Abstract
Bioassay-guided fractionation of the extract of Topsentia sp. led to the identification of two new sulfated sterols, geodisterol-3-O-sulfite (1) and 29-demethylgeodisterol-3-O-sulfite (2), the active constituents reversing efflux pump-mediated fluconazole resistance. Both compounds enhanced the activity of fluconazole in a Saccharomyces cerevisiae strain overexpressing the Candida albicans efflux pump MDR1, as well as in a fluconazole-resistant Candida albicans clinical isolate known to overexpress MDR1. These results provide insight into the clinical utility of combining efflux pump inhibitors with current antifungals to combat the resistance associated with opportunistic fungal infections caused by C. albicans.
In recent decades, the occurrence of systemic fungal infections has been on the rise with the most dramatic increase taking place among immunocompromised individuals whose populations have increased in parallel with the AIDS pandemic, chemotherapy treatment for cancer, and organ transplantation. Among such populations, especially those infected with HIV, low doses of fluconazole (FLU) are often administered prophylactically to prevent the occurrence of opportunistic fungal infections. Such prolonged treatment has resulted in the appearance of azole-resistant phenotypes of Candida albicans.1,2
The azoles are a class of antifungal drugs that include triazoles like FLU and imidazoles such as ketoconazole that inhibit ergosterol biosynthesis by competitive inhibition of the enzyme lanosterol 14α-demethylase. To date, multiple mechanisms of FLU resistance in C. albicans have been identified and include: point mutations in the ERG11 gene that codes for lanosterol 14α-demethylase, alteration of other genes involved in ergosterol biosynthesis (e.g. ERG3), and decreased intracellular accumulation of FLU via membrane-associated efflux pumps.1–3 Of the four major classes of efflux pumps, two have been identified in C. albicans as belonging to either the major facilitator superfamily (MDR) which functions as an H+-antiporter or the ATP-binding cassette superfamily (CDR) which requires ATP for active transport of FLU out of the fungal cell.4
As part of an ongoing effort to discover efflux pump inhibitors to treat FLU-resistant C. albicans, we have employed a whole cell screen similar to that used by Lee et al.5 to identify natural products that reverse efflux pump-mediated FLU resistance.6 Plasmids containing C. albicans genes that encode the efflux pumps CDR1 and MDR1 have been incorporated into Saccharomyces cerevisiae to produce a phenotypic resistance to FLU.7–9 These azole resistant S. cerevisiae strains (along with a null pump strain) were used to evaluate over 10,000 crude extracts of marine organisms from the NCI Open Repository in the presence and absence of a low concentration of FLU. Hits are selected based on lack of inherent antifungal activity but potent growth inhibition when tested in combination with FLU in efflux pump-containing strains (little to no synergy with FLU in the null pump strain). The marine sponge Topsentia sp. was thus selected for bioassay-guided fractionation due to significant enhancement of FLU activity (> 9×) in the genetically-altered S. cerevisiae strain overexpressing the C. albicans MDR1 efflux pump. Subsequent bioassay-guided fractionation of the extract led to the isolation and identification of two new sulfated sterols, geodisterol-3-O-sulfite (1) and 29-demethylgeodisterol-3-Osulfite (2), The isolation, structure elucidation, and biological activities of 1 and 2 are discussed herein.
The initial bioassay-guided fractionation of the extract of Topsentia sp. was carried out by using a Supelco LC-18 cartridge eluted with aqueous MeOH. The actives that demonstrated a synergistic effect with FLU against the S. cerevisiae MDR1 strain were located in the two fractions (60 and 80% aq. MeOH). Subsequent scale-up reversed-phase chromatography of the extract, monitored by bioassay and TLC comparison with the active fractions mentioned above, yielded compounds 1 and 2.
