Abstract
Clostridium perfringens causes gas gangrene and gastrointestinal (GI) diseases in humans. The most common cause of C. perfringens-associated food poisoning is the consumption of C. perfringens vegetative cells followed by sporulation and production of enterotoxin in the gut. Despite the importance of spore formation in C. perfringens pathogenesis, the details of the regulation of sporulation have not yet been defined fully. In this study, microarray and bioinformatic analyses identified a candidate gene (the RNA regulator virX) for the repression of genes encoding positive regulators (Spo0A and sigma factors) of C. perfringens sporulation. A virX mutant constructed in the food poisoning strain SM101 had a much higher sporulation efficiency than that of the wild type. The transcription of sigE, sigF, and sigK was strongly induced at 2.5 h of culture of the virX mutant. Moreover, the transcription of the enterotoxin gene was also strongly induced in the virX mutant. Western blotting confirmed that the levels of enterotoxin production were higher in the virX mutant than in the wild type. These observations indicated that the higher levels of sporulation and enterotoxin production in the virX mutant were specifically due to inactivation of the virX gene. Since virX homologues were not found in any Bacillus species but were present in other clostridial species, our findings identify further differences in the regulation of sporulation between Bacillus and certain Clostridium species. The virX RNA regulator plays a key role in the drastic shift in lifestyle of the anaerobic flesh eater C. perfringens between the vegetative state (for gas gangrene) and the sporulating state (for food poisoning).
INTRODUCTION
Clostridium perfringens is a causative agent of a wide variety of enteric and histotoxic diseases (1, 2). Among five types (A to E), C. perfringens type A is known to cause two types of infectious diseases in humans. The first is gas gangrene (clostridial myonecrosis), in which C. perfringens typically contaminates a skin wound as a spore from the soil and then rapidly grows and produces various histolytic toxins and enzymes that destroy the surrounding muscles and connective tissues. The second type consists of a few strains (∼5% of all C. perfringens type A isolates) that have the ability to produce enterotoxin (CPE) (3). In both diseases, spores play an essential role in the pathogenesis of C. perfringens.
The CPE-producing strains are responsible annually for nearly 1 million cases of C. perfringens food poisoning, which ranks as the second most commonly reported bacterial food-borne disease in the United States (4). When vegetative CPE-producing C. perfringens cells grown in foods are ingested, the cells reach the small intestine and begin to sporulate. During the sporulation process, a large amount of CPE is produced and forms pores in the membranes of the intestinal epithelial cells, which results in watery diarrhea.
The sporulation process in Bacillus and Clostridium is initiated by a wide range of environmental and physiological signals induced by cell density, the Krebs cycle, and nutrient starvation (5). In Bacillus, sporulation-specific orphan histidine kinases integrate these signals and initiate a complex phosphorelay that leads to an increased concentration of phosphorylated Spo0A (Spo0A∼P), the master regulator of sporulation (6, 7). Spo0A∼P directly activates 121 genes, including those required for polar septum formation (8). Once Spo0A becomes an active form (Spo0A∼P), sporulation-specific sigma factors (SigF, SigE, SigG, and SigK) are sequentially activated (7, 9), which then activates hundreds of genes, and spores are formed and released from the mother cells (6, 7). In Clostridium, many genes related to the sporulation function are missing compared with Bacillus subtilis; in particular, the genes for the phosphorelay essential to B. subtilis sporulation are totally absent (10). Although the genes from spo0A to sigK are the same in both genera, recent evidence has suggested that the sporulation-specific RNA polymerase sigma factors are not expressed in a sequential fashion in Clostridium as in Bacillus species (11–13). Following the early forespore-specific gene sigF, the mother cell-specific sigK gene becomes expressed at sufficiently low levels to induce expression of the sigE gene, which in turn provides feedback and allows transcription of sigK (13). Interestingly, the late forespore-specific SigG protein is solely dependent on SigF (12). Sporulation in C. perfringens also seems to be dependent on an Agr-like quorum-sensing (QS) system that positively regulates sporulation and CPE production of C. perfringens (11).
In a previous study, the virX gene was reported to be a regulator for several toxin genes that are expressed at the log phase of vegetative growth (14). The virX gene encodes 51 amino acids that form a zinc-finger-containing peptide, and it positively regulates the theta-, alpha-, and kappa-toxin genes (pfoA, plc, and colA). Moreover, it was shown that virX acts as a small RNA regulator for the control of virulence in C. perfringens (14). In this report, we describe that virX is also a regulatory RNA gene for massive control of the sporulation-related genes and the enterotoxin gene in food-borne C. perfringens strains, which constitutes a unique regulatory system for spore formation that is distinct from that of Bacillus species.
MATERIALS AND METHODS
Strains, culture conditions, and plasmids.
