Abstract
The antibiotic tolerances of bacterial persisters have been attributed to transient dormancy. While persisters have been observed to be growth inhibited prior to antibiotic exposure, we sought to determine whether such a trait was essential to the phenotype. Furthermore, we sought to provide direct experimental evidence of the persister metabolic state so as to determine whether the common assumption of metabolic inactivity was valid. Using fluorescence-activated cell sorting (FACS), a fluorescent indicator of cell division, a fluorescent measure of metabolic activity, and persistence assays, we found that bacteria that are rapidly growing prior to antibiotic exposure can give rise to persisters and that a lack of replication or low metabolic activity prior to antibiotic treatment simply increases the likelihood that a cell is a persister. Interestingly, a lack of significant growth or metabolic activity does not guarantee persistence, as the majority of even “dormant” subpopulations (>99%) were not persisters. These data suggest that persistence is far more complex than dormancy and point to additional characteristics needed to define the persister phenotype.
INTRODUCTION
Bacterial persisters are rare phenotypic variants with the ability to tolerate extraordinary levels of antibiotics (1). This tolerance has commonly been attributed to transient growth inhibition and the resulting inactivity of essential cell functions (2). Convincing evidence in support of this model was obtained by time-lapse fluorescence microscopy, as growth-inhibited cells from high-persister mutants survived prolonged antibiotic treatment and resumed normal replication upon removal of the antibiotic (3). That study, by Balaban and colleagues, undoubtedly demonstrated that persisters can originate from growth-inhibited cells (3), and dormancy has since been deemed a trait of persisters, permeating to aspects of their physiology, such as metabolism, for which only indirect evidence of reduced activity exists (4–6). However, it remains unclear to what extent dormancy is a characteristic of persisters or if persister antibiotic tolerances require growth inhibition. Two notable studies used single-cell growth reporters to study persistence, but neither presented data that answer the question of whether persistence requires dormancy (5, 7). Shah and colleagues segregated an exponential-phase population by using an rRNA reporter and found antibiotic-tolerant cells in the subpopulation that was growth inhibited as well as the subpopulation that was growing normally (5). Unfortunately, the antibiotic treatment used to enumerate tolerant cells was performed in a nonnutritive buffer and yielded persister levels (∼1:50) that were over 2 orders of magnitude higher than those routinely measured in exponential-phase Escherichia coli cultures (<1:5,000) (8). Such a discrepancy suggests that the tolerant cells identified were not representative of normal persister physiology. Roostalu and colleagues segregated growing and nongrowing cells by using a cell division reporter and found that 0.2 to 10% of the nongrowing cells were tolerant to ampicillin treatment (7). Though this result suggested that only a fraction of the nongrowing cells were tolerant to ampicillin, the necessity of growth inhibition for persistence was not queried, since the persistence of the growing subpopulation was not measured.
Recently, persister formation in response to a variety stresses was observed, demonstrating that nonpersister cells can be transformed into the persistent state through the action of signal transduction cascades (9–13, 37). While supportive of the hypothesis that persister populations are heterogeneous (14), these studies do not provide evidence regarding the growth state of the persisters formed and whether growth inhibition prior to antibiotic challenge is a requirement of persistence. Dorr and colleagues demonstrated that ciprofloxacin can induce persistence in a subpopulation of exponential-phase cells through the action of the SOS response, but they did not determine whether those cells that became persisters were growth inhibited prior to antibiotic exposure (9, 10). Moker and colleagues found that a quorum sensing molecule increased persistence in Pseudomonas aeruginosa, but they also did not establish the growth state of persisters prior to antibiotic exposure (13). Vega and colleagues found that indole induces persister formation, and they elegantly demonstrated with a microfluidic device that the strength of indole sensing is predictive of tolerance to 1 h of ampicillin treatment (11). However, the tolerant subpopulation in the microfluidic device was much higher than that obtained when exponential-phase cells were exposed to indole (20% compared to 1%), suggesting that tolerance measured in the device might not be translatable to normal culture conditions. Wu and colleagues found that paraquat at a concentration close to that which completely inhibits growth induces persister formation, but the growth characteristics of the culture and persister subfractions were not reported (12). Interestingly, despite the growing body of literature on persister formation and physiology (14–16), we cannot definitively say whether growth inhibition is necessary or sufficient for persistence. Analogously, and to a greater extent, we do not know whether persister antibiotic tolerances require metabolic dormancy. All insights into persister metabolism have been obtained indirectly from gene expression of persister-enriched samples (4–6) or potentiation of aminoglycoside activity (17).
