Abstract
Stress-induced mutation is a collection of molecular mechanisms in bacterial, yeast and human cells that promote mutagenesis specifically when cells are maladapted to their environment, i.e. when they are stressed. Here, we review one molecular mechanism: double-strand break (DSB)-dependent stress-induced mutagenesis described in starving Escherichia coli. In it, the otherwise high-fidelity process of DSB repair by homologous recombination is switched to an error-prone mode under the control of the RpoS general stress response, which licenses the use of error-prone DNA polymerase, DinB, in DSB repair. This mechanism requires DSB repair proteins, RpoS, the SOS response and DinB. This pathway underlies half of spontaneous chromosomal frameshift and base substitution mutations in starving E. coli [Proc Natl Acad Sci USA 2011;108:13659–13664], yet appeared less efficient in chromosomal than F′ plasmid-borne genes. Here, we demonstrate and quantify DSB-dependent stress-induced reversion of a chromosomal lac allele with DSBs supplied by I-SceI double-strand endonuclease. I-SceI-induced reversion of this allele was previously studied in an F′. We compare the efficiencies of mutagenesis in the two locations. When we account for contributions of an F′-borne extra dinB gene, strain background differences, and bypass considerations of rates of spontaneous DNA breakage by providing I-SceI cuts, the chromosome is still ∼100 times less active than F. We suggest that availability of a homologous partner molecule for recombinational break repair may be limiting. That partner could be a duplicated chromosomal segment or sister chromosome.
Key Words: DNA repair, Double-strand break repair, Evolution, Mutation, Stress-induced mutation, Stress response
Introduction
Initial models of mutagenesis that drives evolution imagined random processes, constant with time [Mayr, 1985]. By contrast, stress-induced mutagenesis is a collection of mechanisms observed in bacterial, yeast and human cells in which cells activate mutation pathways under the control of stress responses [Forche et al., 2011; Galhardo et al., 2007; Mittelman and Wilson, 2010; Rosenberg, 2011]. These mechanisms increase genetic diversity in populations, and potentially the ability to evolve, specifically when cells are maladapted to their environment: when they are stressed. Molecular mechanisms of stress-induced mutation may provide superior models for genetic changes that fuel pathogen-host interactions, antibiotic resistance, aging, cancer progression and resistance, and evolution generally [Galhardo et al., 2007], and so are important to understand.
One molecular mechanism of stress-induced mutagenesis is double-strand break (DSB)-dependent stress-induced mutagenesis, described in starving Escherichia coli [Galhardo et al., 2007]. In this mechanism, the otherwise high-fidelity (non-mutagenic) process of DSB repair by homologous recombination is switched to an error-prone mode using DinB error-prone DNA polymerase, and other low-fidelity DNA polymerases, under the control of the RpoS general or starvation stress response [Ponder et al., 2005; Shee et al., 2011]. This occurs either when cells encounter an RpoS-inducing stress or if the RpoS transcriptional activator is expressed inappropriately in unstressed cells undergoing DSB repair [Ponder et al., 2005; Shee et al., 2011]. This mutation mechanism has been studied using a few different assay systems in E. coli: (1) The Lac assay measures reversion of a lac +1-bp frameshift allele in an F′ conjugative plasmid in Lac− cells starving on lactose medium [Cairns and Foster, 1991] and mutations occur via the RpoS-controlled switch to mutagenic break repair [Ponder et al., 2005]. In the same F′-lac-bearing cells starved on lactose, chromosomal (2) tet +1-bp frameshift [Bull et al., 2001] or (3) ampD loss-of-function [Petrosino et al., 2009] mutations also show DSB-, DinB- and RpoS-dependent mutagenesis. (4) In plasmid-free starved cells not under selection for either resistance, reversion of a tet +1-bp frameshift allele, creating tetracycline resistance, or base-substitution mutation of the gyrB gene to nalidixic acid resistance, occurs by the RpoS-controlled switch to error-prone DSB repair [Shee et al., 2011].
Error-prone DSB repair also appears to underlie stress-induced mutagenesis in circumstances other than starvation and in organisms other than E. coli. E. coli under antibiotic stress induce a similar DSB repair protein-, SOS- and DinB-dependent mutagenesis pathway [Cirz and Romesberg, 2007]. Pathogenic Salmonella induce DSB repair protein-, SOS-, DinB- and RpoS-dependent mutation in response to bile, a membrane irritant [Prieto et al., 2006; Casadesus, pers. commun.]. Pseudomonas aeruginosa biofilms show DSB- and DSB-repair-protein-dependent generation of genetic diversity which may arise by a similar mechanism [Boles and Singh, 2008]. DinB- and RpoS-dependent mutagenesis is not seen in non-pathogenic Salmonella strain LT2 [Koskiniemi et al., 2010; Quinones-Soto and Roth, 2011], but this is likely to reflect the fact that LT2 is a natural variant that is nonpathogenic because it is RpoS defective [Lee et al., 1995; Swords et al., 1997; reviewed in Shee et al., 2011]. DSB-dependent mutation was found first in E. coli [Harris et al., 1994; Rosenberg et al., 1994], then described in baker's yeast [Deem et al., 2011; Hicks et al., 2010; Strathern et al., 1995; Yang et al., 2008], in which the mutagenicity of DSB repair is not known be stress inducible and may be constitutive. However, DSB repair itself appears to be stress inducible in the pathogenic yeast Candida [Forche et al., 2011], making it possible that yeasts also have stress-inducible mutagenesis caused by error-prone DSB repair, but with the stress inducibility controlled at the step of DSE creation or repair rather than the mutagenicity of repair as in E. coli [Rosenberg, 2011].
Here, we focus on DSB-dependent stress-induced mutation in starving E. coli. Because the RpoS-controlled switch to mutagenic break repair is general to both the F′-lac assay [Ponder et al., 2005] and the chromosomal tet assay without selection for Tet resistance mutations during mutagenesis [Shee et al., 2011], we outline the mechanism generally, then discuss quantitative and qualitative contributions to mutagenesis of the specific assay systems. We then consider what factors influence the contribution of this mechanism to spontaneous mutagenesis in various circumstances.