C. perfringens strains 13 (15), TS186/pJIR418, and TS186/pTS907 (14) were cultured in GAM broth (Nissui, Japan) at 37°C under anaerobic conditions as described previously (16). The CPE-positive C. perfringens strain SM101 (17) and its derivative strains were cultured in fluid thioglycolate (FTG, Difco, BD) or TGY (3% tryptic soy broth [Becton Dickinson], 2% glucose, 1% yeast extract, and 0.1% cysteine). Escherichia coli strain DH5α and plasmid pUC118 were used for general cloning procedures. Plasmid pJIR418 (18) was used as an E. coli-C. perfringens shuttle vector and was transformed by electroporation-mediated transformation as previously described (16, 17).
Construction of a virX mutant strain.
A DNA fragment containing the virX region of SM101 (19) was amplified by a PCR using primer 1 (5′-TGGCCGGTCCTAAAGCTGTATCCA-3′) and primer 4 (5′-TGCCTTCATCAACTAATTGCT-3′). The amplified fragment was inserted into the HincII site of pUC118, which was used as a template for an inverse PCR using primers virX-null-2 (5′-TCTGACGCTTAAAATAAGCTAGT-3′) and virX-null-3 (5′-TAGAGCAATCTAACTTATGATT-3′). The erythromycin resistance gene (ermBP) was then cloned into the space between primers virX-null-2 and virX-null-3. The resultant plasmid was transformed into wild-type SM101. The transformants resulting from homologous recombination were screened on a brain heart infusion (BHI) plate containing erythromycin (25 μg/ml). Allelic-exchange mutation of the virX gene due to a double-crossover recombination was confirmed by PCR (see Fig. S1 in the supplemental material).
Construction of a virX-complemented strain.
Single-copy complementation of the virX mutant with wild-type and mutated virX genes was performed by single-crossover insertion of virX derivatives into the chromosome, using the plc gene (encoding alpha-toxin) for the insertion locus. First, the chloramphenicol resistance gene (catP) was amplified from pJIR418 by PCR, and the fragment was inserted into the HincII site of pUC118 (pCM118). The PCR-amplified plc fragment was then inserted into the HindIII site of pCM118 (pKO11). Finally, the intact virX fragment was amplified and inserted into the SmaI site of pKO11 (pKO12). The resulting plasmids, pKO11 and pKO12, were transformed into SM101 (wild type) and KO101 (virX mutant), respectively, by electroporation. The transformants resulting from single-crossover homologous recombination were screened on a BHI plate containing both erythromycin (25 μg/ml) and chloramphenicol (25 μg/ml). Using the same method, we also constructed KO101 derivatives that were complemented with mutated virX genes: KO103 and KO104. KO103 and KO104 were complemented with virX genes mutated at the 4th codon (AAA for Lys) to a stop codon (TAA) and at the 42nd codon (AAA for Lys) to a stop codon, derived from pTS913 and pTS915, respectively, in a previous study (14). All single-copy complementations of virX inserted into the chromosomal plc locus were confirmed by PCR (see Fig. S1 in the supplemental material).
Sporulation assay.
The wild-type strain (SM101), the virX mutant strain (KO101), and the complemented strains (KO102, KO103, and KO104) were inoculated into FTG medium, and the inoculated tubes were heat shocked at 70°C for 20 min. The strains were cultured at 37°C for 24 h, and 0.1 ml of each culture was inoculated and cultured into a second 10 ml of FTG medium at 37°C for 15 h. Four milliliters of the second precultured strain was inoculated into 200 ml of Duncan-Strong (DS) sporulating medium (1.5% proteose peptone [Becton Dickinson], 1% sodium phosphate, 0.4% soluble starch, 0.4% yeast extract, 0.1% sodium thioglycolate). Spores were examined at 6 or 24 h of culture. To count the number of heat-resistant spores, the 24-h DS-grown cultures were heated at 70°C for 20 min, serially diluted with phosphate-buffered saline (PBS), plated onto BHI agar, and incubated anaerobically at 37°C for 18 to 24 h, and then the colonies were counted.
Northern blot hybridizations.
Total RNA from C. perfringens vegetative cells was extracted according to a previously described method (14). From sporulating cultures in DS medium, RNA was isolated by a modified method. Briefly, cell pellets from sporulating cultures were suspended with solution A without SDS (20 mM sodium acetate, pH 5.0, 1 mM EDTA, pH 8.0), and then sodium acetate-saturated phenol (pH 5.0) was added. Zirconia beads were added to the cell suspension, and the tubes were vigorously shaken with a Beads crusher (Taitec, Japan) for 5 min. After removal of the beads, the cells were centrifuged, and a routine RNA extraction method was applied. Northern hybridization and signal detection were done with an AlkPhos-Direct kit (GE Healthcare).
Western blotting.
To examine the production of CPE, the C. perfringens strains were cultured by the same method as that used for the sporulation analysis (see above). Cells were collected from 8-h cultures and washed with PBS twice. The cells were then resuspended with PBS and disrupted by sonication until more than 95% of the cells were lysed. The lysed cells were centrifuged, and the supernatant was recovered. The protein concentration of the supernatant was measured by the Bradford protein assay method, and 1 μg of protein was used for Western blotting using rabbit anti-CPE antiserum. ECL Plus Western blotting detection reagent (GE Healthcare) was used for detection of immunoreactive bands.
