Abstract
Type IV pili are important for microcolony formation, biofilm formation, twitching motility, and attachment. We and others have shown that type IV pili are important for protein secretion across the outer membrane, similar to type II secretion systems. This study explored the relationship between protein secretion and pilus formation in Vibrio cholerae. The toxin-coregulated pilus (TCP), a type IV pilus required for V. cholerae pathogenesis, is necessary for the secretion of the colonization factor TcpF (T. J. Kirn, N. Bose, and R. K. Taylor, Mol. Microbiol. 49:81–92, 2003). This phenomenon is not unique to V. cholerae; secreted virulence factors that are dependent on the presence of components of the type IV pilus biogenesis apparatus for secretion have been reported with Dichelobacter nodosus (R. M. Kennan, O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood, J. Bacteriol. 183:4451–4458, 2001) and Francisella tularensis (A. J. Hager et al., Mol. Microbiol. 62:227–237, 2006). Using site-directed mutagenesis, we demonstrated that the secretion of TcpF is dependent on the presence of selected amino acid R groups at position five. We were unable to find other secretion determinants, suggesting that Y5 is the major secretion determinant within TcpF. We also report that proteins secreted in a type IV pilus biogenesis apparatus-dependent manner have a YXS motif within the first 15 amino acids following the Sec cleavage site. The YXS motif is not present in proteins secreted by type II secretion systems, indicating that this is unique to type IV pilus-mediated secretion. Moreover, we show that TcpF interacts with the pilin TcpA, suggesting that these proteins are secreted by the type IV pilus biogenesis system. These data provide a starting point for understanding how type IV pili can mediate secretion of virulence factors important for bacterial pathogenesis.
INTRODUCTION
In Gram-negative bacteria, the outer membrane forms a barrier for the passive transport of proteins. Pathogens secrete virulence factors inside their hosts, and specific mechanisms have evolved for proteins to traverse the outer membrane. It has been proposed that there is potential for the development of antimicrobial agents that target these secretion systems (1). There are six well-defined secretion systems in Gram-negative bacteria that vary in complexity and mechanism. Although cryo-electron microscopy and other methods have elucidated how assembly occurs in other apparatuses (e.g., type III secretion) (2–4), it remains to be shown how the apparatus is responsible for secreting proteins for type II secretion systems (T2SS). Genetic and structural studies have demonstrated that type II secretion is dependent on the assembly of a complex apparatus that spans the inner and outer membranes of Gram-negative bacteria (for a review, see reference 5), yet the exact mechanism of how secretion occurs is not understood. Moreover, it has been suggested that transient properties of type II secretion make it difficult for cryo-electron microscopy to elucidate how the protein complex responsible for secretion forms, as the entire complex has not been successfully stabilized or localized using this method (2). Recent work strongly favors a piston model of protein secretion first proposed by Hobbs and Mattick (6), with the pseudopilus being a piston that pushes a processed substrate through the outer membrane secretin (5, 7, 8). This model is similar to the proposed mechanism of type IV pilus (T4P) assembly, where the pilus assembles at the inner membrane and traverses the outer membrane through a secretin (9–11).
T4P are cellular appendages that have been shown to be important in biofilm formation, twitching motility, DNA uptake, attachment to surfaces, and bacterium-bacterium interactions. T4P can be subdivided into type IVa pili, type IVb pili, and flp pili. Type IVa pili are important for surface adhesion and are present in a wide variety of Gram-negative bacteria. Type IVb pili, which are found in enteric pathogens such as Vibrio cholerae and Escherichia coli, are important for bacterium-bacterium interactions and pathogenesis. The recently discovered flp pilus may be a subset of type IVb pili. The flp pilus is present on Yersinia enterocolitica (12), Pseudomonas aeruginosa (13), and Actinobacillus actinomycetemcomitans (also known as Aggregatibacter actinomycetemcomitans) (14) and mediates microcolony formation (12) and surface adhesion (13, 14).
In both T2SS and T4P there are major and, in some systems, minor pilins that form the structure of the pilus (15–17). Overexpression of the major T2SS pseudopilins PulG in Klebsiella oxytoca and XcpT in P. aeruginosa have led to the assembly of a pilus-like structure on the surface of the bacterium (18, 19). Additionally, the organization of the genes encoding the apparatus as well as proteins that make up the apparatus are similar in T4P and in T2SS (for a review, see references 9 and 20). The two systems also have a secretin (in Gram-negative bacteria), an inner membrane platform, and ATPases that generate the energy for extension and retraction of the pilus (9, 19–24). Crystallographic evidence has recently demonstrated similarities in the proteins that make up these components, even in those with little sequence homology (25, 26). Moreover, it has been shown in P. aeruginosa that a single biogenesis apparatus can support both T4P generation and T2SS pseudopilus generation (18). For these reasons, it is thought that the two systems share an ancestor.
Thus, it is not surprising that T4P biogenesis apparatuses have been shown to also mediate protein secretion in Myxococcus xanthus (13), Francisella tularensis (27, 28), Dichelobacter nodosus (29, 30), and V. cholerae (31, 32). In these systems the pilin is required for protein secretion, similar to how pseudopilins are required for protein secretion by T2SS (9, 19, 22), despite the fact that structures of the pilus show no pore for a protein to exit through in either type IVa (11, 33) or type IVb (10) pili. Proteins secreted by T2SS require processing by the Sec or Tat apparatus to traverse the inner membrane (34). There are Sec signal sequences in proteins reported to be secreted by a T4P biogenesis apparatus-mediated mechanism. We have shown that in V. cholerae, processing by the Sec apparatus is necessary for protein secretion by the T4P biogenesis apparatus (31).
V. cholerae is a Gram-negative curved rod that causes the disease cholera. We and others have characterized the major virulence factors required for pathogenesis, which include cholera toxin, the toxin-coregulated pilus (TCP), and the secreted colonization factor TcpF (31, 32, 35, 36). Cholera toxin is secreted by a T2SS upon entry of V. cholerae into the small bowel. The toxin binds to GM1 gangliosides on lipid rafts and, after endocytosis (37), initiates a cascade of cell signaling events at the small intestine epithelial brush border that leads to the classical symptom of cholera: secretory diarrhea (38). TCP is a type IVb pilus that mediates autoagglutination in a test tube, phage transduction, secretion of TcpF, and colonization in vivo (31, 39, 40). TCP is required for the bacteria to cause disease in humans and in animal models (36). The various functions of the pilus are dependent on the presence of both the pilin, TcpA, and the pilus biogenesis apparatus, which is encoded within the vibrio pathogenicity island (40–42). The gene tcpF is located within the tcp operon. tcpF encodes a protein that is required for colonization in animal models (31). The kinetics of clearance through the intestine of a tcpF mutant resembles that for a pilus mutant, in which both are similarly 4 to 5 logs defective in colonization. This is noteworthy because a tcpF mutant is able to autoagglutinate and transduce phage, while a pilus mutant is unable to do so (31, 32). Point mutants with changes in tcpA that secrete TcpF but do not autoagglutinate demonstrate this 4- to 5-log defect in colonization, indicating that the functions of TcpF and the pilus are separate but interrelated (31; T. J. Kirn and R. K. Taylor, unpublished data). We have shown through structure-function studies and the generation of monoclonal antibodies that distinct areas in the C terminus of TcpF are required for function (43, 44). The study of the function of this protein has remained elusive: the structure of TcpF shows little similarity to known proteins (44), and no in vitro phenotype has been found for TcpF (32; S. J. Krebs and C. J. Megli, unpublished data).