HRESIMS data for compound 1 contained a pseudomolecular ion at m/z 551.2397 corresponding to a molecular formula of C28H41Na2O6S. The presence of sulfur was supported by the intensity of the [M + 2]+ peak which was about 7.7% of the pseudomolecular ion peak. Evidence for the sulfate group came from the strong asymmetric S=O stretch absorption at 1226 cm−1 in the IR spectrum of 1.10
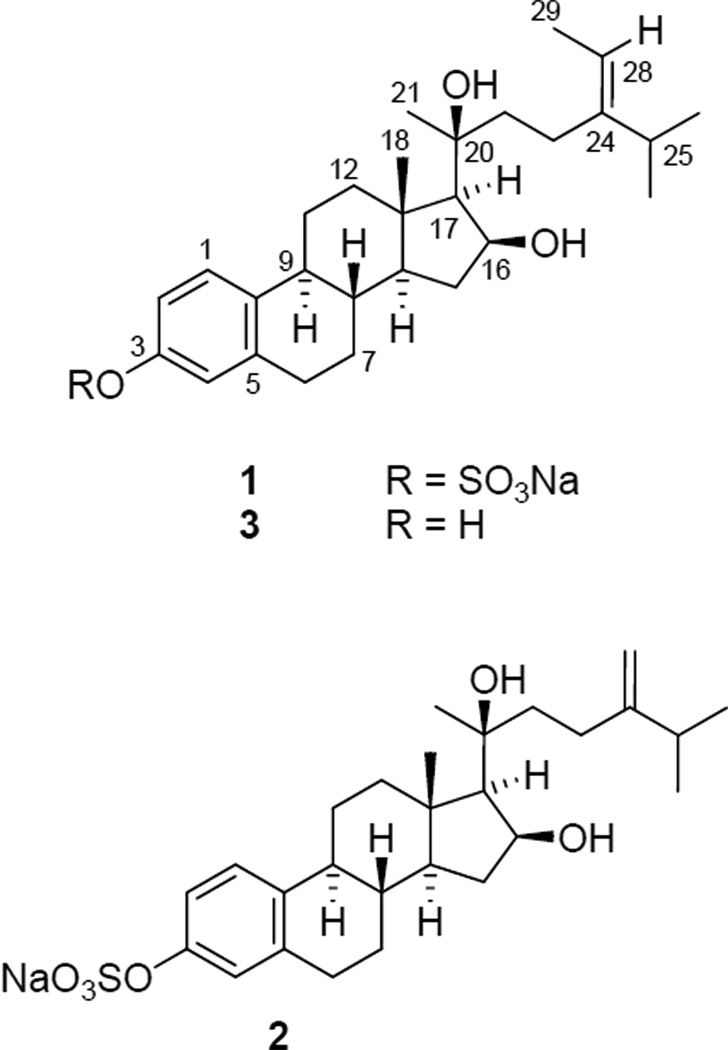
The 13C NMR data of 1 indicated the presence of 28 carbons comprising five methyl, seven methylene, 10 methine, and six quaternary carbons (C28H39). Thus, the remaining two hydrogens were assigned to two hydroxy groups attached to the carbons with resonances at δ 74.5 (CH) and 78.0 (qC). The five methyls resonated at δ 1.04 (6H, d, J = 7.2 Hz), 1.19 (3H, s), 1.37 (3H, s), 1.39 (1H, d, J = 7.8 Hz), and 1.63 (3H, d, J = 6.6 Hz) in the 1H NMR spectrum. This indicated that compound 1 might possess a steroidal skeleton as in the sulfated sterols isolated from the same Topsentia genus.11 A distinguishing feature of the 1H NMR spectrum of 1 was the presence of the resonances at δ 7.02 (brs), 7.05 (d, J = 8.4 Hz), and 7.23 (d, J = 8.4 Hz) attributable to a 1,2,4-trisubstituted aromatic system. The six aromatic carbon resonances were identified by HMQC and HMBC at δ 119.7 (CH), 122.4 (CH), 126.8 (CH), 138.1 (C), 138.6 (C), and 151.6 (C) from the eight aromatic/olefinic carbon resonances in the downfield region of the 13C NMR spectrum. Naturally occurring sterols with an aromatic ring system from marine organisms are rare. A literature survey showed that there was only one compound, geodisterol (3), that was isolated from the tropical marine sponge Geodia sp. and possessed an aromatic A ring.12 On comparison of the NMR data of 1 with those of 3, the close resemblance indicated that 1 was an O-sulfited derivative of 3.
Upon solvolysis with pyridine and 1,4-dioxane, 1 afforded the known geodisterol (3).12 The placement of the single sulfate group in 1 was determined at C-3 since C-2, C-4, and C-10 resonances were shifted downfield (Δδ + 7.2, 7.5, and 7.6 ppm, respectively) compared to 3, while the C-3 resonance was shifted upfield (Δδ − 2.7 ppm). This is consistent with the substitution effects of a sulfate group on the aromatic ring.13 Analysis of the 2D NMR data including COSY, HMQC, HMBC, and NOESY of 1 assigned its 1H and 13C NMR chemical shifts as shown in Table 1. Thus, the structure of 1 was elucidated as sodium (16S,20S,24E)-16,20-dihydroxy-19-norstigmasta-1,3,5(10),24(28)-tetraen-3-yl sulfate (geodisterol-3-O-sulfite).