Analysis of morphology.
Samples (0.5 ml) of 6-h cultures of C. perfringens strains in DS medium were centrifuged, and the cells were collected. The cells were washed with PBS twice and then resuspended with 15 μl of PBS. A final concentration of 0.1 μg/μl of FM4-64 was added to the cells and mixed. The cells were incubated at room temperature for 10 min and then subjected to phase-contrast and fluorescence microscopic analyses using a Biozero 8000 microscope (Keyence, Japan).
Microarray assays.
Microarray experiments using wild-type strain 13 and the virX mutant TS186 (14) were performed according to previously described methods (20).
Microarray data accession number.
The microarray data have been deposited in NCBI's Gene Expression Omnibus under GEO Series accession number GSE37998.
RESULTS
virX negatively regulates sporulation-specific sigma factors in C. perfringens under vegetative and sporulation conditions.
Our previous study indicated that the regulatory RNA virX positively regulates transcription of alpha-toxin (plc), kappa-toxin (colA), and theta-toxin (pfoA) (14). In an attempt to identify further genes that are regulated by virX in the C. perfringens strain 13 genome (21), we used a custom DNA microarray that contained PCR-generated strain 13 DNA (20) and compared the transcriptional levels of each chromosomal gene between wild-type strain 13 and the virXstrain13 mutant strain TS186 (14) in 3-h nutrient-rich GAM cultures. The virXstrain13 gene appeared to control the transcription of 53 and 110 genes, positively (see Table S1 in the supplemental material) and negatively (Table S2), respectively, at 3 h of growth of strain 13. Strikingly, we found that the genes encoding all four sporulation-related RNA polymerase σ factors (sigF, sigE/sigG, and sigK) were upregulated in TS186 (Table 1). To confirm these results, we performed Northern analyses on total RNAs isolated from 4-h GAM cultures. Indeed, the results showed that transcription of all four sporulation-specific σ factors was upregulated in TS186 (Fig. 1A). Note that sigE and sigG are part of the spoIIGA-sigE-sigG operon (12), and therefore the sigE and sigG transcript can be detected with a sigG probe. The upregulation of σ factors in TS186 decreased to wild-type levels upon complementation with the wild-type virXstrain13 gene (Fig. 1A). Collectively, these results suggest that virXstrain13 negatively regulates all four sporulation-specific σ factors during vegetative growth of C. perfringens strain 13.
Table 1.
Sporulation-related genes negatively regulated by virX
| CPE no. | Gene | Product | Fold change (log2) | t test P value |
|---|---|---|---|---|
| CPE1753 | spoIVA | Stage IV sporulation protein A | 3.02 | 8.74E−04 |
| CPE1761 | sigG | RNA polymerase sigma G factor | 2.00 | 1.06E−04 |
| CPE1762 | sigE | RNA polymerase sigma E factor | 3.28 | 2.64E−04 |
| CPE1763 | spoIIGA | Sporulation protein SpoIIGA | 2.98 | 1.16E−03 |
| CPE1812 | spo0A | Transcription factor Spo0A | 2.23 | 1.12E−03 |
| CPE2048 | sigF | RNA polymerase sigma F factor | 4.53 | 6.72E−04 |
| CPE2049 | spoIIAB | Anti-sigma F factor antagonist | 4.61 | 1.29E−04 |
| CPE2050 | spoIIAA | Anti-sigma F factor antagonist | 4.41 | 5.62E−05 |
| CPE2473 | spoIIE | Stage II sporulation protein E | 5.40 | 2.06E−04 |
Fig 1.