Mechanism
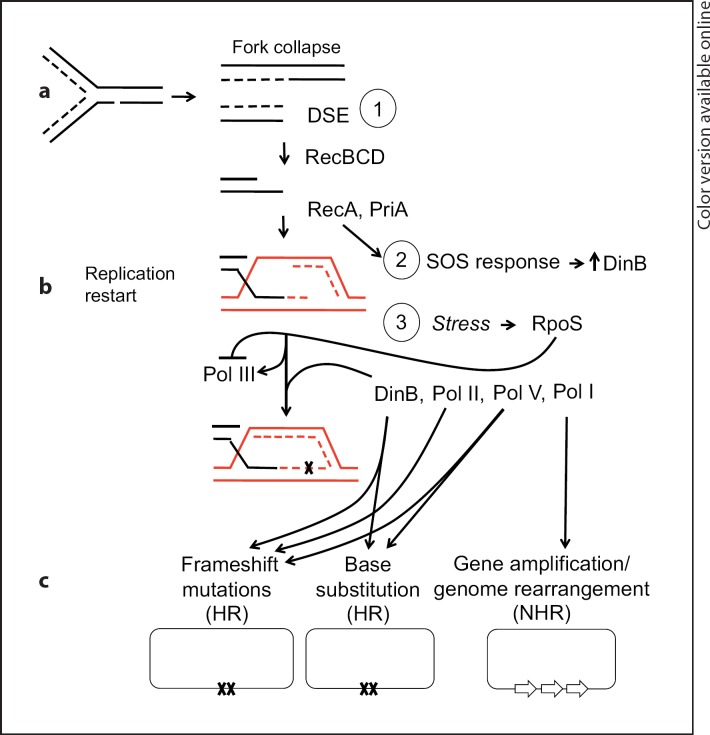
DSB-dependent stress-induced mutation occurs when three events occur simultaneously [Ponder et al., 2005; Shee et al., 2011] (fig. 1): (1) a double-stranded DNA break or double-stranded end (DSB/DSE) and its repair by homologous recombination; (2) activation of the SOS DNA-damage response, which DSBs/DSEs induce [McPartland et al., 1980; Pennington and Rosenberg, 2007], and (3) a second stress, unrelated to the DSB/DSE, that activates RpoS. These events occur and promote mutagenesis as follows.
Fig. 1.
DSB-dependent stress-induced point mutagenesis requires three events: (1) a DSB or double-strand end (DSE) and its repair; (2) activation of the SOS response, which upregulates DinB errorprone DNA polymerase, and (3) a second stress that activates RpoS, which allows the use of DinB and other mutation-causing DNA polymerases Pol II, Pol V and Pol I in repair, instead of/in addition to high-fidelity DNA Pol III. We hypothesize that RpoS licenses use of these alternative DNA polymerases by downregulation of their competitor Pol III [Frisch et al., 2010]. a Creation of a DSE by replication fork collapse at an ssDNA nick. Lines represent single DNA strands, dashed lines newly synthesized DNA. b Replication restart. Xs represent DNA polymerase errors that become mutations. c Mutated chromosomes. Single lines represent double-stranded DNA. HR = Homologous recombination; NHR = nonhomologous or microhomologous recombination.
Origin of DSBs/DSEs and Their Repair
DSBs/DSEs occur spontaneously in just over 10−3 of growing E. coli cells [Pennington and Rosenberg, 2007], and induce the SOS response about 25% of the time that they are repaired [Pennington and Rosenberg, 2007]. Thus, event number 2, the SOS response, is a consequence of event 1, formation of a DSB.
In the Lac assay, most of the DSEs are instigated at a higher-than-spontaneous level by the F-encoded TraI endonuclease, which makes single-strand (ss)DNA nicks at the F origin of transfer. That is, TraI is required for DSB repair protein-, SOS-, DinB- and RpoS-dependent lac reversion in the F′, but is not required if an I-SceI endonuclease-generated DSB is made in vivonear lac in the F′ [Ponder et al., 2005]. Whereas an I-SceI cleavage near lac substituted for TraI and increased mutagenesis an additional 70-fold, one made in a different plasmid (intrans to lac) increased mutagenesis only 3-fold, not the ∼6,000-fold seen when a TraI-defective F′ was cleaved by I-SceI[Ponder et al., 2005]. The DSE made intrans to lac could, however, activate lac reversion if the DNA next to lac contained sequences homologous to one end of the trans-cut plasmid. That is, a DSE in another molecule provoked mutation at lac if that DSE could interact by homologous recombination with the DNA near lac, demonstrating that DSE repair by homologous recombination causes the mutations, and the mutations happen at the sites of repair.
TraI-generated ssDNA nicks in F presumably become DSEs when replicated over, causing fork collapse (fig. 1a) [Kuzminov, 1995]. For the TraI-dependent DSE to occur in the F′, activation of the RpoE membrane protein stress response is also required [Gibson et al., 2010]. An RpoE stress-response-off mutant is mutagenesis defective, and this is overcome by I-SceI cleavage near lac, indicating that RpoE, like TraI, acts upstream of creation of DSEs required for mutation. RpoE may promote local replication that causes fork collapse at the TraI-generated ssDNA nick [Gibson et al., 2010].
In starved F− cells, DSBs created by I-SceI provoke chromosomal tet reversion [Shee et al., 2011]. Moreover, half of the spontaneous frameshift and base substitution mutagenesis in starved E. coli with no I-SceI also required SOS, DinB, RpoS, and DSB-repair proteins, including the highly DSE-specific RecBCD enzyme, indicating that spontaneous DSBs/DSEs instigate DSB-dependent stress-induced mutagenesis in the chromosome [Shee et al., 2011].
Stress Response Control of Mutagenesis: Mechanism and Significance
DSEs induce the SOS DNA-damage response [Pennington and Rosenberg, 2007], which upregulates ∼40 DNA damage-inducible genes transcriptionally [Courcelle et al., 2001; Fernandez De Henestrosa et al., 2000]. Of them, only dinB is required at SOS-induced levels for mutagenesis [Galhardo et al., 2009]. This was shown in experiments in which the induction of the SOS genes was blocked, and only DinB was supplied at its SOS-induced level, and mutagenesis was restored to stressed cells [Galhardo et al., 2009]. However, even with DinB upregulated 10-fold to its SOS-induced level, DSE repair is not mutagenic and does not appear to use DinB unless the RpoS response is also induced [Lombardo et al., 2004; Ponder et al., 2005; Shee et al., 2011].