TcpF is one of the most abundant proteins secreted into the culture supernatant, and it is secreted in a TCP biogenesis apparatus-mediated manner (31). TcpF is processed by the Sec apparatus before secretion across the outer membrane and requires both the presence of the pilin TcpA and the biogenesis apparatus for secretion across the outer membrane (31). The structure-function studies previously mentioned also demonstrate areas that are important for secretion (43). These data provide a focus for examining T4P biogenesis apparatus-mediated protein secretion in V. cholerae.
In this study, we further examined TcpF for secretion determinants, which has led us to discover that there is a determinant in the N terminus that is required for secretion. Moreover, we found a conserved motif in the N termini of other proteins reported to be secreted in a T4P-mediated manner. Additionally, we report an interaction between TcpF and the pilin TcpA, which provides additional information regarding the requirement of the presence of the TCP apparatus for secretion of this protein.
MATERIALS AND METHODS
Strains and constructs.
The strains used in this study are shown in Table 1. Briefly, bacteria were grown for 16 h under inducing (LB medium, pH 6.5, with rotation at 30°C) or noninducing (LB medium, pH 7.0, with rotation at 37°C) conditions. Mutations were introduced into the chromosome as previously described (46). Briefly, 500 bp upstream and downstream of the mutation was amplified by PCR, digested, and subsequently ligated into pKAS32. Small deletions were introduced into a plasmid carrying 500 bp upstream and downstream of the area of interest by using QuikChange mutagenesis (Stratagene). The resulting plasmids carrying the mutation were electroporated into Escherichia coli strain S17-1 λpir. The plasmids were confirmed by sequencing, and the resulting strain was mated with V. cholerae O395 pMIN1 (CJM035). Allelic exchange was carried out as previously described (46), and all strains were confirmed by sequencing.
Table 1.
Bacterial strains used in this study
| Strain | Characteristics | Source or reference |
|---|---|---|
| O395 | Classical Ogawa Smr | Lab collection |
| RT4031 | O395ΔtcpA | 45 |
| RT4369 | O395 ΔtcpC | 21 |
| RT4372 | O395 ΔtcpF | 31 |
| CJM035 | O395 pMin1 | 46 |
| CJM046 | O395 TcpF:ΔAA5-14 | This study |
| CJM066 | O395 TcpF:ΔAA5-9 | This study |
| CJM087 | O395 TcpF:ΔAA−1-−19 | This study |
| CJM100 | O395 TcpF:Y5A | This study |
| CJM101 | O395 TcpF:SS6-7AA | This study |
| CJM103 | O395 TcpF:S7A | This study |
| CJM104 | O395 TcpF:S6A | This study |
| CJM105 | O395 TcpF:YSST5-8AAAA | This study |
| CJM149 | O395 TcpF:E217A | This study |
| CJM150 | O395 TcpF:Y292A | This study |
| CJM151 | O395 TcpF:L298S | This study |
| CJM190 | O395 TcpF:E17Myc | This study |
| CJM191 | O395 TcpF:E25Myc | This study |
| CJM209 | O395 TcpF:Y5S | This study |
| CJM212 | O395 TcpF:T8A | This study |
| CJM217 | O395 TcpF:ΔAA28-31 (RYPY) | This study |
| CJM224 | O395 TcpF:ΔAA99-100 (YL) | This study |
| CJM234 | O395 TcpF:Y5F | This study |
| CJM235 | O395 TcpF:ΔAA59-60 (FY) | This study |
| CJM239 | O395 TcpF:Y5W | This study |
| CJM240 | O395 TcpF:Y5R | This study |
| CJM258 | O395 TcpF:ΔAA38-39 (GM) | This study |
| CJM265 | O395 TcpF:ΔAA160-168 | This study |
| CJM266 | O395 TcpF:ΔAA106-107 | This study |
| CJM267 | O395 TcpF:ΔAA109-111 | This study |
| KSK258 | O395 ΔlacZ | K. Skorupski |
Infant mouse cholera model.
Strains were grown overnight at 30°C and mixed with the ΔlacZ reference strain KSK258 at a ratio of 1:1. Tenfold serial dilutions were made. The 1:100 dilution was mixed with blue food coloring and used to inoculate 50 μl intragastrically into 5- to 6-day-old CD1 mice. Ten microliters of the serial dilutions was plated on LB agar plates containing streptomycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). At 24 h, the intestines were removed, suspended in 4 ml of LB containing 10% glycerol, and homogenized with a tissue homogenizer. A dilution series was again plated on LB agar plates with streptomycin and X-Gal. The competitive index was reported as the ratio of the output and input of the experimental strain in comparison to the reference strain as previously described (32).
Cellular fractionation.
Cellular fractionation was carried out as previously described (21, 47). Briefly, strains were grown overnight under inducing conditions and centrifuged. The resulting supernatants were passed through a 0.22-μm filter (Millipore). The cell pellet was suspended in 1 ml of phosphate-buffered saline (PBS), 8.1 × 104 U of polymyxin B was added, and the suspension was incubated on ice for 10 min and centrifuged in a tabletop centrifuge at 13,000 rpm. The supernatant was included as the periplasmic fraction. The resulting pellet was passed through a French press and centrifuged at 100,000 × g for 1 h to isolate the cytoplasm and membrane fractions (21).
Western blot analysis.
Whole cells and cellular fractions were suspended in sodium-dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer and quantitated using a bicinchoninic acid (BCA) protein quantification kit (Pierce). Culture supernatants were collected and run through a 0.22-μm filter and mixed with SDS-PAGE sample buffer containing bromophenol blue and beta-mercaptoethanol. Twenty microliters of supernatant or 16 μg of protein was loaded onto a polyacrylamide gel (Invitrogen) and subjected to electrophoresis. Proteins were then transferred to a nitrocellulose membrane using the iBlot system (Invitrogen). The membranes were blocked using 3% bovine serum albumin (BSA) in Tris-buffered saline, pH 7.5 (TBS), with 0.05% Tween 20 (blocking buffer). Primary antibodies that recognize TcpA and TcpF were added to the blocking buffer at a 1:10,000 dilution and were incubated for 1 h, with rocking, at room temperature. The membranes were washed 3 times for 5 min each in TBS with 0.05% Tween 20, and horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Bio-Rad) was added at a 1:10,000 dilution to blocking buffer and incubated for 1 h, with rocking, at room temperature. The membranes were washed 3 times for 5 min each in TBS with 0.05% Tween 20 and subsequently washed twice with TBS. Antibody binding was visualized using ECL Pico Western blotting detection reagents (Pierce), followed by autoradiography.
Cross-linking and immunoprecipitation.