Table 1.
NMR Data for Compounds 1 and 2 in methanol-d4a
| 1 |
2 |
|||
|---|---|---|---|---|
| δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) | |
| 1 | 126.8, CH | 7.23, d (8.4) | 126.5, CH | 7.24, d (8.5) |
| 2 | 119.7, CH | 7.05, d (8.4) | 119.4, CH | 7.05, d (8.5) |
| 3 | 151.6, qC | 151.1, qC | ||
| 4 | 122.4, CH | 7.02, brs | 122.2, CH | 7.02, brs |
| 5 | 138.6, qC | 138.3, qC | ||
| 6 | 30.5, CH2 | 2.87, a/b | 30.6, CH2 | 2.87, a/b |
| 7 | 28.6, CH2 | 1.34, α | 28.7, CH2 | 1.49, α |
| 1.91, β | 1.92, β | |||
| 8 | 39.0, CH | 1.48, | 39.1, CH | 1.49, |
| 9 | 45.2, CH | 2.22, | 45.2, CH | 2.23, |
| 10 | 138.1, qC | 137.8, qC | ||
| 11 | 27.5, CH2 | 1.56, β | 27.6, CH2 | 1.55, β |
| 2.29, α | 2.32, α | |||
| 12 | 41.7, CH2 | 1.41, α | 41.7, CH2 | 1.44, α |
| 2.25, β | 2.28, β | |||
| 13 | 44.4, qC | 44.4, qC | ||
| 14 | 54.6, CH | 1.12 | 54.6, CH | 1.15, |
| 15 | 37.8, CH2 | 1.40, a | 37.8, CH2 | 1.40, a |
| 2.34, b | 2.35, b | |||
| 16 | 74.5, CH | 4.67 | 74.4, CH | 4.63, |
| 17 | 61.0, CH | 1.39, d (7.8) | 61.1, CH | 1.38, |
| 18 | 15.3, CH3 | 1.19, s | 15.5, CH3 | 1.19, s |
| 20 | 78.0, qC | 77.9, qC | ||
| 21 | 26.2, CH3 | 1.37, s | 26.4, CH3 | 1.34, s |
| 22 | 43.9, CH2 | 1.70, dt (4.2, 13.2) a | 43.9, CH2 | 1.82, dt (5.0, 12.3) a |
| 1.87, b | 1.93, b | |||
| 23 | 24.9, CH2 | 2.08, dt (3.6, 12.6) a | 30.6, CH2 | 2.12, a/b |
| 2.15, dt (5.4, 12.6) b | ||||
| 24 | 147.5, qC | 157.2, qC | ||
| 25 | 36.1, CH | 2.27 | 35.1, CH | 2.32 |
| 26 | 22.7, CH3 | 1.04, d (7.2) | 22.5, CH3 | 1.08, d (7.4) |
| 27 | 22.6, CH3 | 1.04, d (7.2) | 22.5, CH3 | 1.08, d (7.4) |
| 28 | 117.0, CH | 5.25, q (6.6) | 106.7, CH2 | 4.74, brs, a |
| 4.77, brs, b | ||||
| 29 | 13.4, CH3 | 1.63, d (6.6) | ||
Data obtained at 600 MHz for 1H NMR and 125 MHz for 13C NMR for 1 and at 500 MHz for 1H NMR and 125 MHz for 13C NMR for 2. For 1H NMR, well-resolved couplings are expressed with coupling patterns and coupling constants in Hz in parentheses. For overlapped signals, only chemical shift values are given. Some geminal protons were denoted with α- or β-orientation based on NOE evidence.