The RpoS response throws a switch that allows error-prone DNA polymerases to be used in DSE repair and thus limits mutagenesis to times of RpoS-inducing stress. RpoS upregulates DinB an additional 2-fold [Layton and Foster, 2003], and somehow licenses the use of DinB [Ponder et al., 2005; Shee et al., 2011], as well as DNA polymerases (Pols) II [Frisch et al., 2010], V [Petrosino et al., 2009; Shee et al., 2011], and I [Hastings et al., 2004; Slack et al., 2006], all of which cause mutations, during DSE repair [Frisch et al., 2010] (fig. 1). Whereas DinB promotes 85% and Pol II the remaining 15% of ±1-bp frameshift mutations [Frisch et al., 2010; McKenzie et al., 2001] with Pol V contributing slightly to the DinB-dependent component [Shee et al., 2011], DinB and Pol V promote DSE- and RpoS-dependent base substitutions [Petrosino et al., 2009], and Pol I is required for DSE- [Ponder et al., 2005; Slack et al., 2006] and RpoS-dependent [Lombardo et al., 2004] stress-induced gene amplifications [Hastings et al., 2004; Slack et al., 2006]. By contrast, DSB repair in unstressed cells requires the high-fidelity major replicative DNA polymerase Pol III [Motamedi et al., 1999] and is non-mutagenic [Ponder et al., 2005; Shee et al., 2011]. Perhaps RpoS promotes mutagenic DSB/DSE repair by inhibiting Pol III so that the other four E. coli DNA polymerases have access to the DSB repair replisome [Frisch et al., 2010] (fig. 1). All of the DNA pols appear to compete [Hastings et al., 2010], and if cells are not stressed, or do not have RpoS activated, Pol III appears to win and repair is non-mutagenic. Thus, DSE-dependent stress-induced mutagenesis is controlled critically by RpoS, which switches DSE repair from a high-fidelity mode using Pol III to an error-prone mutagenic process, using error-prone DNA polymerases when cells are stressed, potentially accelerating evolution specifically then.
Mutagenesis associated with the SOS DNA-damage response was argued to be an unavoidable consequence of induced DNA repair [e.g. Andersson et al., 2010; Lynch, 2010], not an evolutionary engine as initially suggested [McPartland et al., 1980; Radman, 1975]. Interestingly, neither RpoS nor DinB is required for survival/repair of DSBs during stress, which works as efficiently [Shee et al., 2011] or more efficiently [Ponder et al., 2005] without them. Therefore, though both RpoS and DinB promote survival of other problems [Battesti et al., 2011; Nohmi, 2006], their roles in mutation during DSE repair are not an unavoidable consequence of the need to repair DSEs and so may be an accelerator of evolution.
Chromosome versus F
Here, we address the efficiency of DSB-dependent stress-induced mutagenesis in starved E. coli. Previous studies indicated that lac in the E. coli chromosome experienced less DSB-dependent stress-induced mutation than lac in an F′ plasmid [Foster and Trimarchi, 1995; Radicella et al., 1995], yet our recent work shows that the chromosome undergoes this mechanism, and that F plasmids are unnecessary [Shee et al., 2011]. We also found that (1) the F′ lac increases chromosomal DSB-dependent stress-induced mutagenesis 2-fold, solely because of an extra dinB gene borne by this F′. The F′ with dinB deleted has no effect on chromosomal DSB-dependent stress-induced mutagenesis [Shee et al., 2011]. (2) Even when the differing rates of DSE formation in F, which has TraI-promoted DSEs [Ponder et al., 2005], and the chromosome, which does not, are transcended by use of I-SceI endonuclease to provide a local DSB, the F appears to be more active for DSB-dependent stress-induced mutation. First, I-SceI promoted chromosomal tet mutation 50-fold compared with uncut (enzyme-only or cutsite-only) controls [Shee et al., 2011], whereas I-SceI promoted lac reversion in the F′ ∼6,000-fold compared with the TraI-defective uncut control [Ponder et al., 2005]. Second, the overall frequency of I-SceI-promoted stress response-dependent mutation in starved E. coli was about 10−4– 10−5 Lac+ mutants in the F′ per cell by day 5 of starvation on lactose plates [Ponder et al., 2005] versusabout 10−7 chromosomal TetR mutants per cell after about 3 days’ starvation in liquid. Obviously, the different protocols in the F′ Lac versuschromosomal Tet assays and different DNA sequences in the lac and tet genes mutated prevent meaningful comparison of these mutant frequencies. Here, we use the same protocol and gene as used previously in the F′, the lac gene, but measure its reversion in the chromosome, and compare the efficiency of mutation and fold increase with I-SceI-generated DSBs in the two locations.
Methods
Strains, Media and Growth
E. coli strains used in this study are given in table 1. Bacteria were grown in LBH [Torkelson et al., 1997] or M9 minimal medium [Miller, 1992] supplemented with 10 μg/ml thiamine (vitamin B1) and 0.1% glucose or 0.1% glycerol as carbon source. Strain constructions were made by standard methods [Miller, 1992] except where noted. I-sites were moved into specific locations using Tn7 per Ponder et al. [2005]. Other additives were used at the following concentrations (μg/ml): ampicillin, 100; kanamycin, 50; rifampicin, 100; tetracycline, 10; sodium citrate 20 mM; 5-fluorocytosine, 20; 5-fluorouracil, 10; 5-bromo-4-chloro-3-indolyl β-D-galactoside, 40.
Table 1.
E. coli K12 strains and plasmids
| Plasmid/strain | Relevant genotype | Reference or source |
|---|---|---|
| DH5α | SuII+ recA hsdrK–mK+ | Hanahan [1983] |
| pCW15(C127) | pACYC184 derivative, expresses Tn7 transposition proteins TnsA, B, and CA225V, CamR | Stellwagen and Craig [1997] |