Bacterial strains were grown overnight under inducing conditions. Five-milliliter overnight cultures were centrifuged, and bacterial cells were suspended in 1 ml of 150 mM NaCl–200 mM NaHPO4, pH 7.2 (cross-linking buffer). This was repeated 3 times to wash the cells. Cells were then suspended in 250 μl of cross-linking buffer, 25 μl of 10 mM dithiobis[succinimidyl propionate] (DSP) was added, and cells were incubated at 37°C for 1 h. The reaction was quenched by the addition of 40 mM Tris-HCl, pH 8.5, and cells were incubated for 30 min at 37°C. The mixture was centrifuged and the loose pellet was suspended in 500 μl of a 10% Triton X-100–50 mM Tris–20% glycerol solution with protease inhibitors (Roche) (nondenaturing sample buffer). Lysates were boiled for 5 min, and 5 μl (2 U/μl) of DNase was added. Lysates were incubated on ice for 1 h. For immunoprecipitation, the lysates were incubated with 2 mg/ml of polyclonal anti-TcpF and rotated overnight at 4°C. Fifty microliters of protein A Dynabeads (Invitrogen) was added, and lysates were incubated at 4°C for 4 h and vortexed every 30 min. The tubes were placed on a magnet and rinsed 3 times with PBS before elution. Proteins were eluted with 500 μl of SDS-PAGE sample buffer, and after protein quantification, bromophenol blue and beta-mercaptoethanol were added.
Protein sequence analysis.
Proteins reported to be secreted in a T4P biogenesis apparatus-dependent manner were searched on NCBI. The protein sequences were entered into the Center for Biological Sequence Analysis Signal P 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/), and the Sec cleavage site was noted. The first 30 amino acids of the mature protein were then visually scanned for a YXS motif.
RESULTS
Characterization of N-terminal secretion determinants of TcpF.
We have previously demonstrated that the mature N terminus of TcpF is important for secretion into the supernatant (43). Specifically, the deletion of amino acids (aa) 6 to 29 after the Sec cleavage site rendered the protein incapable of being secreted across the outer membrane (43). To understand which amino acids are important for secretion, smaller deletions of this hydroxyl-rich region were made.
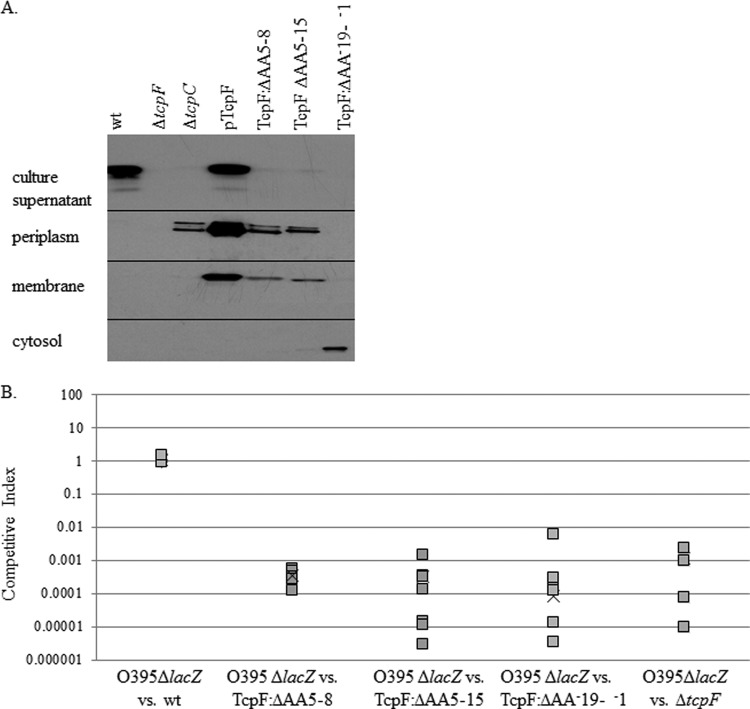
In Fig. 1A, we show that, as previously demonstrated, wild-type O395 TcpF is secreted, while a strain lacking tcpC, the gene encoding the secretin, is unable to secrete TcpF across the outer membrane; thus, protein accumulates in the periplasmic space (31, 43). As previously shown, overexpression of TcpF leads to saturation of secretion, which leads to its accumulation in various subcellular compartments (43, 44). TcpF derivatives missing amino acids 5 to 8 and 5 to 15 are unable to be secreted extracellularly and therefore accumulate in the periplasm, localizing to the same fractions as in a tcpC deletion strain. As an additional control, we constructed a strain where the TcpF derivative is unable to be secreted into the periplasm by deleting the region encoding the Sec signal sequence (aa −19 to −1). Because this TcpF derivative is unable to be secreted across the inner membrane, it is present only in the cellular cytoplasm (Fig. 1A). These data demonstrate that the N terminus of mature TcpF, likely through amino acids 5 to 8, is important for secretion of the protein.
Fig 1.
TcpF secretion is required for function. (A) Immunoblot analysis of cellular fractionation products demonstrates that TcpF localization is dependent on the presence of the biogenesis apparatus and the N terminus of TcpF. The culture supernatant, periplasm, cytosol, and membrane fractions were separated by SDS-PAGE analysis and transferred to a nitrocellulose membrane as described in Materials and Methods. The membrane was probed with TcpF antisera, and corresponding bands at 36 kDa are shown. (B) In vivo competitive indices of wild-type, ΔAA5-8, ΔAA5-15, ΔAA−19-−1, and ΔtcpF strains. Strains were competed with the reference ΔlacZ strain. Squares represent data from each individual mouse, while “×” represents the average for the group of six mice. A value less than 1 indicates that there is a defect in colonization.
TcpF secretion is required for function as a colonization factor.
Next, we wanted to test if TcpF secretion is important for TcpF function. TcpF from the wild-type strain does not rescue the defect in colonization of a strain lacking TcpF when coinoculated in competitive-index experiments (31), which could indicate that TcpF functions in conjunction with the bacterium that secretes this protein (in cis). In order to determine whether secretion across the inner or outer membrane is required for TcpF function, we tested mutants that are unable to secrete TcpF into the periplasm or extracellularly in the infant mouse cholera model. Our results demonstrate that strains that do not secrete TcpF into the culture supernatant in vitro display a defect in colonization, identical to a tcpF deletion strain (Fig. 1B). These results suggest that TcpF must be secreted to function as a colonization factor. It is possible that the TcpF derivatives that are defective in secretion are defective for function in vivo because the mutation has rendered TcpF nonfunctional in the periplasmic space. However, we believe that this is unlikely because (i) the N-terminal deletion is very small, and (ii) the deletion is in a flexible region of the protein (44), such that a disruption in this region would be unlikely to affect the conformation of the entire protein. Therefore, these data indicate that the TcpF derivatives are deficient in colonization because they are deficient in secretion, demonstrating that TcpF must function outside the bacterial cell.
TcpF:Y5A is defective for secretion, and TcpF:S7A cannot mediate colonization.
Because the smallest deletion of TcpF that displays a defect in secretion and colonization is missing amino acids 5 to 8 (YSST), we used alanine substitution to identify which residues in this region are important for secretion. Figure 2A demonstrates that although TcpF:Y5A and TcpF:YSST5-8AAAA are unable to be secreted, all of the other TcpF derivatives (TcpF:S6A, TcpF:S7A, TcpF:SS6-7AA, and TcpF:T8A) are secreted into the culture supernatant at wild-type levels. These data indicate that Y5 is required for secretion.
Fig 2.