The absolute configuration of C-16 in 3 was determined as S by Mosher’s method, while the absolute configuration of C-20 was assigned as S on the basis of the NOE correlation between Me-21 and H-12.12 We have further examined the issue concerning the C-20 configuration in 1. According to the results reported by Nes and Varkey,14,15 free rotation around the C-17 − C-20 bond in 20-hydroxysteroids is hindered when C-20 is tetrahedral but is unhindered when C-20 is trigonal planar. Reasons for these observations were based on steric interactions between the hydrogens of Me-21 and C-22 with the hydrogens of C-16 and Me-18, which in the case of 20S-hydroxysteroids results in a predominant conformation with the C-20 hydroxy group arranged pseudoaxially in front and parallel to Me-18, and Me-21 arranged pseudoequatorially to the rear. Such a conformation sets up a 1,3-diaxial interaction between the C-20 hydroxy group and Me-18 which would be reflected in a downfield shift for the Me-18 hydrogens relative to cholesterol.15 Indeed, such a downfield shift for the Me-18 hydrogens of 1 is observed (δ 1.19 vs. 0.68 in cholesterol). Along similar lines, hindered rotation around the C-17 − C-20 bond would put Me-21 in a different chemical environment depending on the configuration of C-20 which, for example, would be reflected in the chemical shift of the methyl hydrogens, as well as placing Me-21 in close spatial proximity to the equatorial C-12β hydrogen in 20Shydroxysteroids. Based on this observation, the configuration of C-20 was deduced as S by comparison of the chemical shift for Me-21 of 1 (δ 1.37) with that of similar 20S- and 20R-hydroxysteroids. For 20S- and 20R-hydroxysteroids, Me-21 resonates at ca. δ 1.28 and ca. δ 1.13, respectively.15 Again, a NOE correlation between Me-21 and the equatorial H-12β at δ 2.25 was observed in the NOESY spectrum of 1, indicating that such NOE evidence is reliable for the assignment of the C-20 absolute configuration of this compound.
The HRESIMS spectrum for 2 showed a pseudomolecular ion at m/z 537.2252 corresponding to the molecular formula C27H39Na2O6S. As for 1, the presence of sulfur was indicated from the intensity of the [M + 2]+ ion (8%). The IR spectrum also showed the same strong asymmetric stretch absorption of the S=O bond at 1229 cm−1 seen in 1. Analysis of the 1H NMR spectrum of 2 showed it to have marked similarities with that of 1 except for the distinct absence of the methyl doublet at δ 1.63 present in the 1H NMR spectrum of 1. Instead, the presence of a 1,1-disubstituted double bond was evident from the proton resonances at δ 4.74 (brs) and 4.77 (brs). Further analysis of the 13C, COSY, HMQC, and HMBC NMR data verified the structural similarities and difference between 1 and 2, leading to the proposed structure of 2, i.e., lacking the C-29 methyl group in the side chain of 1. The similarity of the 1H and 13C NMR chemical shifts of the hydrogens and carbons around the stereogenic centers of compounds 1 and 2 (Table 1) as well as their similar specific rotation values supports the proposal that they have the same absolute configuration. Thus, the structure of 2 was established as sodium (16S,20S)-16,20-dihydroxy-19-norergosta-1,3,5(10),24(28)-tetraen-3-yl sulfate (29-demethylgeodisterol-3-Osulfite).