| pMCB40 | pBW30 derivative, miniTn7Kan donor plasmid, KanR, has R6Kλ origin that requires π protein for replication | Biery et al. [2000] |
| pRP1 | pMCB40 mini Tn7Kan(I-SceI site) donor plasmid, KanR, requires π protein for replication | Ponder et al. [2005] |
| pTGV express | pBR322-derived chromosomal expression cassette-integration vector, has PBAD and a multiple cloning site flanked by gal and bio sequences homologous to DNA surrounding att λ AmpR, Tets | Gumbiner-Russo et al. [2001] |
| MG1655 | Wild type | Blattner et al. [1997] |
| BW23474, or MB69 | endA(BT333) hsdR514 recAl ΔuidA(MluI)::pir-116 (expresses π protein for R6Kλ plasmid replication) | Biery et al. [2000] |
| FC36 | δ(lac-proB)XIII thi ara RifRF- λ- | Cairns and Foster [1991] |
| FC40 | FC36[F’ laclq lacI33ΩllacZ proAB+] | Cairns and Foster [1991] |
| K12 [F+] | Wild type [F+] | Bachmann [1972] |
| MB75 | BW23474 [pMCB40] | Biery et al. [2000] |
| SMR696 | proC zaj3053::Tn10 Δ lacX74 | Lab collection |
| SMR1316 | MG1655 RhR | Spontaneous rifampicin resistance |
| SMR1698 | MG1655 RifR proC zaj3053:Tn10 ΔlacX74 | SMR1316 × PI SMR696 |
| SMR4453 | FC40 5FCR 5FUS codA/B::Tn10dTet | FC40 × XNK1323 |
| SMR4514 | MG1655 RifR proC zaj3053:Tnl0 ΔlacX74 | SMR1698 × P1 MG1655 |
| SMR4522 | MG1655 cod::Tn10 lacq lacI33ΩlacZ | MG1655 × PI SMR4453 |
| SMR4562 | Independent construction of FC40 | McKenzie et al. [2000] |
| SMR4615 | MG1655 lacq lacI33ΩlacZ RifR | SMR4514 × PI SMR4522 |
| SMR4874 | SMR4615 lacq lacI33ΩlacZ RifR [F+] | SMR4615 mated with K12 [F+] |
| SMR5078 | recB21 recC22 sbcB15 sbcC201 hsdrK−mK+ (λ xis1 cIt857)-mono lysogen | Gumbiner-Russo et al. [2001] |
| SMR5405 | FC40 [pCW15(C127)] | Ponder et al. [2005] |
| SMR5448 | DH5Α [pTGV express] | Gumbiner-Russo et al. [2001] |
| SMR5739 | BW23474 [pRPl-13] | BW23474 × ligation mix of pMCB40 DNA with oligos |
| SMR5741 | BW23474 [pRPl-13-1] | BW23474 × digested and religated pRP1-13, selected KanR, confirmed by sequencing |
| SMR5744 | recB21 recC22 sbcB15 sbcC201 Δattλ::PBADI-SceI | Gumbiner-Russo et al. [2001] |
| SMR5746 | recB21 recC22 sbcB15 sbcC201 Δattλ::PBAD | Gumbiner-Russo et al. [2001] |
| SMR5973 | SMR4562 [F’ mhpA32::miniTn7Kan(I-SceI site)] [pCW15(C127)] | Ponder et al. [2005] |
| SMR5974 | SMR4562 [F’ mhpA33:: miniTn7Kan(I-SceI site)] [pCW15(C127)] | SMR5405 × pRPl, selected KanR, screened 5-FCR; 5-FUs, Tn orientation determined by PCR; insertion between F’ equivalents of chromosomal bp 369,382 and 369,383 |
| SMR5975 | SMR4562 [F’ codA21::miniTn7Kan(I-SceI site)] [pCW15(C127)] | Ponder et al. [2005] |
| SMR5976 | SMR4562 [F’ codA22::miniTn7Kan(I-SceI site)] [pCW15(C127)] | Ponder et al. [2005] |
| SMR6267 | SMR4615 leu::Tn10 [F+] (λ xisl cIts857) | SMR5995 × PI SMR6263 |
| SMR6278 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI | SMR6274 × PI SMR5744 |
| SMR6279 | SMR4615 ΔaraBAD567 Δattλ::PBAD | SMR6274 × PI SMR5746 |
| SMR6292 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI mhpA32::miniTn7Kan(I-SceI site) | SMR6278 × PI SMR5973 |
| SMR6293 | SMR4615 ΔaraBAD567 Δattλ::PBAD mhpA32::miniTn7Kan(I-SceI site) | SMR6279 × PI SMR5973 |
| SMR6294 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI mhpA33::miniTn7Kan(I-SceI site) | SMR6278 × PI SMR5974 |
| SMR6295 | SMR4615 ΔaraBAD567 Δattλ::PBAD mhpA33::miniTn7Kan(I-SceI site) | SMR6279 × PI SMR5974 |
| SMR6296 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI codA21::miniTn7Kan(I-SceI site) | SMR6278 × PI SMR5975 |
| SMR6297 | SMR4615 ΔaraBAD567 Δattλ::PBAD codA21::miniTn7Kan(I-SceI site) | SMR6279 × PI SMR5975 |
| SMR6298 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI codA22::miniTn7Kan(I-SceI site) | SMR6278 × PI SMR5976 |
| SMR6299 | SMR4615 ΔaraBAD567 Δattλ::PBAD codA22::miniTn7Kan(I-SceI site) | SMR6279 × PI SMR5976 |
| SMR6323 | SMR4615 ΔaraBAD567 Δattλ::PBAD [F+] | SMR6279 × SMR4874 |
| SMR6324 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI [F+] | SMR6278 × SMR4874 |
| SMR6325 | SMR4615 ΔaraBAD567 Δattλ::PBAD mhpA32::miniTn7Kan(I-SceI site) [F+] | SMR6293 × SMR4874 |
| SMR6326 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI mhpA32::miniTn7Kan(I-SceI site) [F+] | SMR6292 × SMR4874 |
| SMR6327 | SMR4615 ΔaraBAD567 Δattλ::PBAD mhpA33::miniTn7Kan(I-SceI site) [F+] | SMR6295 × SMR4874 |
| SMR6328 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI mhpA33::miniTn7Kan(I-SceI site) [F+] | SMR6294 × SMR4874 |
| SMR6329 | SMR4615 ΔaraBAD567 Δattλ::PBAD codA21::miniTn7Kan(I-SceI site) [F+] | SMR6297 × SMR4874 |
| SMR6330 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI codA21::miniTn7Kan(I-SceI site) [F+] | SMR6296 × SMR4874 |
| SMR6331 | SMR4615 ΔaraBAD567 Δattλ::PBAD codA22::miniTn7Kan(I-SceI site) [F+] | SMR6299 × SMR4874 |
| SMR6332 | SMR4615 ΔaraBAD567 Δattλ::PBADI-SceI codA22::miniTn7Kan(I-SceI site) [F+] | SMR6298 × SMR 4874 |
Stress-Induced Mutation Assays
Stress-induced lac reversion assays were performed as described previously [Ponder et al., 2005]. Bacterial strains carrying the inducible I-SceI system consisting of a chromosomal I-SceI gene controlled by the glucose-repressible and arabinose-inducible PBAD promoter inserted into the phage lambda attachment site (Δattλ::PBADI-SceI [Gumbiner-Russo et al., 2001]) and an I-SceI cutsite were maintained on medium containing 0.1% glucose to repress I-SceI synthesis [Ponder et al., 2005]. Stress-induced mutation assays were performed by growing the cells to saturation in minimal glycerol medium with 0.1% glucose to repress I-SceI, then plating on minimal lactose medium, in which there is no glucose to repress I-SceI, but also no arabinose. This results in very low level expression of I-SceI [Ponder et al., 2005].