Point mutations demonstrate the requirement of Y5 for secretion and S7 for function of TcpF. (A) Immunoblot with TcpF antisera demonstrates that TcpF:Y5A and TcpF:YSST5-8AAAA are not secreted. Twenty microliters of supernatant and 16 μg of whole-cell lysates were subjected to SDS-PAGE. (B) Competitive indices of tcpF point mutants. Strains were coinoculated with a ΔlacZ reference strain into mice. Blue and white colonies were counted, and data are reported with respect to the reference strain. Squares represent data from individual mice, and “×” represents the average for each group of five mice.
In the infant mouse cholera model, strains expressing TcpF:Y5A, TcpF:S7A, TcpF:SS6-7AA, and TcpF:YSST5-8AAAA are 4 to 5 logs defective in colonization, similar to the ΔtcpF strain (Fig. 2B). Interestingly, TcpF:T8A has an intermediate phenotype in colonization, at 2 to 3 logs defective, making this the only intermediate phenotype seen thus far for TcpF derivative strains. Wild-type levels of secretion and colonization were seen with the TcpF:S6A strain, indicating that S6 is not important for secretion or for function. Consistent with the results in Fig. 1, the strains with TcpF derivatives that are not secreted (TcpF:Y5A and TcpF:YSST5-8AAAA) were also unable to compete effectively in the infant mouse, further supporting the concept that TcpF functions outside the bacterial cell. Interestingly, the strains with secreted TcpF derivatives TcpF:S7A and TcpF:SS6-7AA were also unable to compete, indicating that S7 is important for TcpF function as a colonization factor. Importantly, we did not see any difference in TcpF derivative secretion at earlier time points (data not shown), indicating that a delay in secretion was not be causing the defect in colonization.
Screening for additional TcpF secretion determinants.
We have previously shown that environmental strains of V. cholerae have TcpF homologues that are secreted but are nonfunctional in colonization (43). In order to examine other areas of TcpF important for secretion, we used site-directed mutagenesis to target all areas conserved between the environmental homologues and pathogenic TcpF that were on the surface of the crystal structure of TcpF (44). We chose not to target glycine residues at positions 175 and 154, because the R group of glycine does not lend itself to interaction with other amino acids. As seen in Fig. 3A and in Fig. S2 in the supplemental material, none of the 10 regions that were chosen displayed a complete defect in secretion, and the majority of these mutations resulted in proteins that were less stable. As previously seen, degradation products of 19 kDa (43) are seen in culture supernatants in the majority of these strains, indicating that these deletions render the protein unstable and subject to degradation but have no effect on protein secretion (Fig. 3A; see also Fig. S2). None of these TcpF derivatives accumulated in the whole-cell fraction (Fig. 3A) to the same degree as TcpF:Y5A. TcpF:ΔAA99-100 is present in larger amounts in whole-cell lysate than is the wild type, yet this TcpF derivative is still able to be secreted into the culture supernatant. This may represent an area of TcpF that is a minor determinant for secretion. Nonetheless, TcpF:ΔAA28-31, TcpF:ΔAA99-100, TcpF:E217A, TcpF:Y292A, TcpF:L298S are secreted at near wild-type levels and have protein products that migrate equivalently to full-length TcpF in the supernatant. However, strains containing TcpF:ΔAA28-31, TcpF:ΔAA99-100, TcpF:Y292A, and TcpF:L298S TcpF derivatives were unable to colonize in the infant mouse model (Fig. 3B), indicating that these regions may also contain determinants important in colonization or regions that are required for protein stability. There is abundant TcpF in the culture supernatant for all of these strains (Fig. 3A); the defect in colonization of TcpF:ΔAA28-31, TcpF:ΔAA99-100, TcpF:Y292A, and TcpF:L298S is particularly interesting because these residues are conserved with the nonfunctional environmental homologues, and thus, their role in TcpF function may be due to structural changes of the three-dimensional protein. Because none of these areas were found to be required for protein secretion, in combination with the results from Fig. 2, these results indicate that Y5 is the major secretion signal for TcpF.
Fig 3.
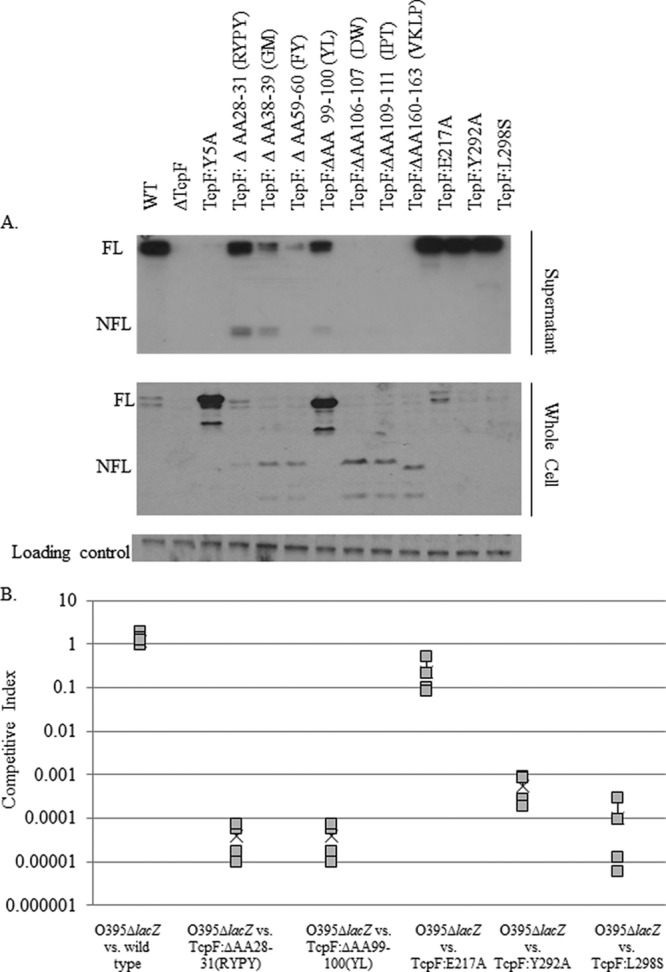
Mutations in additional regions of tcpF predicted to encode secretion determinants by linker scanning mutagenesis and alignment with environmental homologues result in TcpF derivatives secreted into culture supernatant. (A) Immunoblot of supernatant from strains with TcpF derivatives predicted to be defective in secretion. Twenty microliters of culture supernatant or 16 μg of whole cells was run on SDS-PAGE. Immunoblotting with anti-TcpF was performed. The loading control is a cross-reactive band. “FL” indicates full-length TcpF at 36 kDa. “NFL” indicates a 19-kDa product which is likely a major degradation product of TcpF (36). A 5×-longer exposure of the region of NFL TcpF is shown in Fig. S2 in the supplemental material. (B) Competitive indices of TcpF:ΔAA28-31, TcpF:ΔAA99-100, TcpF:E217A, TcpF:Y292A, and TcpF:L298S strains. Strains were coinoculated with a ΔlacZ reference strain into four mice. Blue and white colonies were counted, and data are reported with respect to the reference strain. TcpF:ΔAA28-31, TcpF:ΔAA99-100 TcpF: Y292A, and TcpF:L298S strains are defective in colonization similarly to a ΔtcpF strain at a 4- to 5-log defect.
Identification of a YXS motif in proteins secreted in a T4P biogenesis apparatus-mediated manner.