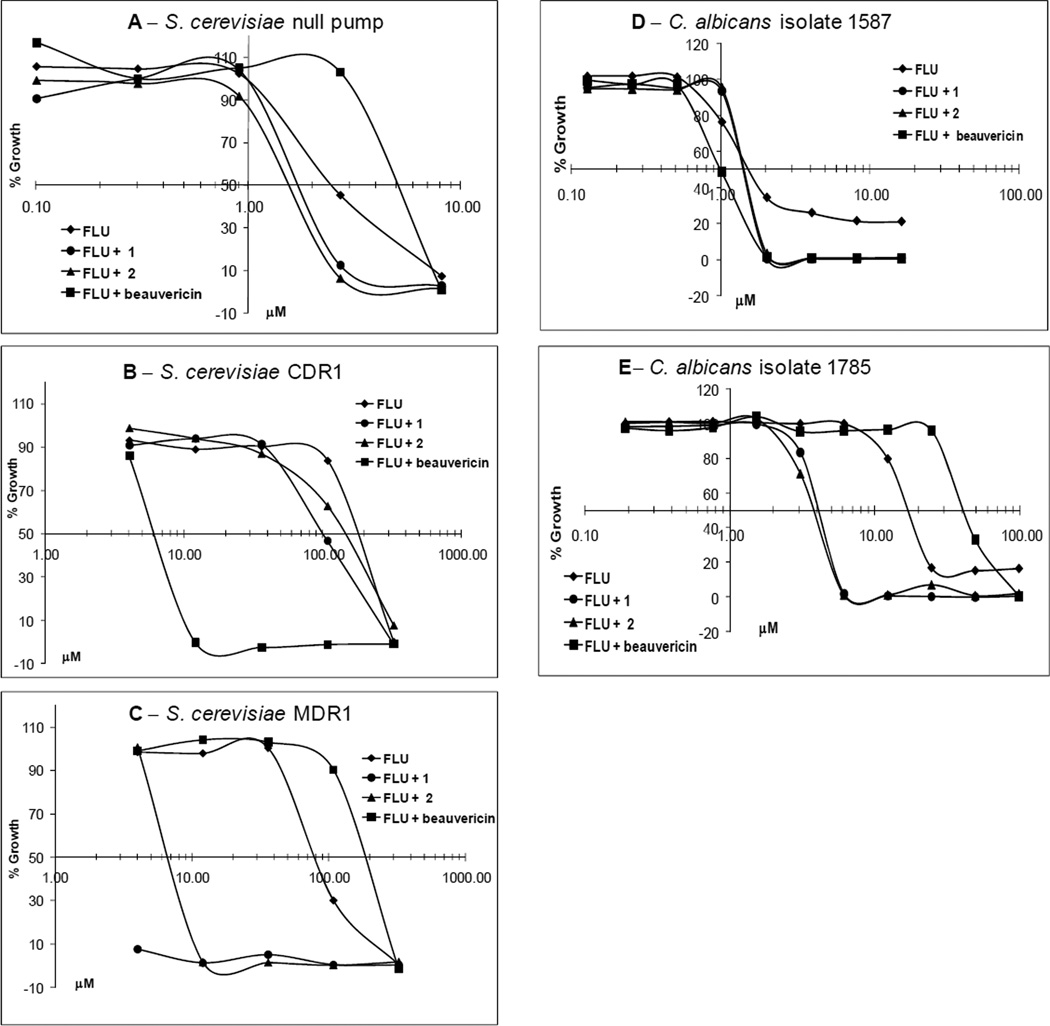
Compounds 1 and 2 were evaluated for their FLU-resistance reversal activity using a checkerboard assay6 against the null pump, CDR1 and MDR1 S. cerevisiae strains. Figure 1 shows the selectivity of 1 and 2 against the MDR1 efflux pump strain, with 1 exhibiting stronger synergistic activity with FLU compared to 2. Compound 3 was also assayed for FLU-resistance reversal activity and was found to have no activity, showing the sulfate group to be necessary for MDR1 inhibition and synergy with FLU (data not shown). The fractional inhibitory concentrations (FICs), which are used to determine the extent of synergy or antagonism in combination treatment,16 are shown in Table 2.
Figure 1.
Dose-response curves for FLU alone (diamond) and in combination with 13 µM 1 (circle), 13 µM 2 (triangle), or 25 µM beauvericin (square), in the null pump (A), CDR1 (B), and MDR1 (C) S. cerevisiae strains and for FLU alone (diamond) and in combination with 189 µM 1 (circle), 194 µM 2 (triangle), or 5.7 µM beauvericin (square) in C. albicans isolates 1587 (FLU-susceptible) (D) and 1758 (FLU-resistant via MDR1 efflux pumps) (E).
Table 2.
Fractional Inhibitory Concentrations (FICs)a of Combination Treatment FLU with 1, 2, and Beauvericin
|
S. cerevisiae Null pumpb |
S. cerevisiae CDR1b |
S. cerevisiae MDR1b |
C. albicans #1b,c |
C. albicans #17b,d |
C. albicans 1587e,f |
C. albicans 1758e,g |
|
|---|---|---|---|---|---|---|---|
| 1 | 1.10 | 1.04 | 0.08 | 1.5 | 1.3 | 1.7 | 0.2 |
| 2 | 0.75 | 1.34 | 0.15 | 1.7 | 1.8 | 1.6 | 0.2 |
| beauvericin | 3.38 | 0.04 | 2.28 | 0.5 | 0.7 | 0.5 | 2.8 |
FIC [(IC50 of test compound in combination with FLU/IC50 of test compound alone) + (IC50 of FLU in combination with test compound/IC50 of FLU alone)] ≤ 0.5 = synergistic; 0.51–1.0 = additive; 1.1–2.0 = indifferent; > 2.0 = antagonistic.
Concentration of 1 = 13 µM, 2 = 13 µM, beauvericin = 26 µM.
FLU-susceptible isolate.
FLU-resistant isolate (multiple resistant mechanisms).
Concentration of 1 = 189 µM, 2 = 194 µM, beauvericin = 5.7 µM.
FLU-susceptible isolate.
FLU-resistant isolate overexpressing MDR1 efflux pump.
In addition, both compounds were evaluated for their ability to reverse FLU resistance in two different matched FLU-susceptible and FLU-resistant patient isolates of C. albicans. FLU-resistant C. albicans #17 (matched FLU-susceptible isolate #1) is reported to possess several mechanisms of FLU resistance, including CDR1 and MDR1 overexpression, as well as overexpression of ERG11 (formerly ERG16), the gene coding for FLU’s target, lanosterol 14α-demethylase.17 FLU-resistant C. albicans 1758 (matched FLU-susceptible isolate 1587) is reported to possess overexpression of the MDR1 efflux pump as the sole mechanism of resistance.18 Significant synergy with FLU was observed for both 1 and 2 (FIC = 0.2 for both compounds) only in strain 1758 (Figure 1 and Table 2). Enhancement of FLU activity was not observed with strains #1, 17 and 1587 suggesting that inhibition of the MDR1 efflux pump alone may not be enough to counteract FLU resistance in strains with multiple mechanisms of resistance.