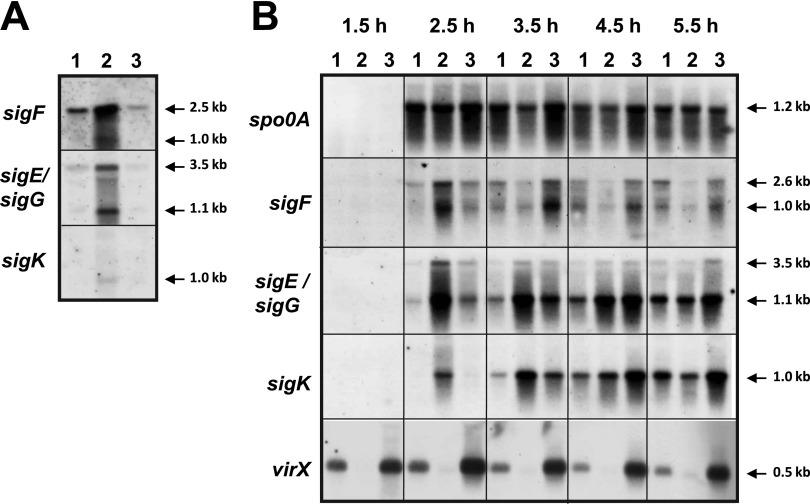
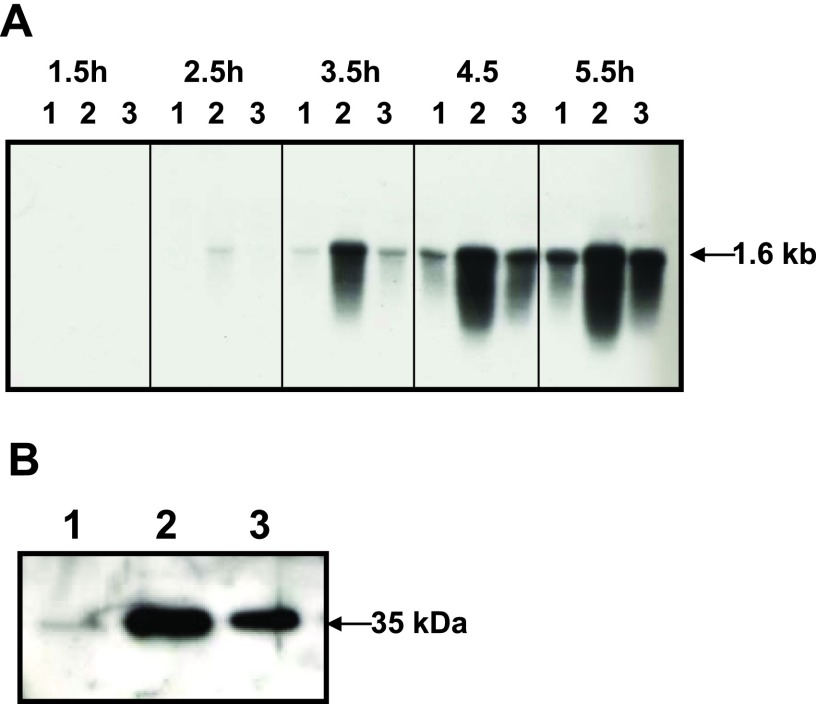
Effects of a virX mutation on transcription of sporulation-specific sigma factors of C. perfringens strains. (A) The levels of sigF, sigE/sigG, and sigK transcripts in total RNAs isolated from 4-h nonsporulating GAM cultures were analyzed by Northern blotting. Lanes: 1, strain 13(pJIR418) (wild type); 2, TS186(pJIR418) (virXstrain13 mutant); 3, TS186(pTS907) (TS186 complemented with wild-type virXstrain13). (B) Northern blot results are shown for the transcription of spo0A, sigF, sigE/sigG, sigK, and virX during early logarithmic growth (1.5 and 2.5 h) and logarithmic growth (3.5, 4.5, and 5.5 h) of DS sporulation cultures. Lanes: 1, SM101 (wild type); 2, KO101 (virXSM101 mutant); 3, KO102 (KO101 complemented with wild-type virXSM101).
Since a previous report (21) indicated that C. perfringens strain 13 has a frameshift mutation in the gene of the master regulator, spo0A (22, 23), we examined the ability of spore formation of strain 13 under the same conditions as those used for SM101. As expected, strain 13 did not form spores (data not shown). Therefore, we newly constructed a virX-knockout mutant (KO101) and a complemented strain (KO102) from the sporulation-proficient food poisoning strain SM101 to evaluate whether virX plays a role in the sporulation of C. perfringens. These strains were cultured in DS medium, and they all showed similar growth curves, which could be divided into early-log (∼2.5 h), mid- to late-log (∼6 h), and stationary (∼7 h) phases.
Northern analysis of the master regulator of sporulation, spo0A, showed that virX seemed to have no regulatory effect on spo0A transcription during the sporulation process of SM101 (Fig. 1B). Much as in B. subtilis, σF was the earliest and most sporulation-specific σ factor to be expressed (12, 24). In wild-type SM101, sigF was expressed after 2.5 h of initiation of sporulation as a 2.6-kb sigF transcript and at very low levels (Fig. 1B). Interestingly, an ∼1.0-kb transcript that also reacted with the sigF probe appeared after 2.5 h of sporulation, which indicates that sigF is transcribed from two promoters, giving 2.6-kb spoIIAA-spoIIAB-sigF and ∼1.0-kb sigF transcripts (Fig. 1B). For the virXSM101 mutant KO101, strong expression of both the 2.6- and ∼1.0-kb transcripts was observed in a 2.5-h DS culture (Fig. 1B) but decreased to levels lower than those in SM101 after 3.5 h (Fig. 1B). The negative regulatory effect of virX was more pronounced on the small sigF transcript (∼1.0 kb), with significantly more overexpression of sigF during early sporulation and lower sigF levels than at later stages compared with the large (2.6 kb) spoIIAA-spoIIAB-sigF transcript (Fig. 1B).