Several proteins which have a Sec recognition signal peptide are thought to be secreted across the cytoplasmic membrane in a Sec-dependent manner and then across the outer membrane in a T4P biogenesis apparatus-mediated manner: the proteases AprV2, AprV5, and BprV in D. nodosus (29, 30) and PepO in F. tularensis (27, 28). As seen in Table 2, the N terminus of the mature form of these proteins contains a YXS motif near the Sec cleavage site. In contrast, 25 proteins secreted by T2SS from various organisms were examined for this motif at the N terminus, and none were found to have this motif (data not shown). These data suggest that the tyrosine may be conserved as a signal for T4P biogenesis apparatus-dependent secretion.
Table 2.
YXS in the N termini of proteins secreted by the T4P biogenesis apparatus
| Organism | Protein | Sequence after Sec cleavage site |
|---|---|---|
| Vibrio cholerae | TcpF | FNDNYSSTSTVYATSNEATDSRGSEHLRYPY... |
| Dichelobacter nodosus | BprV | AESIVNYESANAISKQPEGSVRFIVKYKDGTPSS... |
| AprV2 | ETMVNYASAKAIGKQPAGSVRFIVKYKDNSQSSKDL... | |
| AprV5 | AVNYESANYIGSQPEGSVRFIIKYKDKSQSQQMM... | |
| Francisella tularensis | PepO | SPIIVESFVEEKKYSSNKIGIDTQYIDGSINEKDDF... |
The Y5 aromatic ring allows for TcpF secretion.
Because Y5 is important for the secretion of TcpF and is conserved in other proteins secreted in a T4P biogenesis apparatus-dependent manner, we were interested in understanding what properties of tyrosine are important for secretion. We replaced tyrosine with serine to preserve the hydroxyl group and with phenylalanine in order to preserve the aromatic ring. Tryptophan was also substituted for tyrosine to investigate whether another larger aromatic R group would preserve secretion. As a control, tyrosine was replaced with arginine as a means of completely changing the charge and structure of the R group. As seen in Fig. 4, replacement of Y5 with other amino acids containing an aromatic ring (as present in TcpF:Y5F and TcpF:Y5W) allows for protein secretion, whereas the hydroxyl-containing TcpF:Y5S and the positively charged TcpF:Y5R are not secreted. These results indicate that the aromatic ring at position 5 may facilitate secretion of TcpF.
Fig 4.
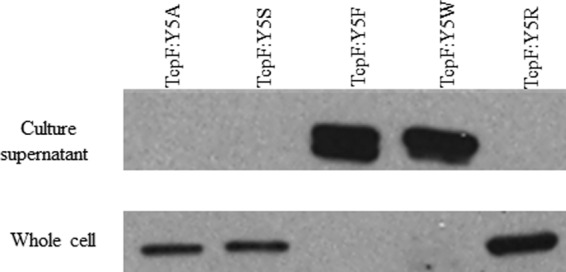
Effects of amino acid substitutions for tyrosine at position 5 on TcpF secretion. Culture supernatants and whole-cell lysates were subjected to SDS-PAGE as described in Materials and Methods. Immunoblotting was performed using TcpF antisera.
TcpF interacts with the TcpA pilin during secretion.
Cross-linking and immunoprecipitation were used to test the hypothesis that TcpF interacts with one or more of the components of the TCP biogenesis apparatus during the extracellular secretion process. We used the cross-linking agent DSP prior to immunoprecipitation in order to preserve transient interactions in the cell as TcpF is secreted. Cross-linked whole-cell lysates were subjected to electrophoresis and transferred to a nitrocellulose membrane. The membrane was probed with anti-TcpF, and chemiluminescent detection revealed a 56-kDa band recognized by the anti-TcpF antibody, indicating that TcpF was in complex with a 20-kDa protein (data not shown).
In order to identify the protein bound to TcpF, we then performed immunoprecipitation on the cross-linked lysates. We wanted to test the hypothesis that the Y5A substitution caused aberrant association with a protein in the TCP biogenesis apparatus which resulted in the defect in secretion. Immunoprecipitation was performed using the TcpF polyclonal antibody (32) as well as TcpF monoclonal antibodies (44) (data not shown), with the same results. As seen in Fig. 5, a 20-kDa band appears in wild-type TcpF and TcpF:Y5A which is absent in the immunoprecipitation from the strain lacking TcpF, indicating that TcpF interacts with this protein in both strains. Figure 5 also shows a second band from a protein bound to TcpF:Y5A around 26kDa that does not bind to wild-type TcpF. Mass spectrometry analysis followed by protein sequencing indicated that the 20-kDa protein was TcpA and the 26-kDa protein was the protein encoded by VCO395_A0815. Immunoprecipitation in the absence of a cross-linking agent only demonstrated heavy and light chains of antibody and TcpF (data not shown), suggesting that the observed interaction (described below) is transient. Loss of the hypothetical OmpA-like protein encoded by VCO395_A0815 displayed no phenotype with respect to TcpF secretion (see Fig. S1 in the supplemental material). It is likely that the protein immunoprecipitated with TcpF due to a nonspecific interaction caused by the accumulation of the TcpF:Y5A derivative that is unable to cross the outer membrane because of this alteration.
Fig 5.
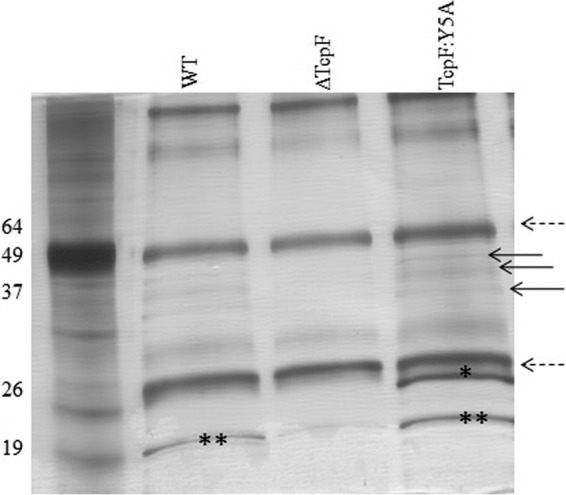
Immunoprecipitation of cross-linked whole-cell lysates with anti-TcpF antibody. After immunoprecipitation, antibody-bound proteins were loaded onto an SDS-PAGE gel and subjected to electrophoresis. Proteins in the gel were silver stained. The dashed arrows correspond to heavy- and light-chain immunoglobulin. The bands with the asterisks were removed and subjected to trypsin digestion and mass spectrometry analysis. “*” corresponds to VCO395_A0815; “**” corresponds to TcpA. The arrows point to TcpF and its degradation products. WT, wild type.
To determine if the 20-kDa TcpF coimmunoprecipitation product was TcpA, we performed immunoprecipitation on a strain lacking TcpA (Fig. 6). TcpF immunoprecipitation of the ΔtcpA strain failed to coimmunoprecipitate the 20-kDa product present in the protein profiles of immunoprecipitated wild-type O395 (Fig. 6). Moreover, Western blot analysis of the immunoprecipitation with anti-TcpA demonstrated that TcpA immunoprecipitates with both wild-type TcpF and TcpF:Y5A (Fig. 7), indicating that TcpA is the protein interacting with TcpF. Nevertheless, the data showing that TcpF interacts with TcpA suggest that TcpF is secreted by the pilus biogenesis apparatus. Although we were unable to detect other components of the apparatus by Western blotting (data not shown), it is possible that there are other transient interactions or weak interactions that occur as TcpF is secreted across the outer membrane that we were unable to detect with this cross-linking agent.