Here we answer two fundamental questions about bacterial persistence. (i) Can bacteria that are normally replicating prior to antibiotic treatment be persisters? (ii) Are persisters metabolically dormant prior to antibiotic treatment, as most research assumes (3, 5, 7, 18), or can they exhibit a range of metabolic activities? To answer these questions, we employed fluorescence-activated cell sorting (FACS), a fluorescent indicator of cell division (7), a fluorescent measure of metabolic activity (19, 20), and persistence assays to rigorously quantify the metabolic and cell division distributions of persisters within an exponentially growing E. coli culture. Using the cell division reporter, we found that the nongrowing subpopulation was far more enriched with persisters (∼1%) than the growing subpopulation (∼0.01%), but due to the relative abundances of the growing and nongrowing subpopulations, ∼20% of all persisters originated from growing cells. Surprisingly, even the most rapidly growing subpopulation gave rise to persisters, demonstrating that growth inhibition prior to antibiotic exposure is not required for persistence. Using a fluorescent measure of metabolic activity, redox sensor green (RSG), we found that within an exponential-phase E. coli population, cells with low reductase activity were ∼40 times more likely to be persisters than cells with high reductase activity. These are the first direct measurements of metabolic activity in persisters, and interestingly, persisters appear to display a range of metabolic activities. We subsequently performed a two-dimensional (2D) FACS experiment (red channel, cell division; and green channel, metabolism) and found that persisters were enriched in the subpopulation with low reductase activity due to the prevalence of persisters in the nongrowing subpopulation, whose metabolic activity was significantly less than that of the growing subpopulation. Overall, the experimental evidence presented in this study supports the emerging hypothesis that persister subpopulations are highly diverse (14–16) and provides a more holistic view of the persistence phenomenon, one in which inhibition of replication or metabolism enhances the likelihood of a cell to be a persister but in itself cannot adequately explain the phenotype.
MATERIALS AND METHODS
Bacterial strains.
All strains were derived from E. coli MG1655. Strain MO001 contained a chromosomally integrated lacIq promoter in place of the lacI promoter and a chromosomally integrated T5p-mCherry cassette in place of lacZYA. This was done in order to eliminate plasmid copy number as a variable. The T5 promoter (T5p) is a strong, IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter under the control of two lac operator sites (21). Using the forward and reverse primers indicated in Table S1 in the supplemental material, the T5p was amplified from the pQE-80L plasmid (Qiagen, Valencia, CA). Both lacIqp and T5p-mCherry were integrated into the chromosome by using the method of Datsenko and Wanner (22). All primers used, as well as a description of the cloning steps, are presented in Table S1. All mutations were confirmed using PCR and/or DNA sequencing (Genewiz, South Plainfield, NJ).
Chemicals, media, and growth conditions.
All chemicals, unless noted below, were purchased from Fisher Scientific or Sigma-Aldrich. RSG and a Live/Dead (Syto9/PI) staining kit were purchased from Life Technologies, Invitrogen (Grand Island, NY), and IPTG was purchased from Gold Biotechnology (St. Louis, MO). LB medium (10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl) and LB-agar plates (LB plus 15 g/liter agar) were used for planktonic growth and enumeration of CFU, respectively. Antibiotics were used at the following concentrations for selection or translational inhibition: 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, and 100 μg/ml ampicillin. For persister assays, 200 μg/ml ampicillin (23) and 5 μg/ml ofloxacin (8) were used. Unless otherwise noted, overnight cultures were prepared by culturing cells from a 25% glycerol, −80°C stock in 2 ml LB medium at 37°C with shaking (250 rpm) for 24 h. Overnight cultures were then diluted 1,000-fold in 50 ml of fresh LB medium in 500-ml baffled flasks and incubated for 2.5 h at 37°C and 250 rpm, at which time an optical density at 600 nm (OD600) of ∼0.1 was achieved. CFU were enumerated by washing and diluting samples in phosphate-buffered saline (PBS), plating them on LB-agar plates, and incubating them at 37°C for 16 h. MICs of antibiotics for both wild-type MG1655 and MO001 at exponential phase were determined by serial 2-fold dilution of antibiotics in LB broth (24). MIC ranges for both strains were found to be 1.5 to 3 μg ampicillin/ml and 0.075 to 0.15 μg ofloxacin/ml.
Staining with redox sensor green.