In C. perfringens, the low levels of transcription of sigK required for sigE activation precede sigE transcription, which in turn activates sigK at a later stage of sporulation (13). In this study, no early (i.e., 1.5 and 2.5 h of DS culture) transcription of sigK was detected by Northern analysis of wild-type SM101, and this transcription was detectable only after 3.5 h (Fig. 1B). In contrast, the earliest mother cell-specific σ factor is σE, which was expressed as a 3.5-kb spoIIGA-sigE-sigG transcript after 2.5 h of sporulation, making the ∼1.1-kb transcript encoding only sigG the most predominant at later stages of sporulation (i.e., 4.5 and 5.5 h) (Fig. 1B). A detectable level of a 1.0-kb sigK transcript was present in wild-type SM101 after only 3.5 h of sporulation (Fig. 1B).
More strikingly, in the virX mutant KO101, 3.5-, 1.1-, and 1.0-kb transcripts, specific for spoIIGA-sigE-sigG, sigG, and sigK, respectively, were upregulated during early (2.5 h) sporulation. We also assayed the transcription levels of virX during sporulation, and the results showed that virX transcription decreased only slightly during the progression of sporulation in SM101 (Fig. 1B), indicating that virX transcription remained active throughout the sporulation process. Importantly, the effect of the absence of virX in KO101 on the transcription of all four sporulation-specific σ factors was restored to wild-type levels in the complemented strain KO102 (Fig. 1B). Collectively, these results indicate that (i) virX is well transcribed during the C. perfringens sporulation process and (ii) virX negatively regulates the expression of sigF, sigE, sigK, and sigG in wild-type C. perfringens.
virX mutation increases the sporulation efficiency of C. perfringens.
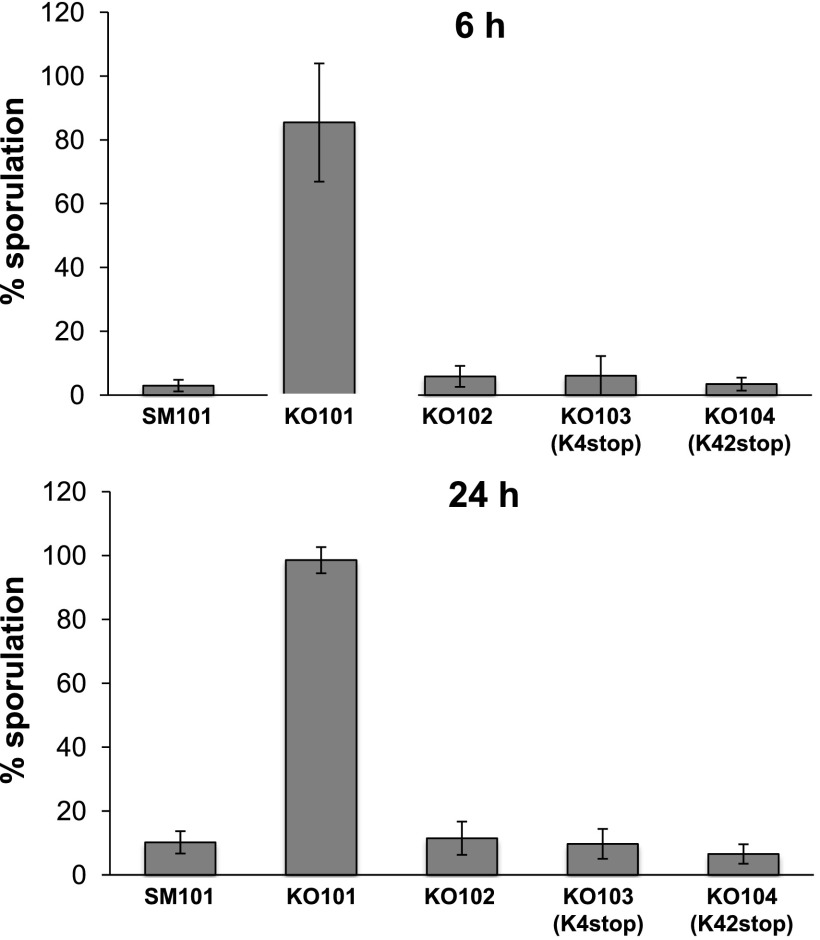
To quantify the effect of a virX mutation on the sporulation efficiency of C. perfringens, 6- and 24-h DS cultures were assayed for heat-resistant CFU. Interestingly, 6-h cultures of the virXSM101 mutant KO101 had ∼28-fold more heat-resistant CFU than the SM101 cultures (Fig. 2). After 24 h, the KO101 cultures had nearly 10-fold more heat-resistant CFU than the SM101 cultures, and they reached a sporulation efficiency of ∼95% (Fig. 2), indicating that sporulating cultures of KO101 not only sporulate with a high efficiency but also begin to sporulate earlier than SM101 spores. Moreover, complementation with intact virX (KO102) resulted in a drastic decrease of heat-resistant spores, almost to the same levels as those of SM101, at both 6 and 24 h (Fig. 2), which clearly indicates that virX is a strong repressor of C. perfringens sporulation.
Fig 2.
