lane B. G. Wheat-embryo ribonucleates. III. Modified nucleotide constituents in each of the 5.8S, 18S and 26S ribonucleates. Can J Biochem. 1974 Dec;52(12):1110–1123. doi: 10.1139/o74-155. [DOI] [PubMed] [Google Scholar]
- Mackay R. M., Spencer D. F., Doolittle W. F., Gray M. W. Nucleotide sequences of wheat-embryo cytosol 5-S and 5.8-S ribosomal ribonucleic acids. Eur J Biochem. 1980 Dec;112(3):561–576. doi: 10.1111/j.1432-1033.1980.tb06122.x. [DOI] [PubMed] [Google Scholar]
- Martin T. E., Bicknell J. N., Kumar A. Hybrid 80S monomers formed from subunits of ribosomes from protozoa, ungi, plants, and mammals. Biochem Genet. 1970 Oct;4(5):603–615. doi: 10.1007/BF00486098. [DOI] [PubMed] [Google Scholar]
- Martin T. E., Wool I. G. Active hybrid 80 s particles formed from subunits of rat, rabbit and protozoan (Tetrahymena pyriformis) ribosomes. J Mol Biol. 1969 Jul 14;43(1):151–161. doi: 10.1016/0022-2836(69)90085-0. [DOI] [PubMed] [Google Scholar]
- Morales N. M., Roberts J. F. The ribonucleic acids of Crithidia fasciculata. J Protozool. 1978 Feb;25(1):140–144. doi: 10.1111/j.1550-7408.1978.tb03886.x. [DOI] [PubMed] [Google Scholar]
- Palmer F. B. Biosynthesis of choline and ethanolamine phospholipids in Crithidia fasciculata. J Protozool. 1974 Feb;21(1):160–163. doi: 10.1111/j.1550-7408.1974.tb03631.x. [DOI] [PubMed] [Google Scholar]
- Parish J. H., Kirby K. S. Reagents which reduce interactions between ribosomal RNA and rapidly labelled RNA from rat liver. Biochim Biophys Acta. 1966 Dec 21;129(3):554–562. doi: 10.1016/0005-2787(66)90070-0. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne P. I., Loening U. E. RNA breakdown accompanying the isolation of pea root microsomes. An analysis by polyacrylamide gel electrophoresis. Biochim Biophys Acta. 1970 Nov 12;224(1):128–135. doi: 10.1016/0005-2787(70)90626-x. [DOI] [PubMed] [Google Scholar]
- Perry R. P. Processing of RNA. Annu Rev Biochem. 1976;45:605–629. doi: 10.1146/annurev.bi.45.070176.003133. [DOI] [PubMed] [Google Scholar]
- Sherton C. C., Di Camelli R. F., Wool I. G. Separation of large quantities of eukaryotic ribosomal subunits by zonal ultracentrifugation. Methods Enzymol. 1974;30:354–367. doi: 10.1016/0076-6879(74)30038-9. [DOI] [PubMed] [Google Scholar]
- Sinclair J. H., Brown D. D. Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry. 1971 Jul 6;10(14):2761–2769. doi: 10.1021/bi00790a017. [DOI] [PubMed] [Google Scholar]
- Spencer R., Cross G. A. Lability of RNA from the large cytoplasmic ribosomal subunit of the protozoon Crithidia oncopelti. J Gen Microbiol. 1976 Mar;93(1):82–88. doi: 10.1099/00221287-93-1-82. [DOI] [PubMed] [Google Scholar]