The antimicrobial and antiprotozoal activities of 1−3 were evaluated against Candida albicans, C. glabrata, C. krusei, Cryptococcus neoformans, Aspergillus fumigatus, methicillin-resistant Staphylococcus aureus (MRSA), Mycobacterium intracellulare, Plasmodium falciparum (D6 and W2 clones),19 and Leishmania donovani.20,21 Of the compounds tested none exhibited any inherent antimicrobial or antiprotozoal activities, and all were shown to be non-toxic against Vero cells up to the highest concentration tested [9.00 µM (1), 9.25 µM (2), 11.2 µM (3)].22 The lack of antimicrobial/cytotoxic activity exhibited by 1 and 2 coupled with the fact that they are only active in the presence of FLU in the MDR1 S. cerevisiae and FLU-resistant C. albicans 1758 strains further suggest that their site of action is indeed the major facilitator efflux pump MDR1. Such specific activity provides a sound argument for the clinical utility of efflux pump inhibitors in combination with currently used azole antifungals to approach azole-resistant populations of C. albicans whose sole mechanism of resistance is overexpression of the MDR1 efflux pump.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Rudolph Autopol IV polarimeter. The IR spectra were obtained on a Bruker FTIR OPUS spectrometer. A Varian UV/VIS spectrometer was used to acquire UV spectra. The 1D and 2D NMR data for 1 were acquired on a Varian Inova 600 spectrometer operating at 600 MHz for 1H and 150 MHz for 13C. 1D and 2D NMR data for 2 were acquired on a Bruker AVX 500 spectrometer operating at 500 MHz for 1H and 125 MHz for 13C. The 1H and 13C NMR spectra for 3 were acquired on a Bruker DRX 400 spectrometer operating at 400 MHz for 1H and 100 MHz for 13C and processed using MestReLab’s MNova software (v. 2.2). The chemical shifts are relative to the NMR solvent methanol-d4 (δH, 3.33; δC, 49.0) for 1 and 2 and to the TMS for 3. The HRESIMS data was acquired on a Bruker-Magnex BioAPEX 30es ion cyclotron high resolution HPLC-FT spectrometer by direct injection. A Supelco Supeclean LC-18 cartridge (C18 reversed-phase silica gel, 10 g) was used for initial fractionation. Vacuum liquid chromatography (VLC) was done with reversed-phase silica gel (Bakerbond ODS, 40 µm, J.T. Baker). TLC was conducted on Si gel sheets (Si gel 60 F254, Merck, Germany) and reversed-phase glass plates (RP-18, F254s, Merck, Germany). General procedures for the antimicrobial and cytotoxicity assays have been described previously.6,20–22
Sponge Material
The marine sponge Topsentia sp. (Halichondriidae) was collected in December 1992, from Chuuk, Federated States of Micronesia (latitude: 07°08.70′ N, longitude: 151°53.15′ E) by divers using SCUBA. The sponge was identified by Michelle Kelly Borges. A voucher specimen was deposited in the Smithsonian Institute, Washington, D.C. (voucher # OCDN0879).
Extraction and Isolation
The extraction process was carried out at the NCI. Briefly, the sponge was allowed to thaw to 4 °C and extracted with H2O. The aqueous mixture was centrifuged and the pellet freeze-dried. The pellet was then extracted overnight at room temperature with CH2Cl2/MeOH (1:1). The organic layer was removed, residual solvent evaporated in vacuo and the crude extract stored at −20 °C in the NCI Open Repository at the Frederick Cancer Research and Development Center (Frederick, MD). The sponge extract was obtained through the NCI Open Repository Program.