Sporulation efficiencies of various C. perfringens strains. To measure sporulation efficiency, 6- and 24-h sporulating cultures of SM101 (wild type), KO101 (virX mutant), KO102 (complemented with wild-type virX), KO103 (complemented with virX-K4stop), and KO104 (complemented with virX-K42stop) were directly plated onto BHI agar to determine the total numbers of CFU. The cultures were then heat treated (70°C, 20 min) and plated onto BHI agar to determine the numbers of CFU from heat-resistant spores. Both were incubated overnight under anaerobic conditions at 37°C. Sporulation efficiency (%) was calculated as follows: (heat-resistant CFU ml−1/total CFU ml−1) × 100. The data are presented as means ± standard deviations (SD) calculated from at least 3 independent experiments.
Since virX has been reported to act as an RNA regulator on toxin genes (14), we also made complemented strains of KO101 with virX genes that had a nonsense (stop codon) mutation at the codon for K4 (K4stop) or K42 (K42stop) in the protein coding region of virX. The sporulation efficiencies of the resultant complemented strains (KO103 and KO104) had no significant difference from that of KO102 (Fig. 2). These data indicate that virX also act as a RNA regulator but not as a regulatory protein/peptide and that the RNA molecule negatively controls sporulation in C. perfringens, in just the same manner as that of toxin regulation in strain 13 (14).
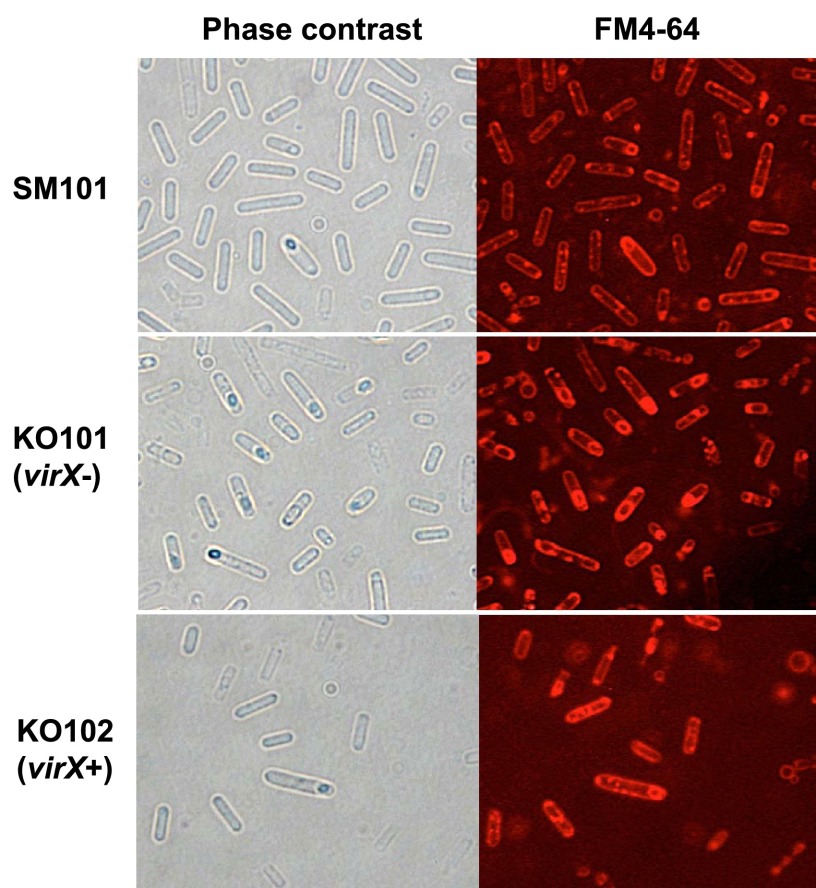
We further evaluated whether the absence of virX would affect the morphology of sporulating cells grown in DS medium by using phase-contrast microscopy. C. perfringens wild-type SM101 grown for 6 h in DS medium showed some cells harboring refractive polar prespores (Fig. 3). By fluorescence microscopy analysis using the fluorescent dye FM4-64 (specific for membrane lipids), it was possible to delimit the membrane of the polar prespore in SM101 (Fig. 3). Similarly, for sporulating KO101 cells, the prespore and mother cell compartments were clearly separated by a membrane visible by FM4-64 staining (Fig. 3), indicating that the virXSM101 mutant KO101 has an asymmetrical sporangium phenotype similar to that of wild-type cells under sporulation conditions. Complementation of KO101 with wild-type virX (KO102) had no effect on the FM4-64-staining cell morphology compared to that of SM101 and KO101 (Fig. 3). These results indicate that virX is not involved in cell morphology during sporulation of C. perfringens cells.
Fig 3.
Cell morphologies of C. perfringens SM101 and its derivatives grown under sporulation conditions. Morphologies of 6-h DS sporulation cultures of C. perfringens SM101 (wild type), KO101 (virX mutant), and KO102 (KO101 complemented with wild-type virX) were analyzed by phase-contrast microscopy and fluorescence microscopy with FM4-64.
virX represses cpe transcription during sporulation of C. perfringens.