Results and Discussion
DSBs Activate Chromosomal Stress-Induced Lac Reversion
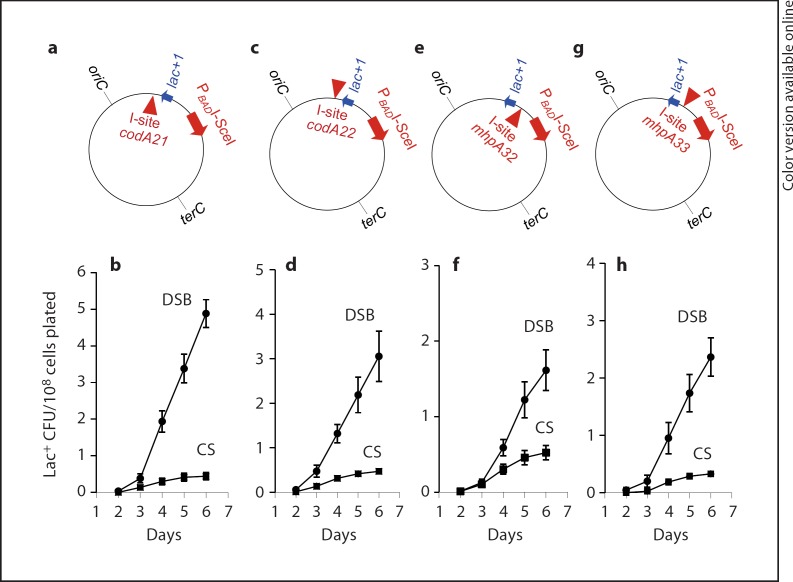
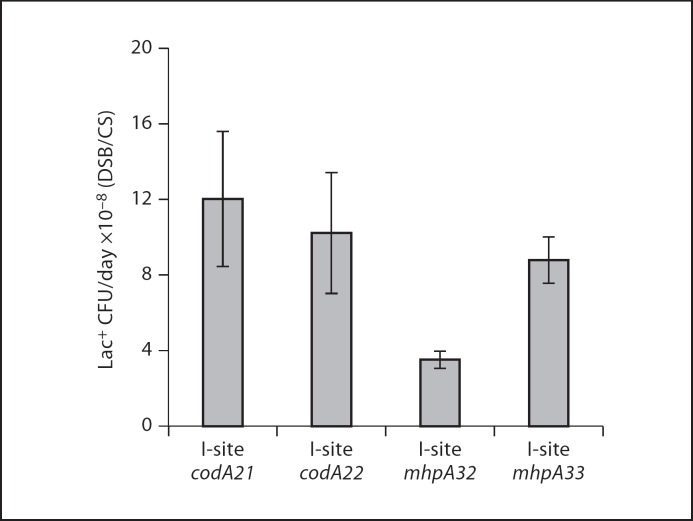
To study chromosomal DSB-dependent stress-induced lac reversion stimulated by I-SceI, we replaced the lac operon of E. coli wild-type strain MG1655 with the lacI33ΩlacZ frameshift allele from F′128, the F′ in the Lac-assay strain. We also engineered in the chromosomal regulatable I-SceI endonuclease gene of Ponder et al. [2005]. We placed four different I-SceI cutsites near the lac frameshift allele in separate strains (fig. 2a, c, e, g). Three of these sites were used previously in our study of I-SceI-induced lac reversion in the F′ [Ponder et al., 2005]. I-sites codA21 and codA22 are in opposite orientations in the codA gene, about 10 kb downstream of the lac allele and I-sites mhp32 and mhp33 are in opposite orientations in the mhpA gene, about 2 kb upstream of the lac allele [Ponder et al., 2005]. For historical reasons, all strains carry an F+ plasmid that contains no E. coli chromosomal genes, which is now known to have no effect on mutagenesis, in that an F plasmid without an extra copy of dinB had no effect on chromosomal mutagenesis [Shee et al., 2011]. We find that strains carrying each independent cutsite and the regulatable I-SceI endonuclease show increased mutations over time starving on lactose compared with control strains that carried cutsite only (fig. 2b, d, f, h) or enzyme only (data not shown), indicating DSB-dependent mutation. The I-site codA21-carrying strain exhibited the highest mutation rate, about 12-fold higher than the control cutsite-only strain (fig. 3). Strains carrying I-sites codA22, mhp32, andmhp33 showed about 10-, 4- and 9-fold increases in mutation rate compared with cutsite-only control strains, respectively (fig. 3).
Fig. 2.
Chromosomal Lac reversion mutations are stimulated by I- Sce I-induced DSBs over time during starvation. a, c, e, g Cartoon of E. coli chromosome showing position of PBADI-SceI, lacI33ΩlacZ allele, (lac+1), and I- Sce I cut sites (I-sites). Arrows denote direction of transcription. b, d, f, h I-SceI-induced DSBs increase Lac+ colonies compared with cutsite-only control strains (CS). Cultures were grown at 37°C. DSB strains (•) carry both PBADISceI and one I- Sce I cut site; and CS (▪) carry I-SceI cutsite only. a, b I-site codA21. c, d I-site codA22. e, f I-site mhpA32. g, h I-site mhpA33. All cutsites except mhp33 were used by Ponder et al. [2005]. Strains used: SMR6330, SMR6329 (a, b); SMR6332, SMR6331 (c, d); SMR6326, SMR6325 (e, f); SMR6328, SMR6327 (g, h).
Fig. 3.
Fold effect of I-SceI-induced DSBs on mutation rate. Mutation rates are averaged from three independent mutation assays based on Lac+ colonies arising per day from days 4 to 6 ± SEM. DSB strains carry both PBADI-SceI and one I-SceI cutsite and CS carry I-SceI cutsite only. Strains as in figure 2.
Higher Efficiency of I-SceI-Stimulated Mutation in F than Chromosome
We can compare these data to the data of Ponder et al. [2005] in which the same lac gene underwent DSB-dependent stress-induced mutagenesis during starvation on lactose medium stimulated by I-SceI cuts at three of the same sites. Overall, induction of mutation by I-SceI in those experiments was ∼6,000-fold, compared with the ∼10-fold observed here (fig. 3). Also, the overall mutant frequencies ranged from 3,000 to 25,000 Lac+ mutants per 108 cells by day 5 (or 3–25 × 10−5 mutants per cell) in the F experiments of [Ponder et al., 2005] versus∼2 Lac+ mutants per 108 cells by day 5 (or 2 × 10−8 mutants per cell; fig. 2), ≥1,000-fold fewer. Parts of that 1,000-fold difference can be accounted for as follows: first, in the experiments of Ponder et al. [2005], the F′ dinB+ was present, and this extra dinB copy increases DSB-dependent stress-induced mutagenesis 2-fold [Shee et al., 2011]. Second, Ponder et al. [2005] conducted their experiments in the FC36 strain background, which we showed is about 4- to 5-fold more active for DSB-dependent stress-induced mutation than MG1655, the strain background used here [Shee et al., 2011]. These two factors together can account for about 10-fold of the difference in frequencies in the F experiments of Ponder et al. [2005] and the chromosomal lac reversions measured here. We conclude that there is still about 100-fold more I-SceI-stimulated DSB-dependent stress-induced mutation in the F than in the chromosome when the same lactose starvation protocol and lac gene are compared.