Cell staining was performed according to the manufacturer's instructions, with some modifications. Briefly, cells were cultured as described above, and after 2.5 h at 37°C and 250 rpm, 1 ml of exponential-phase culture was stained with 1 μM RSG and incubated in the dark at room temperature for approximately 30 min before sorting. As controls, 1-ml samples were first incubated with carbonyl cyanide m-chlorophenylhydrazone (CCCP) or potassium cyanide (KCN) at 10 μM or 1 mM, respectively, for 5 min prior to addition of RSG. To analyze the effect of RSG on cell viability, CFU in samples were measured before staining and 30 min after staining.
Single-cell division assay.
MO001 cells were cultured as described above, except that 1 mM IPTG was included overnight to express mCherry, and following the overnight incubation, IPTG was removed by centrifugation (3 min at 15,000 rpm), removal of the supernatant, and resuspension of the culture in fresh LB. Washed cells were then diluted 1,000-fold in 50 ml fresh LB and cultured as described above. One milliliter of the culture was analyzed by flow cytometry to sort the growing and nongrowing cells. To compare MO001 background fluorescence to that of the parent strain (without mCherry), MO001 was incubated without IPTG during overnight growth and then inoculated into fresh medium with or without IPTG, and fluorescence intensities of exponential-phase cultures were measured by flow cytometry (see Fig. S1 in the supplemental material). To verify that mCherry was not degraded during the time frame of the experiment, the washed overnight culture was diluted in LB with 50 μg/ml chloramphenicol, and 1-ml samples were analyzed by flow cytometry (see Fig. S2).
Microscopy imaging.
Phase-contrast and fluorescence images of induced MO001 were taken by a Nikon Eclipse 90i microscope equipped with a 100×/1.40-numerical-aperture (NA) phase objective, a Q Imaging Rolera-XR camera, and NIS Elements software. Cells were stabilized on 1% agarose pads during the imaging process. To do this, 200 μl of fully dissolved warm agarose was transferred to the middle of a clean microscope slide, and then another slide was placed on top of the agarose to attain a flat surface (air bubbles were avoided). After the agarose solidified, the upper glass slide was carefully removed, and 2.5 μl of cell culture was loaded on the solid medium and covered with a coverslip to be analyzed with the microscope.
Flow cytometry analysis.
All samples, including controls, were analyzed by an LSRII flow cytometer (BD Biosciences, San Jose, CA). Microorganisms were identified using forward and side scatter parameters (FSC and SSC, respectively). Stained bacteria were assayed with a laser emitting at 488 nm for RSG and 561 nm for mCherry; fluorescence was collected using green and red fluorescence filters (525/50- and 610/20-nm-band-pass filters, respectively). Data were acquired and analyzed using FACSDiVa software (BD Biosciences, San Jose, CA).
Cell sorting.
Cells were sorted using either a FACSVantage SE w/DiVa (BD Biosciences, San Jose, CA) cell sorter at 16 lb/in2 with a 70-μm nozzle or a Reflection (Sony-iCyt Mission Technology, Champaign, IL) cell sorter at 30 psi with a 70-μm nozzle. Microorganisms were determined using forward and side scatter parameters (FSC and SSC), and physiologically distinct subpopulations were identified by measuring green and red fluorescence (488-nm excitation with 530/30-nm-band-pass filter and 561-nm excitation with 615/30-nm-band-pass filter, respectively) and by running unstained samples, RSG-only-stained cells, mCherry-expressing cells, and mCherry-expressing cells stained with RSG. Cells were sorted using sterile 1× PBS as sheath fluid in the sorter.
As a test to quantify the number of nongrowing cells (C* region in Fig. 1B) that were improperly sorted into the fastest-growing quantile (A* region in Fig. 1B), approximately 5 × 105 cells were sorted from the A* or C* region. The sorter sample line was then immediately washed first with 10% bleach for 5 min and then with PBS for 5 min to remove any cell remnants from the previous sample. The sorted cells from A* and C* were reanalyzed separately by the same sorter to assess the proportion of persisters in the fastest-growing subpopulation that may have arisen from nongrowing cells (see Fig. S3 in the supplemental material). The sorter was washed as described above between analyzing A* and C*.
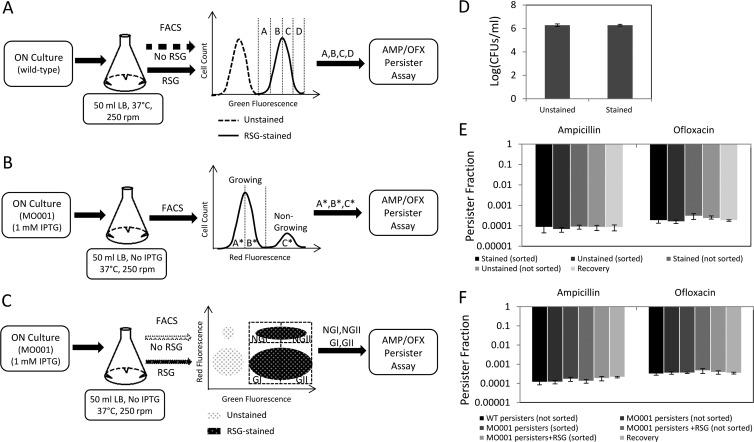
Fig 1.