Fig 6.
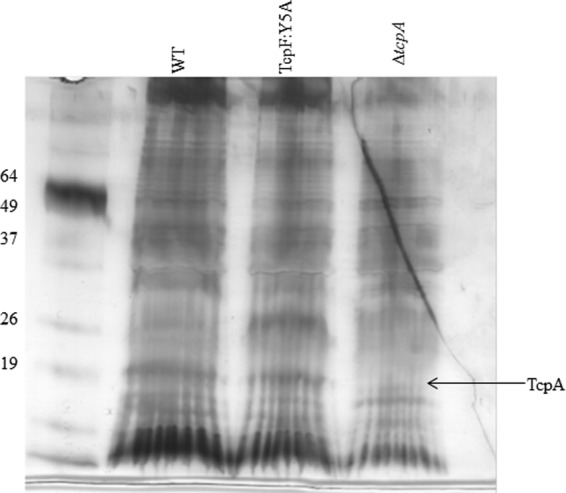
Anti-TcpF immunoprecipitation in a ΔtcpA strain does not coimmunoprecipitate a 20-kDa protein. Anti-TcpF coimmunoprecipitation products of cross-linked whole-cell lysates were loaded on an SDS-PAGE gel and subjected to electrophoresis. Silver staining demonstrates an absence of the 20-kDa band in the ΔtcpA strain.
Fig 7.
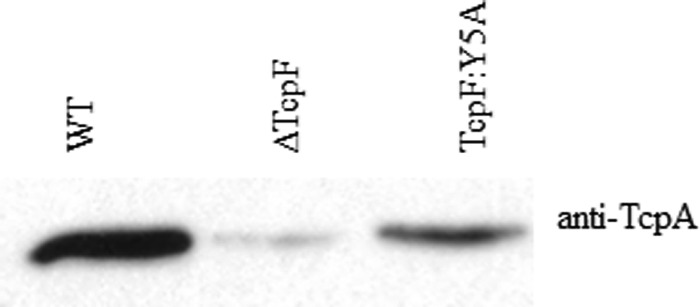
Immunoblotting confirms that TcpA is the 20-kDa protein that binds TcpF. Shown is an immunoblot analysis of anti-TcpF immunoprecipitation products of cross-linked whole-cell lysates.
DISCUSSION
In this study, we identified the tyrosine at position 5 as a TcpF secretion determinant. We had previously reported that various regions identified by linker-scanning mutagenesis were important for secretion (43), yet when we introduced point mutations in these regions, we were unable to recapitulate any phenotypes in secretion. We hypothesize that this is because linker-scanning mutants result in the insertion of amino acids, which may have perturbed the three-dimensional structure of TcpF and resulted in the secretion phenotype, but the regions where the insertions occurred are not important for secretion (as demonstrated by mutation). Additionally, we created alterations in all the surface-exposed regions that were conserved within the secreted environmental homologues (43) and saw no phenotype with respect to TcpF secretion. Combined, these results suggest that Y5 is a major secretion determinant for TcpF secretion.
The N terminus of TcpF is predicted to be in a flexible conformation. The first 25 amino acids of TcpF were not resolved in the electron density map (44). Moreover, TcpF constructs containing a Myc tag after residues E17 and E25 were also secreted (data not shown), indicating that the process of TcpF secretion tolerates variability in sequence length between the N terminus (with Y5) and the rest of the structured protein. Taken together, these data suggest that the proximal N terminus is flexible and thus able to interact with proteins within the T4P biogenesis apparatus or with other regions of TcpF in a variety of ways. We were unable to define a protein that Y5 interacts with by immunoprecipitation. The TcpF:Y5A mutant can bind to TcpA, indicating that this region of TcpF is important for recognition or interaction with a different protein that participates in secretion.
Our data suggest that the aromatic properties of tyrosine in position 5 may mediate protein secretion, likely due to an interaction with TcpA or another protein of the biogenesis apparatus. There are only a few potential types of protein-protein interactions that are mediated by an aromatic residue. The aromatic ring of tyrosine could also transiently interact with a lysine or arginine residue on a protein within the TCP biogenesis apparatus via a cation-π interaction (48), or interact with other aromatic residues through hydrophobic stacking. These types of interactions are classified as weak and thus could be missed by the DSP cross-linker used in this study. Moreover, if the TcpF three-dimensional structure is recognized by components of the biogenesis apparatus by similar weak interactions, then it is possible that we missed selected areas to identify secretion determinants, since we only examined surface-exposed residues that were conserved between pathogenic TcpF and secreted TcpF environmental homologues (43).
We have shown that TcpF secretion is important for its function as a colonization factor (Fig. 1 and 2). This result was a surprise to us because TcpF does not rescue mutants coinoculated in competitive index experiments. We have previously shown that monoclonal and polyclonal antibodies that recognize TcpF are protective against V. cholerae challenge (32, 44). Taken together, this evidence suggests that TcpF functions proximal to a target that is external to the bacterial cell. Although it could be postulated that TcpF is present in a cell-associated form by loosely associating either with the outer surface of the bacterium or with outer membrane vesicles, we have been unable to show that TcpF functions in colonization in this location. TcpF has been shown to be highly upregulated when V. cholerae is adjacent to intestinal epithelium and downregulated in the intestinal lumen (49). This has led us to hypothesize that TcpF may function at the epithelial cell directly, potentially as an adhesin.
We also report additional domains of TcpF important for function in colonization. Mutations in S7, Y99L100, R28Y29P30Y31, Y292, and L298 resulted in strains that secreted full-length TcpF but were defective for colonization in the infant mouse cholera model. R28Y29P30Y31 and Y99L100 are in the N-terminal domain in the crystal structure, whereas all previously reported functional domains were in the C-terminal domain (43, 44). These data indicate that both domains of TcpF are important for in vivo function. We hypothesize that each domain may interact with a different binding partner, both of which are important for function, or that the proposed binding pocket made up of both domains (44) is required for function.
In this study, we found that the Y5 secretion determinant, in combination with S7, is repeated in other proteins reported to be secreted by T4P, such that this YXS motif appears to be conserved. TcpF:S7A is secreted but not functional; therefore, the conservation of this residue in other proteins secreted in a similar manner is puzzling. It is possible that this serine is modified by a protein in the biogenesis apparatus during the process of TcpF secretion, and this modification is important for TcpF function as a colonization factor. Supporting this, Western blotting with anti-TcpF shows a double band for TcpF in culture supernatants. However, TcpF:S7A also appears as a doublet, indicating that if the double band is due to modification, there is no difference between wild-type TcpF and the nonfunctional S7A derivative. Additionally, we have been unable to identify serine phosphorylation of the N terminus of TcpF (data not shown). Although this residue may be important for proteins secreted by the type IV pilus biogenesis apparatus to facilitate bacterial pathogenesis, as is seen with TcpF, we are unable to explain the significance of this conserved residue in these proteins.