Previously, using a protocol of shorter-term generic starvation conditions in liquid medium in the assay for chromosomal mutation of a tet gene [Shee et al., 2011], overall frequencies and fold-induction by I-SceI were ∼10-fold higher than observed here: about 2 × 10−7 TetR mutants per cell and 50- to 60-fold induction by I-SceI, respectively. This probably reflects the differences in strain background (FC36 for tet experiments versusMG1655 here, the former of which shows 4–5 times more DSE-dependent mutagenesis [Shee et al., 2011]) and starvation-stress protocol, rather than use of the tet versus the lac allele, because a comparison of stress-induced reversion of these two alleles in the F′ indicated that lac was somewhat more mutationally active than tet [Foster, 1997]. That is the opposite of our results with the two alleles used in two different starvation protocols for chromosomal stress-induced reversion (fig. 2, 3) [Shee et al., 2011]. Although the lactose starvation protocol for the chromosomal reversion assays presented here is clearly not optimized for efficient DSB-dependent stress-induced mutation, compared with the protocol of Shee et al. [2011], the results obtained with it allow direct comparison with the data of Ponder et al. [2005] in F.
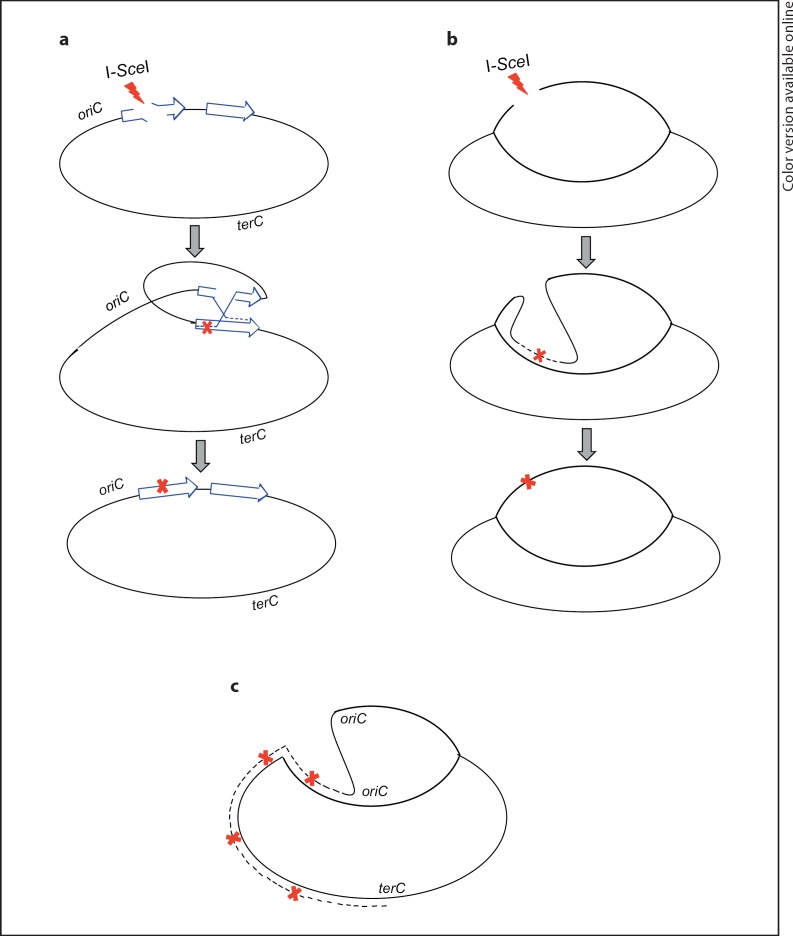
What might cause the ∼100-fold lower I-SceI-induced lac reversion in the chromosome (fig. 2, 3) compared with the F′ [Ponder et al., 2005]? Copy-number differences between the chromosome and F plasmid could be important. In both sets of experiments, mutations are formed during acts of homologous recombinational repair of I-SceI-induced DSBs. Such repair requires an identical DNA sequence that has not also been cut by I-SceI. In the haploid E. coli genome, the homologous partner for repair could be either a sister chromosome or duplicated DNA segment (fig. 4a, b). The experiments here and of Ponder et al. [2005] were done using conditions of low-level leaky expression of I-SceI from the PBAD promoter [Ponder et al., 2005] to maximize the possibility that not all I-sites present in a cell would be cleaved simultaneously. Copy number could be critical and different between F and the chromosome. Most stationary-phase cells contain one chromosome, but about 40% carry two [Akerlund et al., 1995], and tandem duplications are even less frequent in the chromosome (≤10−3) [Roth et al., 2006]. Presumably, successful homologous recombinational DSB repair events occur in the sub-fraction of cells with a sister or duplication, and the sub-fraction of those in which only one of the two I-sites has been cleaved. This is expected to be a smaller sub-fraction for chromosomal than F′ mutagenesis because F copy number is slightly higher than that of the chromosome, and was estimated to be two to three F plasmids per cell [Foster and Rosche, 1999]. We suggest that perhaps the 100-fold difference might be that F′s more often have an uncleaved sister with which to repair (fig. 4b), but that chromosomal repair might rely on the less frequent duplications of chromosomal segments or sisters (fig. 4a, b). Although the estimates of sister chromosomes in stationary phase (40% of cells [Akerlund et al., 1995]) are about 400 times higher than estimates of chromosomal segment duplication frequency (10−3), neither of those estimates was made in the strain background or specific stationary-phase conditions used here, and moreover the estimate of duplications was made in a different bacterium: Salmonella rather than E. coli. Thus, there is a lot of room for the difference in frequency to be caused by use of a frequent sister in F-located mutagenic DSB repair and use of a less frequent duplication in chromosomal mutagenic DSB repair, as argued by Roth and colleagues [e.g. Kugelberg et al., 2006], particularly when I-SceI cuts are the source of the DSBs that activate mutagenesis.
Fig. 4.
Possible templates for mutagenic DSB repair by homologous recombination during stress. a An I-SceI-induced DNA DSB that occurs within a duplicated region (blue arrows) can be repaired by homologous recombination using the intact duplicated segment as homology donor. Because duplications are rapidly lost by homologous recombination, the two copies could subsequently be reduced to one, either mutant or not (not shown). b I-SceIinduced DSBs could also be repaired by homologous recombination using a sister DNA molecule. This route may be more prevalent in conjugative plasmids than chromosomes because of their somewhat higher copy number. c Error-prone repair of a collapsed replication fork during stress differs from I- Sce I cuts in having one DSE, whereas I- Sce I creates two. Also, although in principle either a sister (shown) or duplication (not shown) could be used for repair, the sister is always present. Single lines represent double-stranded DNA, dashed lines newly synthesized DNA, and red Xs DNA polymerase errors that become mutations. For figure in color, please refer to the online version of the article.