FACS methods and control experiments. (A to C) Exponential-phase cells were sorted from the indicated regions (gates) in order to quantify the metabolic and cell division distributions of persisters. Gates were determined by using a fluorescent measure of metabolic activity (RSG) (A), a cell division reporter (mCherry with an IPTG-inducible promoter) (B), or both (C). ON, overnight. (D) RSG staining did not affect cell viability (P = 0.94 by the t test). (E) RSG staining, cell sorting, and segregation did not change the persister levels of exponential-phase E. coli cultures sorted as illustrated in panel A (P = 0.97 for the ampicillin group and P = 0.52 for the ofloxacin group, using ANOVA). (F) RSG staining, cell sorting, and segregation did not alter persister levels of mCherry-expressing cells sorted as in panel C (P = 0.31 for the ampicillin group and P = 0.75 for the ofloxacin group, using ANOVA). Recovery is the calculated frequency of persisters in the total population, based on the persister frequencies measured from the segregated quantiles (see Materials and Methods).
Persister assays.
After determining the fluorescence quantiles to be sorted (Fig. 1A to C), approximately 1 × 106 cells from each subpopulation were collected in 1 ml of PBS. Cells were sorted at room temperature, and each sorting procedure lasted 15 min or less, depending on the percentage of the population corresponding to the region to be sorted. The collected cells were then mixed 1:1 with a rich medium (2× tryptone, 2× yeast extract, and 1× NaCl) to produce a medium similar to LB. One milliliter of this culture, containing 5 × 105 cells, was immediately treated with either ampicillin (200 μg/ml) or ofloxacin (5 μg/ml). Cells were then incubated in a shaker at 37°C and 250 rpm for 5 h. Samples at designated time points were collected by centrifugation for 3 min at 15,000 rpm, washed with PBS twice, and resuspended in 100 μl of PBS. Next, 10 μl of the sample was serially diluted in PBS and spotted on LB agar to measure the CFU. The remaining 90-μl sample was also plated on LB agar in case the CFU from the 10-μl sample might be under the limit of detection. To analyze the effect of RSG on persister levels, unstained cells (negative control) were sorted and treated with antibiotics as described above to enumerate the level of persisters, which was compared to the persister levels obtained from RSG-stained samples. To determine the effect of flow through the sorter on persister levels, cells that were not sorted were diluted in 1 ml fresh LB to obtain approximately 5 × 105 cells and then were treated with antibiotics, and CFU were measured at the indicated time points.
To quantitatively estimate the percentage of nongrowing cells that started to grow during the antibiotic treatment period, the sorted cells from the C* region (Fig. 1B) were treated with or without antibiotics for 5 h in LB as described above, and the number of cells retaining high mCherry levels was quantified by LSRII flow cytometry with the use of fluorescent counting particles (Spherotech Inc., Lake Forest, IL). To determine the number of dead cells in the nongrowing subpopulation, stationary-phase cultures diluted 1,000-fold in 1 ml 0.85% NaCl buffer were stained with Syto9 and propidium iodide (PI) simultaneously at concentrations of 5 μM and 30 μM, respectively, and then incubated at room temperature for approximately 15 min before flow cytometry analysis. This method enumerates dead cells in the nongrowing subpopulation, under the assumption that dead cells within this subpopulation arose from 24-h stationary-phase inoculums. As a control, stationary-phase cells were incubated in 1 ml of 70% ethanol solution for 1 h.
The dilution/growth experiment described by Keren et al. (8) was applied in order to demonstrate that the persister levels measured with the FACS procedure were identical to those measured on the bench. Cells with mCherry protein from overnight cultures were diluted 1:1,000 in 50 ml fresh medium in a 500-ml baffled flask without inducer and cultured at 37°C and 250 rpm. When the cell culture reached an OD600 of 0.1, the cells were diluted 1:50 in fresh medium and cultured in the same way until the OD600 reached 0.1 again. This dilution/growth cycle was repeated twice, thus resulting in three rounds in total (R0, R1, and R2). At the end of each round, cells were analyzed by FACS and sorted to determine the persister levels as described above.