It has been suggested that T4P and T2SS have a common evolutionary ancestor (9, 50, 51). In T2SS, a universal protein sequence that signals a protein for secretion by this system has not been elucidated. Additionally, proteins secreted by one organism are often unable to be secreted by a T2SS of another organism, suggesting that there is organism-specific diversion and selection of T2SS. The conserved YXS motif suggests that the T4P systems evolved from a common ancestor and that divergence into the type IVa and type IVb subtypes occurred after. It is possible that T4P biogenesis systems are responsible for protein secretion in more organisms than the three that have been reported, and this YXS motif could be used in a bioinformatics search that combines this motif with a Sec recognition site to reveal additional, yet undiscovered, proteins that are secreted in a T4P biogenesis apparatus-dependent manner. Since these proteins that have thus far been identified are virulence factors, there is potential to design therapeutics that would bind to this motif in the periplasm of mature proteins and prevent secretion or that would compete with this motif for recognition by the biogenesis apparatus and block secretion.
Combining what has been previously published on T2SS with the results from this study, there are various possibilities of how T4P biogenesis apparatus-dependent secretion occurs. The machinery involved in T2SS and the machinery involved in T4P biogenesis have very similar architectures, and crystallography has demonstrated that proteins with little or no sequence identity between these systems are structural homologues (25). It is believed that the mechanics of T2SS protein secretion are similar to those of T4P pilus biogenesis (25, 52, 53). Recent evidence supports the piston model for T2SS protein secretion (7, 25, 52). In this model, each extension of the pseudopilus results in a single protein being secreted. Thus, it is possible that secretion occurs as the pilus is synthesized, identical to T2SS, except that the pilus continues to be polymerized. We believe that this is unlikely because TcpF is as abundant as cholera toxin in culture supernatants, and there are likely more molecules of TcpF in the supernatant than there are individual pili (31). This has led us to reason that in the V. cholerae TCP system, it is improbable that an entire pilus would be made for each molecule of TcpF to be secreted. Another possible hypothesis to explain the mechanism by which the type IV pilus biogenesis apparatus mediates protein secretion is that there is additional space in the pore of the secretin that allows for TcpF to be secreted across the outer membrane along with the extrusion of the pilus. However, crystallographic evidence suggests that TCP has a maximum diameter of 80 Å (11, 54), whereas secretins have an inner diameter of 65 to 75 Å, with a smaller constriction (7, 25, 55). It has been proposed that the pilus or pseudopilus interacts with the secretin and overcome this constriction by causing a conformational change to allow the pilus to pass through the pore. These data and model lead us to believe that it is unlikely that TcpF, with dimensions of 24 by 36 Å (44), would fit through the secretin along with the pilus. It is possible that there are more monomers that make up the secretin and lead to a wider pore that allows both the pilus to extrude and TcpF to be secreted; however, this mechanism is quite different from the piston mechanism proposed for T2SS. As T4P and T2SS are thought to be closely related, it seems unlikely that secretion would occur in a completely different way.
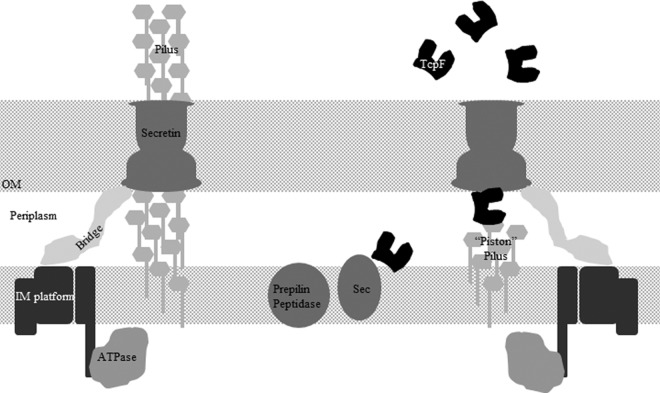
We propose that TcpF is secreted by TCP biogenesis systems that are not responsible for synthesizing entire pili. We recognize that it is possible that there is some sort of temporal regulation of protein secretion and pilus formation, but previous data suggest that the two systems are not regulated differently (41, 49, 56). Furthermore, we suggest that there are two subtypes of T4P biogenesis systems within each cell, one for protein secretion and the other for production of pili on the surface of the cell (Fig. 8). Because both protein secretion and pilus production require the components of the biogenesis apparatus and the pilin, we hypothesize that secretion is mediated by a T2SS-like piston composed of pilins. In this model, the pilin interacts with the secreted protein to “push” it through the secretin, similar to what has been proposed for T2SS (7). The model is supported by the demonstration that TcpF interacts with the pilin TcpA (Fig. 6), and future studies are aimed at demonstrating that this interaction is functional. However, a major caveat to this hypothesis is that there has not been a retractile ATPase identified for TCP, although it is possible that the apparatus that secretes proteins utilizes a retractile ATPase from type IVa pili. Furthermore, T4P biogenesis apparatus-mediated protein secretion and phage transduction can be separated from pilus biogenesis in a tcpB mutant. In this mutant, there are pili on the surface of the cell but no TcpF secretion or phage transduction (J. M. Marles and R. K. Taylor, unpublished data). This proposed model would accommodate the tcpB phenotype in that there are different roles for the biogenesis apparatus. Moreover, this model supports the data reported in this study as well as building upon data and models previously reported for T2SS and T4P.
Fig 8.
Model of T4P systems that secrete proteins. The T4P biogenesis system on the left displays typical pilus biogenesis with no protein secretion. The system on the right shows a biogenesis system that functions only to secrete proteins and does not produce an extracellular pilus. Both systems are dependent on the presence of the pilin and potentially would operate simultaneously.
In this study, we have identified an N-terminal determinant for TcpF secretion and identified a motif conserved in other proteins secreted in a T4P-mediated manner. Additionally, we have shown that TcpF interacts with the pilin TcpA, supporting the idea that the pilus contacts monomers of TcpF and pilus polymerization facilitates the secretion of this protein across the outer membrane. Our data have led us to propose a model on how the biological process of the T4P biogenesis apparatus mediates protein secretion (Fig. 8).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by National Institutes of Health grant AI 025096. Funding for C.J.M. was provided by National Institutes of Health training grant T32AI07363.
We thank Raquel Martinez for her input in editing the manuscript. We also acknowledge that discussion with Maria Sandkvist and Wim Hol led to the development of the proposed model.
We have no conflict of interest to report.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01122-12.