DSE-dependent stress-induced mutation also underlies half of spontaneous mutation in starved E. coli cells [Shee et al., 2011], resulting from spontaneous DSEs. Spontaneous DSEs are thought to occur when a replication fork collapses at an ssDNA nick (fig. 1a). If that is the source of most spontaneous DSEs, then repair synthesis might be expected to use a sister as template for error-prone repair synthesis under those circumstances (e.g. fig. 4c).
Further Discussion
Recent work on DSB-, RpoS-, SOS- and DinB-dependent chromosomal tet reversion in starving cells [Shee et al., 2011] showed that DSB-dependent stress-induced mutation is general and not restricted to conjugative plasmids as had been suggested [Foster and Trimarchi, 1995; Godoy et al., 2000; Radicella et al., 1995; Roth et al., 2006; Slechta et al., 2002; Slechta et al., 2003], and not limited to genes under selection while they are mutating. Genes under selection might have mutated independently of stress but the mutants selected by the stress condition. This study also dispelled concern about the extra F-borne dinB copy, which was shown to contribute only 2-fold [Shee et al., 2011]. Thus, the mechanism of DSB-dependent stress-induced mutagenesis is general.
Another area of discussion on the mechanism of DSB-dependent stress-induced mutation is the basis for the requirement of homologous recombinational DSB repair proteins. In one model, homologous recombination events are proposed to promote duplication and amplification of the leaky lac frameshift allele under selection for Lac function, as a prerequisite to mutagenesis [Roth et al., 2006]. In another model, DSB-repair proteins function in error-prone DSE repair under stress [Ponder et al., 2005]. These views need not be opposed. First, the generation of required duplications is compatible with the error-prone DSE-repair model (fig. 4a) and helpful in explaining the frequencies of chromosomal mutation, discussed above.
Second, in the Lac assay, Lac+ colonies can arise by two mechanisms: stress-induced compensatory frameshift or ‘point’ mutation, or stress-induced amplification [Hastings et al., 2000; Hastings et al., 2004]. In stress-induced amplification, the leaky lac allele is amplified to many copies such that sufficient β-galactosidase activity is produced for growth on lactose medium without acquisition of a Lac+ point mutation. Like point mutation, stress-induced amplification requires RpoS [Lombardo et al., 2004] and DSBs/DSEs [Ponder et al., 2005; Slack et al., 2006], but it does not require SOS and DinB [McKenzie et al., 2001] and uniquely requires Pol I [Hastings et al., 2004; Slack et al., 2006], distinguishing it from DSE-dependent stress-induced point mutagenesis. Studies of the stress-induced amplification mechanism are leading thought on models for the generation of copy-number variations in human and other organisms [Hastings et al., 2009a, b]. Clearly, amplification is an important source of evolutionarily and practically significant genomic change.
Third, the primary distinctive feature of stress-induced mutation is its stress inducibility: the requirement for the RpoS general stress response in the mechanism, which controls a switch from high-fidelity to error-prone DNA DSB repair under stress [Ponder et al., 2005; Shee et al., 2011]. This was shown in experiments in which growing, unstressed cells, under no selection for mutations, switched their repair of I-SceI cuts to mutagenic repair using DinB, if RpoS was expressed [Ponder et al., 2005; Shee et al., 2011]. Thus, the RpoS response contribution to mutagenesis is unequivocal. It is also seen generally in many other circumstances. Other pathways of stress-inducible mutation also require RpoS, including transposition/excision of phage Mu [Gomez-Gomez et al., 1997; Lamrani et al., 1999], stress-inducible point mutation [Saumaa et al., 2002] and transposition [Ilves et al., 2001] in Pseudomonas putida, DSB-independent stress-induced mutation in aging colonies of an E. coli natural isolate [Bjedov et al., 2003], DSB-dependent bile-induced resistance mutation in pathogenic Salmonella [Prieto et al., 2006; Casadesus, pers. commun.], and DSB-dependent [Ponder et al., 2005; Slack et al., 2006] stress-induced gene amplification in E. coli [Lombardo et al., 2004]. RpoS is induced by many different stressors including starvation, osmotic, pH, temperature and oxidative stresses [Battesti et al., 2011]. The importance of coupling mutagenesis pathways to a broad general stress response like RpoS might be that genetic diversity may be generated responsively to many different stressors and environments.
Other bacterial starvation and general stress responses also promote mutagenesis during stress as do pathways in yeast and human cancer cells [Galhardo et al., 2007]. These include the stringent and the competence starvation-stress responses in Bacillus subtilis [Robleto et al., 2007], the stringent [Coros et al., 2009; Wright et al., 1999], cyclic AMP [Lamrani et al., 1999; Taddei et al., 1995] responses to starvation, and the RpoE membrane-protein stress response [Gibson et al., 2010] in E. coli. These promote base substitutions [Taddei et al., 1995; Wright et al., 1999], frameshift mutations [Gibson et al., 2010], amplification [Gibson et al., 2010], mobile-intron movement [Coros et al., 2009], and transposon excision [Coros et al., 2009; Lamrani et al., 1999]. Some yeast and mammalian stress-induced mutagenesis pathways are reviewed elsewhere [Bindra et al., 2007; Forche et al., 2011; Galhardo et al., 2007; Mittelman and Wilson, 2010; Rosenberg, 2011]. These examples illustrate the apparently multiple evolutions of mechanisms that couple genomic instability pathways with stress responses and stress. The importance of all of these is that genetic diversity is generated preferentially when cells are maladapted to their environment – when stressed, in contrast with early ideas about constant and gradual mutation underlying evolution.
Although the coupling of mutagenesis with the general/starvation stress responses is a fact and widely accepted by many groups, aspects of the mechanism of DSB-dependent stress-induced mutation are still under discussion, mainly concerning the involvement of duplication and amplification as part of the stress-induced mutation mechanism [Kugelberg et al., 2006]. Per fig ure 4, we agree that use of duplications as a part of the DSB-dependent stress-induced mutation mechanism may be expected, particularly in the chromosome. Limiting homologous DNA for repair, caused by limiting chromosomal duplications, may explain the different efficiencies of DSB-dependent stress-induced mutation in the chromosome versusthe more frequently multi-copy F.
Acknowledgements
We thank P.J. Hastings for comments on the manuscript. Supported by NIH grant R01-GM53158.