During the sporulation of C. perfringens, the expression of the enterotoxin gene (cpe) is under the direct control of sporulation-specific σ factors (σE and σK) that are expressed in the mother cell compartment of the sporulating cell (12, 13, 17, 25). Since virX represses σE and σK during early sporulation, we tested the hypothesis that virX would also repress the expression and production of CPE. Northern analyses of SM101 DS cultures showed that the transcription of cpe began after 3.5 h of sporulation in SM101 (Fig. 4A), while transcription of cpe in the virX mutant KO101 began ∼1 h earlier than that in SM101 (Fig. 4A). At later stages of sporulation, transcriptional levels of cpe remained higher in KO101 than in SM101 (Fig. 4A). Western blot analyses showed that CPE levels in 8-h sporulating cultures of KO101 were approximately 6 times higher than those in SM101 (Fig. 4B). The complemented strain KO102 restored the levels of cpe transcript and CPE toxin to nearly wild-type levels (Fig. 4A and B). These results indicate that virX negatively regulates the transcription and production of CPE during sporulation of C. perfringens.
Fig 4.
Effects of virX mutation on transcription and production of C. perfringens enterotoxin. (A) Total RNA was extracted at various times from DS sporulation cultures and analyzed by Northern blotting. Lanes: 1, SM101 (wild type); 2, KO101 (virX mutant); 3, KO102 (KO101 complemented with wild-type virX). (B) Eight-hour DS sporulation cultures of C. perfringens strains were analyzed by Western blotting using rabbit CPE antiserum. Lanes: 1, SM101 (wild type); 2, KO101 (virX mutant); 3, KO102 (KO101 complemented with wild-type virX).
DISCUSSION
C. perfringens possesses great toxin versatility and is capable of producing up to 18 different toxins (3, 26, 27). The majority of these toxins are under the control of regulatory mechanisms exclusive to the vegetative growth phase (16, 20, 28). Recent evidence has demonstrated that alpha- and theta-toxin production is also dependent on an Agr-like quorum-sensing system that plays roles in toxin regulation under vegetative growth (29, 30) and is also required for efficient sporulation and production of CPE (11). Our results demonstrate not only that the regulatory RNA virX is required for positive regulation of virulence factors (alpha-, kappa-, and theta-toxin) during vegetative growth (14) but, in contrast to the Agr-like quorum-sensing system, that virX RNA acts as a repressor of sporulation in C. perfringens. Here it was shown that the transcription levels of spo0A were not affected in the virX mutant KO101, indicating that the negative effect of virX on sporulation does not occur at the level of spo0A. Instead, it is likely that virX negatively regulates sporulation by repressing transcription of the sporulation-associated and forespore-specific σ factor σF. Recent studies have shown that σF regulates expression of the remaining three sporulation-associated σ factors (σE, σK, and σG) (12, 13). Indeed, in a C. perfringens sigF-knockout mutant, there was no production of σE, σK, or σG (12), supporting our hypothesis that virX negatively regulates the transcription of all four sporulation-specific σ factors, likely through σF.
The expression of cpe during C. perfringens sporulation is under the direct control of the mother cell-specific σ factors σE and σK, since the promoter region of cpe has one σK-dependent promoter and two σE-dependent promoters (17). Given the cross-regulation between σK and σE and the fact that both are directly controlled by σF, our results also suggest that virX negatively regulates the transcription of cpe and production of CPE through the forespore-specific σ factor σF. It was notable that unlike in the previous study indicating that expression of sigK precedes expression of sigE (13), we were unable to detect sigK transcription by Northern blotting at 2.5 h of sporulation. A likely explanation is that the amount of early sigK transcript being produced under our sporulating conditions was much lower than the detection limit of Northern blot analyses. Indeed, we checked the expression of sigK by quantitative reverse transcription-PCR (qRT-PCR), and small amounts of the sigK transcript (5 to 15% of the sigF mRNA level) were detected in sporulating cultures at 2.5 h (data not shown). Overall, the reduction of transcription levels of sigF, sigE, and sigG during late sporulation in the virXSM101 mutant KO101 may have been due to an overall decrease of the sigF transcript.
Sporulating cells have unique morphological properties, primarily asymmetrical septum formation, with the formation of a large progeny called the mother cell and a small progeny termed the forespore. In this study, construction of a C. perfringens SM101 isogenic virX-knockout strain did not have any effect on the sporulating cell morphotype. The lack of a role in the sporulating cell morphotype supports the hypothesis that virX acts uniquely on the earliest forespore sporulation-associated σ factor, σF. virX mutation also led to a striking increase in heat-resistant spores at a very early stage of sporulation, indicating that a quick upregulation of the sporulation-specific σ factor cascade leads to rapid spore formation, a phenomenon that might have certain implications in pathogenesis, since food poisoning strains with low levels of virX might be more prone to sporulate under food poisoning conditions. Further comparative studies on the expression of virX in C. perfringens food poisoning versus non-food-borne isolates should clarify this point.