REFERENCES
- 1. Baron C, Coombes B. 2007. Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect. Disord. Drug Targets 7:19–27 [DOI] [PubMed] [Google Scholar]
- 2. Hazes B, Frost L. 2008. Towards a systems biology approach to study type II/IV secretion systems. Biochim. Biophys. Acta 1778:1839–1850 [DOI] [PubMed] [Google Scholar]
- 3. Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Enninga J, Rosenshine I. 2009. Imaging the assembly, structure and activity of type III secretion systems. Cell. Microbiol. 11:1462–1470 [DOI] [PubMed] [Google Scholar]
- 5. Johnson TL, Abendroth J, Hol WG, Sandkvist M. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175–186 [DOI] [PubMed] [Google Scholar]
- 6. Hobbs M, Mattick JS. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10:233–243 [DOI] [PubMed] [Google Scholar]
- 7. Reichow SL, Korotkov KV, Hol WG, Gonen T. 2010. Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17:1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reichow SL, Korotkov KV, Gonen M, Sun J, Delarosa JR, Hol WG, Gonen T. 2011. The binding of cholera toxin to the periplasmic vestibule of the type II secretion channel. Channels (Austin) 5:215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sandkvist M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271–283 [DOI] [PubMed] [Google Scholar]
- 10. Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT, Tainer JA. 2003. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11:1139–1150 [DOI] [PubMed] [Google Scholar]
- 11. Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651–662 [DOI] [PubMed] [Google Scholar]
- 12. Schilling J, Wagner K, Seekircher S, Greune L, Humberg V, Schmidt MA, Heusipp G. 2010. Transcriptional activation of the tad type IVb pilus operon by PypB in Yersinia enterocolitica. J. Bacteriol. 192:3809–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernard CS, Bordi C, Termine E, Filloux A, de Bentzmann S. 2009. Organization and PprB-dependent control of the Pseudomonas aeruginosa tad locus, involved in Flp pilus biology. J. Bacteriol. 191:1961–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for nonspecific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542–554 [DOI] [PubMed] [Google Scholar]
- 15. Durand E, Michel G, Voulhoux R, Kurner J, Bernadac A, Filloux A. 2005. XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J. Biol. Chem. 280:31378–31389 [DOI] [PubMed] [Google Scholar]
- 16. Alphonse S, Durand E, Douzi B, Waegele B, Darbon H, Filloux A, Voulhoux R, Bernard C. 2010. Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J. Struct. Biol. 169:75–80 [DOI] [PubMed] [Google Scholar]
- 17. Giltner CL, Habash M, Burrows LL. 2010. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 398:444–461 [DOI] [PubMed] [Google Scholar]
- 18. Durand E, Bernadac A, Ball G, Lazdunski A, Sturgis JN, Filloux A. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185:2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sauvonnet N, Vignon G, Pugsley AP, Gounon P. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19:2221–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen JK, Forest KT. 2006. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J. Mol. Microbiol. Biotechnol. 11:192–207 [DOI] [PubMed] [Google Scholar]
- 21. Bose N, Taylor RK. 2005. Identification of a TcpC-TcpQ outer membrane complex involved in the biogenesis of the toxin-coregulated pilus of Vibrio cholerae. J. Bacteriol. 187:2225–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sandkvist M, Michel LO, Hough LP, Morales VM, Bagdasarian M, Koomey M, DiRita VJ. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaufman MR, Shaw CE, Jones ID, Taylor RK. 1993. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene 126:43–49 [DOI] [PubMed] [Google Scholar]
- 24. Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. 2008. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154:114–126 [DOI] [PubMed] [Google Scholar]
- 25. Ayers M, Howell PL, Burrows LL. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5:1203–1218 [DOI] [PubMed] [Google Scholar]
- 26. Yamagata A, Milgotina E, Scanlon K, Craig L, Tainer JA, Donnenberg MS. 2012. Structure of an essential type IV pilus biogenesis protein provides insights into pilus and type II secretion systems. J. Mol. Biol. 419:110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, Guina T. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227–237 [DOI] [PubMed] [Google Scholar]
- 28. Zogaj X, Chakraborty S, Liu J, Thanassi DG, Klose KE. 2008. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology 154:2139–2150 [DOI] [PubMed] [Google Scholar]
- 29. Kennan RM, Dhungyel OP, Whittington RJ, Egerton JR, Rood JI. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han X, Kennan RM, Parker D, Davies JK, Rood JI. 2007. Type IV fimbrial biogenesis is required for protease secretion and natural transformation in Dichelobacter nodosus. J. Bacteriol. 189:5022–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirn TJ, Bose N, Taylor RK. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 49:81–92 [DOI] [PubMed] [Google Scholar]
- 32. Kirn TJ, Taylor RK. 2005. TcpF is a soluble colonization factor and protective antigen secreted by El Tor and classical O1 and O139 Vibrio cholerae serogroups. Infect. Immun. 73:4461–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363–378 [DOI] [PubMed] [Google Scholar]
- 34. Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, Wu LF, Filloux A. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1986. Identification of a pilus colonization factor that is coordinately regulated with cholera toxin. Ann. Sclavo Collana Monogr. 3:51–61 [PubMed] [Google Scholar]
- 36. Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chinnapen DJ, Chinnapen H, Saslowsky D, Lencer WI. 2007. Rafting with cholera toxin: endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol. Lett. 266:129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finkelstein RA, LoSpalluto JJ. 1969. Pathogenesis of experimental cholera. Preparation and isolation of choleragen and choleragenoid. J. Exp. Med. 130:185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84:2833–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karaolis DK, Somara S, Maneval DR, Jr, Johnson JA, Kaper JB. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375–379 [DOI] [PubMed] [Google Scholar]
- 41. Manning PA. 1997. The tcp gene cluster of Vibrio cholerae. Gene 192:63–70 [DOI] [PubMed] [Google Scholar]
- 42. Jude BA, Taylor RK. 2008. Genetics of Vibrio cholerae colonization and motility, p 67–99 In Nair GB, Faruque SM. (ed), Vibrio cholerae genomics and molecular biology. Horizon Scientific Press, Norfolk, United Kingdom [Google Scholar]
- 43. Krebs SJ, Kirn TJ, Taylor RK. 2009. Genetic mapping of secretion and functional determinants of the Vibrio cholerae TcpF colonization factor. J. Bacteriol. 191:3665–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Megli CJ, Yuen AS, Kolappan S, Richardson MR, Dharmasena MN, Krebs SJ, Taylor RK, Craig L. 2011. Crystal structure of the Vibrio cholerae colonization factor TcpF and identification of a functional immunogenic site. J. Mol. Biol. 409:146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896–910 [DOI] [PubMed] [Google Scholar]
- 46. Skorupski K, Taylor RK. 1996. Positive selection vectors for allelic exchange. Gene 169:47–52 [DOI] [PubMed] [Google Scholar]
- 47. Tripathi SA, Taylor RK. 2007. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J. Bacteriol. 189:4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dougherty DA. 1996. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271:163–168 [DOI] [PubMed] [Google Scholar]
- 49. Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 6:e1001102. 10.1371/journal.ppat.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sandkvist M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Forest KT. 2008. The type II secretion arrowhead: the structure of GspI-GspJ-GspK. Nat. Struct. Mol. Biol. 15:428–430 [DOI] [PubMed] [Google Scholar]
- 52. Gray MD, Bagdasarian M, Hol WG, Sandkvist M. 2011. In vivo cross-linking of EpsG to EpsL suggests a role for EpsL as an ATPase-pseudopilin coupling protein in the type II secretion system of Vibrio cholerae. Mol. Microbiol. 79:786–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. 2009. Docking and assembly of the type II secretion complex of Vibrio cholerae. J. Bacteriol. 191:3149–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL, Jr, Craig L. 2008. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure 16:137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Collins RF, Davidsen L, Derrick JP, Ford RC, Tonjum T. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stonehouse EA, Hulbert RR, Nye MB, Skorupski K, Taylor RK. 2011. H-NS binding and repression of the ctx promoter in Vibrio cholerae. J. Bacteriol. 193:979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.