References
- Akerlund T, Nordstrom K, Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI, Koskiniemi S, Hughes D. Biological roles of translesion synthesis DNA polymerases in eubacteria. Mol Microbiol. 2010;77:540–548. doi: 10.1111/j.1365-2958.2010.07260.x. [DOI] [PubMed] [Google Scholar]
- Bachmann BJ. Pedigrees of some mutant strains of Escherichia coli k-12. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biery MC, Stewart FJ, Stellwagen AE, Raleigh EA, Craig NL. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 2000;28:1067–1077. doi: 10.1093/nar/28.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra RS, Crosby ME, Glazer, PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull HJ, Lombardo MJ, Rosenberg SM. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase I.V. dependence. Proc Natl Acad Sci USA. 2001;98:8334–8341. doi: 10.1073/pnas.151009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Romesberg FE. Controlling mutation: intervening in evolution as a therapeutic strategy. Crit Rev Biochem Mol Biol. 2007;42:341–354. doi: 10.1080/10409230701597741. [DOI] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M. Global regulators orchestrate group II intron retromobility. Mol Cell. 2009;34:250–256. doi: 10.1016/j.molcel.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio. 2011;2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Rosche WA. Increased episomal replication accounts for the high rate of adaptive mutation in recD mutants of Escherichia coli. Genetics. 1999;152:15–30. doi: 10.1093/genetics/152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc Natl Acad Sci USA. 1995;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch RL, Su Y, Thornton PC, Gibson JL, Rosenberg SM, Hastings PJ. Separate DNA Pol II- and Pol IV-dependent pathways of stress-induced mutation during double-strand-break repair in Escherichia coli are controlled by RpoS. J Bacteriol. 2010;192:4694–4700. doi: 10.1128/JB.00570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Do R, Yamada M, Friedberg E, Hastings P, Nohmi T, Rosenberg S. DinB up-regulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JL, Lombardo MJ, Thornton PC, Hu KH, Galhardo RS, Beadle B, Habib A, Magner DB, Frost LS, Herman C, Hastings PJ, Rosenberg SM. The sigma(e) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol Microbiol. 2010;77:415–430. doi: 10.1111/j.1365-2958.2010.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy VG, Gizatullin FS, Fox MS. Some features of the mutability of bacteria during nonlethal selection. Genetics. 2000;154:49–59. doi: 10.1093/genetics/154.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez JM, Blazquez J, Baquero F, Martinez JL. H-NS and RpoS regulate emergence of Lac Ara+ mutants of Escherichia coli MCS2. J Bacteriol. 1997;179:4620–4622. doi: 10.1128/jb.179.14.4620-4622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner-Russo LM, Lombardo M-J, Ponder RG, Rosenberg SM. The TGV transgenic vectors for single copy gene expression in the E. coli chromosome. Gene. 2001;273:97–104. doi: 10.1016/s0378-1119(01)00565-0. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Hersh MN, Thornton PC, Fonville NC, Slack A, Frisch RL, Ray MP, Harris RS, Leal SM, Rosenberg SM. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS One. 2010;5:e10862. doi: 10.1371/journal.pone.0010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2004;2:e399. doi: 10.1371/journal.pbio.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves H, Horak R, Kivisaar M. Involvement of sigma(s) in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J Bacteriol. 2001;183:5445–5448. doi: 10.1128/JB.183.18.5445-5448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S, Hughes D, Andersson DI. Effect of translesion DNA polymerases, endonucleases and RpoS on mutation rates in Salmonella typhimurium. Genetics. 2010;185:783–795. doi: 10.1534/genetics.110.116376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR. Multiple pathways of selected gene amplification during adaptive mutation. Proc Natl Acad Sci USA. 2006;103:17319–17324. doi: 10.1073/pnas.0608309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- Lamrani S, Ranquet C, Gama MJ, Nakai H, Shapiro JA, Toussaint A, Maenhaut-Michel G. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Mol Microbiol. 1999;32:327–343. doi: 10.1046/j.1365-2958.1999.01352.x. [DOI] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IS, Lin J, Hall HK, Bearson B, Foster JW. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol. 1995;17:155–167. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Cambridge: Harvard University Press; 1985. [Google Scholar]
- McKenzie GJ, Harris RS, Lee PL, Rosenberg SM. The SOS response regulates adaptive mutation. Proc Natl Acad Sci USA. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Mittelman D, Wilson JH. Stress, genomes, and evolution. Cell Stress Chaperones. 2010;15:463–466. doi: 10.1007/s12192-010-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Szigety SK, Rosenberg SM. Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino JF, Galhardo RS, Morales LD, Rosenberg SM. Stress-induced beta-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J Bacteriol. 2009;191:5881–5889. doi: 10.1128/JB.00732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Prieto AI, Ramos-Morales F, Casadesus J. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics. 2006;174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Soto S, Roth JR. Effect of growth under selection on appearance of chromosomal mutations in Salmonella enterica. Genetics. 2011;189:37–53. doi: 10.1534/genetics.111.130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella JP, Park PU, Fox MS. Adaptive mutation in Escherichia coli: a role for conjugation. Science. 1995;268:418–420. doi: 10.1126/science.7716545. [DOI] [PubMed] [Google Scholar]
- Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- Robleto EA, Yasbin R, Ross C, Pedraza-Reyes M. Stationary phase mutagenesis in B. subtilis: a paradigm to study genetic diversity programs in cells under stress. Crit Rev Biochem Mol Biol. 2007;42:327–339. doi: 10.1080/10409230701597717. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM. Stress-induced loss of heterozygosity in Candida: a possible missing link in the ability to evolve. MBio. 2011;2:e00200-11. doi: 10.1128/mBio.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- Saumaa S, Tover A, Kasak L, Kivisaar M. Different spectra of stationary-phase mutations in early-arising versus late-arising mutants of Pseudomonas putida: involvement of the DNA repair enzyme MutY and the stationary-phase sigma factor repos. J Bacteriol. 2002;184:6957–6965. doi: 10.1128/JB.184.24.6957-6965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc Natl Acad Sci USA. 2011;108:13659–13664. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta ES, Bunny KL, Kugelberg E, Kofoid E, Andersson DI, Roth JR. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci USA. 2003;100:12847–12852. doi: 10.1073/pnas.1735464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta ES, Harold J, Andersson DI, Roth JR. The effect of genomic position on reversion of a lac frameshift mutation (lacIZ33) during non-lethal selection (adaptive mutation) Mol Microbiol. 2002;44:1017–1032. doi: 10.1046/j.1365-2958.2002.02934.x. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Craig NL. Gain-of-function mutations in Tnsc, an Atp-dependent transposition protein that activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords WE, Cannon BM, Benjamin WH., Jr Avirulence of LT2 strains of Salmonella typhimurium results from a defective RpoS gene. Infect Immun. 1997;65:2451–2453. doi: 10.1128/iai.65.6.2451-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Matic I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE, Longacre A, Reimers JM. Hypermutation in derepressed operons of Escherichia coli K12. Proc Natl Acad Sci USA. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]