The initial bioassay-guided fractionation was carried out by using 100 mg of the extract, which was loaded to a Supelco LC-18 cartridge (10 g reversed-phase silica gel). Elution of the cartridge with H2O, 40% MeOH, 60% MeOH, 80% MeOH, MeOH, and acetone (20 mL each) afforded six column fractions. The activity was located in the 60 and 80% MeOH fractions, both showing an IC50 of 1.0 µg/mL, in combination with FLU at 9 μg/mL, against S. cerevisiae MDR1 (the IC50s of FLU and both column fractions were 90 and >14 μg/mL, respectively). Subsequently, the extract (1.78 g) was fractionated using reversed-phase VLC and an isocratic solvent system of 70% MeOH to afford 20 fractions. The fractions were monitored by TLC comparison with the aforementioned active fractions and bioassay. Compound 1 was obtained from fraction 10 (111.4 mg, yield 6.2%) and 2 from fractions 6 and 7 (56.7 mg, yield 3.2%) in a pure state.
Geodisterol-3-O-sulfite (1): amorphous solid; [α]D29 +14.7 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 202 (4.43), 268 (3.00) nm; IR (NaCl) νmax 3219 (br), 2920, 1633, 1226 (s), 1054 (s), 931, 823 cm−1; NMR (methanol-d4), see Table 1; HRESIMS m/z 551.2397 [M − H + 2Na]+ (calcd for C28H41Na2O6S, 551.2419).
29-Demethylgeodisterol-3-O-sulfite (2)
amorphous solid; [α]29D +14.5 (c 0.18, MeOH); UV (MeOH) λmax (log ε) 202 (4.22), 266 (2.73) nm; IR (NaCl) νmax 3254 (br), 2915, 1634, 1229 (s), 1057 (s), 933, 822 cm−1; NMR (methanol-d4), see Table 1; HRESIMS m/z 537.2252 [M − H + 2Na]+ (calcd for C27H39Na2O6S, 537.2262).
Solvolysis of 1
Compound 1 (52.8 mg) was dissolved in 10 mL of 4:1 pyridine:1,4-dioxane under Ar and kept in an oil bath at 80 °C for 1.5 h. The solvents were removed in vacuo and the unpurified reaction product crystallized from MeOH to afford compound 3 (25 mg).
Geodisterol (3)
[α]25D +74.0 (c 0.34, CH2Cl2−MeOH 9:1) [lit.14 [α]D +67 (c 0.31, CH2C12−MeOH 9:1)]; IR (NaCl) νmax 3379 (br), 2919, 1620, 926, 820 cm−1; NMR data (see Supporting Information) is in agreement with the literature values.12
Assay for Reversal of Azole Resistance in S. cerevisiae and C. albicans Strains
Samples were diluted in 0.9% saline and transferred to 96-well microplates. C. albicans and S. cerevisiae inocula were prepared in YPD broth to afford 1 × 104 CFU/mL after addition to the samples. FLU (Sequoia Research Products, UK), dissolved in DMSO was added to the FLU + inocula at a concentration of ~1/10th (S. cerevisiae) or ~1/3rd (C. albicans) the IC50 (an equivalent volume of DMSO alone was added to duplicate inocula for FLU – data). Beauvericin (Sigma, St. Louis, MO) was included in each experiment as a positive control for the CDR1 strain. The microplates were read at 630 nm using an EL-340 Biokinetics microplate reader (Bio-Tek Instruments, Winooski, VT) prior to and after incubation at 30 °C for 40–48 h (S. cerevisiae) or 37 °C for 24 h (C. albicans). For checkerboard assays, FLU and diluted samples were transferred to microplates to afford one compound running vertically and FLU horizontally (all combinations of concentrations were generated). One row or column was reserved for each test compound and saline only. The microbial inocula were prepared as above without FLU or DMSO, and added to microplates. The fractional inhibitory concentration (FIC) is calculated by the formula, (IC50 of test compound in combination with FLU/IC50 of test compound alone) + (IC50 of FLU in combination with test compound/IC50 of FLU alone).
Supplementary Material
Figure 2.
Acknowledgment
We thank the Natural Products Branch Repository Program at the National Cancer Institute for providing marine extracts from the NCI Open Repository, D. Sanglard (Institute of Microbiology, University Hospital, Lausanne, Switzerland) for supplying the S. cerevisiae strains, T. White (Seattle Biomedical Research Institute, Seattle, WA) and S. Redding (The University of Texas Health Science Center at San Antonio) for providing the fluconazole-resistant Candida albicans patient isolates, M. Wright, S. Khan, and J. Trott for biological assays, F. Wiggers for NMR data acquisition, and D. C. Dunbar for HRESIMS data. This work was supported in part by a grant from the Public Health Service, National Institute of Allergy and Infectious Diseases, Grant No. R01 AI027094, and the USDA Agricultural Research Service Specific Cooperative Agreement No. 58-6408-2-0009.