Our previous report indicated that virX is a positive regulator of several toxin genes in C. perfringens strain 13 (the gas gangrene strain) (14). In contrast, this study suggested that virX is also a negative regulator of spore formation in SM101 (a food poisoning strain). These results suggest that virX strongly represses spore formation under nutrient-rich conditions (i.e., in the human body), which leads to the VirR–VirS–VR-RNA-mediated expression of various toxins and enzymes (20) that will degrade environmental nutritional sources in order to obtain multiple nutrition for the organism's growth and survival. In contrast, under sporulation-favoring conditions, the repression of the sporulation-related sigma factors by virX would be loosened up through unknown environmental signals that allow the cells to start sporulation for survival in the environment. The virX RNA regulator would play a key role in the drastic shift in lifestyle of the anaerobic flesh eater C. perfringens in response to various environmental conditions. Most importantly, the elucidation of the signals and/or conditions that affect the regulatory function of virX is a prerequisite to fully understanding the mechanism of spore formation in C. perfringens, which could lead to many applications for prevention of C. perfringens infection and food poisoning.
Lastly, as shown in Table S3 in the supplemental material, virX homologues (based on amino acid sequences) could not be found in any Bacillus species, Clostridium tetani, or Clostridium difficile, while there are multiple copies of virX homologues in limited species of Firmicutes, including Clostridium botulinum, Clostridium acetobutyricum, and C. perfringens (although C. perfringens has four virX homologues, only the virX studied here has regulatory RNA activity for toxin production and sporulation [data not shown]). This highly biased existence of virX homologues among these genera might correspond to the complexity of the world of various spore formers and could determine their lifestyles on earth.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (Applied Genomics) (to K.O. and T.S.) and a Grant-in-Aid for Scientific Research (B) (to T.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a Multi-University Research Initiative award through the U.S. Army Research Laboratory and the Army Research Office, under contract number W911NF-09-1-0286 (to M.R.S.).
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02152-12.
REFERENCES
- 1. Petit L, Gibert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110 [DOI] [PubMed] [Google Scholar]
- 2. Songer JG. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McClane BA. 2007. Clostridium perfringens, p 423–444 In Doyle MP, Beuchat LR. (ed), Food microbiology: fundamentals and frontiers, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 4. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stragier P. 2002. A gene odyssey: exploring the genomes of endospore-forming bacteria, p 519–525 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 6. Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13–39 [DOI] [PubMed] [Google Scholar]
- 7. Sonenshein AL. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561–566 [DOI] [PubMed] [Google Scholar]
- 8. Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 9. Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579–586 [DOI] [PubMed] [Google Scholar]
- 10. Paredes CJ, Alsaker KV, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 11. Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect. Immun. 79:2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, McClane BA. 2010. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect. Immun. 78:4286–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harry KH, Zhou R, Kroos L, Melville SB. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ohtani K, Bhowmik SK, Hayashi H, Shimizu T. 2002. Identification of a novel locus that regulates expression of toxin genes in Clostridium perfringens. FEMS Microbiol. Lett. 209:113–118 [DOI] [PubMed] [Google Scholar]
- 15. Mahony DE, Moore TI. 1976. Stable L-forms of Clostridium perfringens and their growth on glass surfaces. Can. J. Microbiol. 22:953–959 [DOI] [PubMed] [Google Scholar]
- 16. Shimizu T, Ba-Thein W, Tamaki M, Hayashi H. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao Y, Melville SB. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sloan J, Warner TA, Scott PT, Bannam TL, Berryman DI, Rood JI. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27:207–219 [DOI] [PubMed] [Google Scholar]
- 19. Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohtani K, Hirakawa H, Tashiro K, Yoshizawa S, Kuhara S, Shimizu T. 2010. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16:258–264 [DOI] [PubMed] [Google Scholar]
- 21. Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paredes-Sabja D, Sarker MR. 2011. Host serum factor triggers germination of Clostridium perfringens spores lacking the cortex hydrolysis machinery. J. Med. Microbiol. 60:1734–1741 [DOI] [PubMed] [Google Scholar]
- 23. Huang IH, Waters M, Grau RR, Sarker MR. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233–240 [DOI] [PubMed] [Google Scholar]
- 24. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 25. Melville SB, Labbe R, Sonenshein AL. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 62:5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. 10.1371/journal.ppat.0040026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amimoto K, Noro T, Oishi E, Shimizu M. 2007. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 153:1198–1206 [DOI] [PubMed] [Google Scholar]
- 28. Cheung JK, Keyburn AL, Carter GP, Lanckriet AL, Van Immerseel F, Moore RJ, Rood JI. 2010. The VirSR two-component signal transduction system regulates NetB toxin production in Clostridium perfringens. Infect. Immun. 78:3064–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vidal JE, Chen J, Li J, McClane BA. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. 10.1371/journal.pone.0006232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohtani K, Yuan Y, Hassan S, Wang R, Wang Y, Shimizu T. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191:3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.