Footnotes
Supporting Information Available: NMR data of compounds 1−3 are available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Kontoyiannis DP, Lewis RE. Lancet. 2002;359:1135–1144. doi: 10.1016/S0140-6736(02)08162-X. [DOI] [PubMed] [Google Scholar]
- 2.White TC, Marr KA, Bowden RA. Clin. Microbiol. Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanglard D. Curr. Opin. Microbiol. 2002;5:379–385. doi: 10.1016/s1369-5274(02)00344-2. [DOI] [PubMed] [Google Scholar]
- 4.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. Antimicrob. Agents Chemother. 2002;46:1704–1713. doi: 10.1128/AAC.46.6.1704-1713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MD, Galazzo JL, Staley AL, Lee JC, Warren MS, Fuernkranz H, Chamberland S, Lomovskaya O, Miller GH. Il Farmaco. 2001;56:81–85. doi: 10.1016/s0014-827x(01)01002-3. [DOI] [PubMed] [Google Scholar]
- 6.Jacob MR, Hossain CF, Mohammed KA, Smillie TJ, Clark AM, Walker LA, Nagle DG. J. Nat. Prod. 2003;66:1618–1622. doi: 10.1021/np030317n. [DOI] [PubMed] [Google Scholar]
- 7.Sanglard D, Ischer F, Monod M, Billie J. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 8.Calabrese D, Billie J, Sanglard D. Microbiology. 2000;146:2743–2754. doi: 10.1099/00221287-146-11-2743. [DOI] [PubMed] [Google Scholar]
- 9.Sanglard D, Kuchler K, Ischer F, Pagani J, Monod M, Billie J. Antimicrob. Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun HH, Cross SS, Gunasekera M, Koehn FE. Tetrahedron. 1991;47:1185–1190. [Google Scholar]
- 11.(a) Yang SW, Buivich A, Chan TM, Smith M, Lachowicz J, Pomponi SA, Wright AE, Mierzwa R, Patel M, Gullo V, Chu M. Bioorg. Med. Chem. Lett. 2003;13:1791–1794. doi: 10.1016/s0960-894x(03)00260-9. [DOI] [PubMed] [Google Scholar]; (b) Whitson EL, Bugni TS, Chockalingam PS, Concepcion GP, Harper MK, He M, Hooper JNA, Mangalindan GC, Ritacco F, Ireland CM. J. Nat. Prod. 2008;71:1213–1217. doi: 10.1021/np8001628. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Guzii Al G, Makarieva TN, Denisenko VA, Dmitrenok PS, Burtseva YV, Krasokhin VB, Stonik VA. Tetrahedron Lett. 2008;49:7191–7193. [Google Scholar]
- 12.Wang G-Y-S, Crews P. Tetrahedron Lett. 1996;37:8145–8146. [Google Scholar]
- 13.Li XC, Joshi AS, Tan B, ElSohly HN, Walker LA, Zjawiony J, Ferreira D. Tetrahedron. 2002;58:8709–8717. [Google Scholar]
- 14.Nes WR, Varkey TE. J. Org. Chem. 1976;41:1652–1653. doi: 10.1021/jo00871a041. [DOI] [PubMed] [Google Scholar]
- 15.Nes WR, Varkey TE, Crump DR, Gut M. J. Org. Chem. 1976;41:3429–3433. doi: 10.1021/jo00883a023. [DOI] [PubMed] [Google Scholar]
- 16.Li R. Antimicrob. Agents Chemother. 1999;43:1401–1405. doi: 10.1128/aac.43.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White TC. Antimicrob. Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaller MA, Rhine-Chalberg J, Redding SW, Smith J, Farinacci G, Fothergill AW, Rinaldi MG. J. Clin. Microbiol. 1994;32:59–64. doi: 10.1128/jcm.32.1.59-64.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The D6 clone is a chloroquine-sensitive strain and the W2 clone is a chloroquine-resistant strain.
- 20.Kaur K, Patel SR, Patil P, Jain M, Khan SI, Jacob MR, Ganesan S, Tekwani BL, Jain R. Bioorg. Med. Chem. 2007;15:915–930. doi: 10.1016/j.bmc.2006.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muhammad I, Dunbar DC, Kahn SI, Tekwani BL, Bedir E, Takamatsu S, Ferreira D, Walker LA. J. Nat. Prod. 2003;66:962–967. doi: 10.1021/np030086k. [DOI] [PubMed] [Google Scholar]
- 22.Yang CR, Zhang Y, Jacob MR, Khan SI, Zhang YJ, Li XC. Antimicrob. Agents Chemother. 2006;50:1710–1714. doi: 10.1128/AAC.50.5.1710-1714.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.