The frequency of persisters (f) was calculated as the ratio of the number of persisters in a sample to the initial number of sorted cells before antibiotic treatment. Persister frequencies of all sorted regions are provided in Table S2 in the supplemental material. We defined recovery (R) as follows: R = pAfA + pBfB + pCfC + pDfD, where pA is the proportion of the total population in the A quantile and fA is the frequency of persisters in the A quantile (Fig. 1A). Recovery is the frequency of persisters in the total population, as calculated from the segregated quantiles, and thus was used as an internal consistency check, since it should equal the frequency of persisters obtained from a nonsegregated sample. The persister fraction from a region (such as region A) is the ratio of persisters in that region to the total number of persisters in the whole population, which was calculated as pAfA/R. We note that the sum of persister fractions from all quantiles equals 1. To illustrate, persister frequencies of the A*, B*, and C* regions (Fig. 1B; see Fig. 3A) were found to be 4.47 × 10−5, 5.8 × 10−5, and 4.27 × 10−3 (see Fig. 3C), respectively. Since pA* = 0.48, pB* = 0.48, and pC* = 0.04 (see Fig. 3A), the recovery is calculated as follows: R = 4.47 × 10−5 × 0.48 + 5.8 × 10−5 × 0.48 + 4.27 × 10−3 ×0.04 = 2.2 × 10−4, which also corresponds to the frequency of persisters in the total population. The persister fraction of A* was calculated as 4.47 × 10−5 × 0.48/2.2 × 10−4 = 0.1; similarly, the B* and C* fractions were found to be 0.12 and 0.78, respectively (see Fig. 3D). All experiments were repeated 3 times in this study. One-way analysis of variance (ANOVA) was used to test the null hypothesis that the population means for all conditions, including the treatment and control groups, were the same. Pairwise comparisons were performed using the two-tailed t test. A significance P value threshold of 0.05 was selected. Data points in figures are average values for experimental repeats. Error bars represent standard errors.
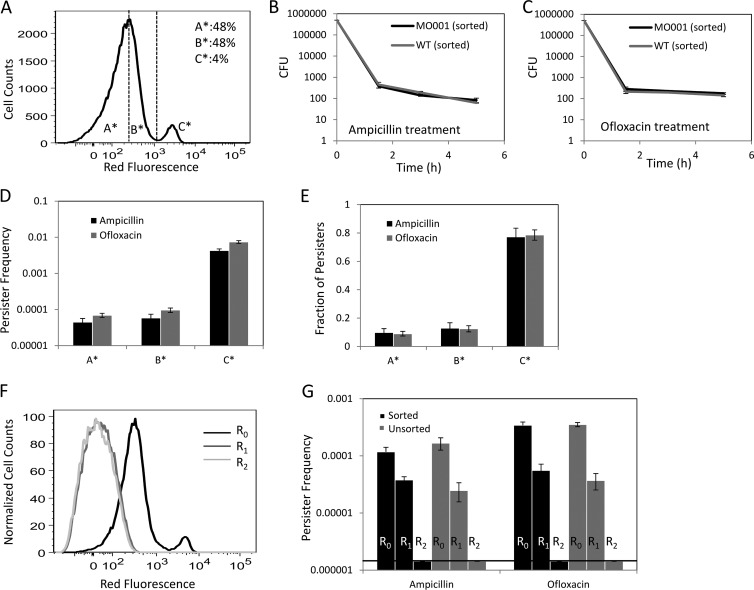
Fig 3.
Cell division properties of persisters. (A) The regions to be sorted (A*, B*, and C*) were determined based on the proliferation rates of exponential-phase cells. A* and B* comprised approximately 96% of the population (48% each), whereas C* contained approximately 4% of all cells. (B and C) CFU were counted at the indicated time points during the ampicillin or ofloxacin treatment of FACS-sorted or nonsorted cells. MO001 had statistically indistinguishable persister levels compared to the wild type (P = 0.38 for ampicillin persisters at the 5-h time point and 0.30 for ofloxacin persisters at the 5-h time point, by the t test). (D and E) Persister frequencies and fractions of A*, B*, and C* were determined 5 h after antibiotic treatment. The frequency is the ratio of persisters to the initial number of FACS-sorted cells. The fraction of persisters of a region such as A* is the ratio of persisters in A* to the total number of persisters in the culture. (F) Repeated inoculation into fresh medium eliminated the nongrowing subpopulation. Overnight cultures of cells with mCherry protein were inoculated (1:1,000) into fresh medium without inducer and cultured until the OD600 reached 0.1. The cells were then diluted 1:50 in fresh medium and cultured identically until the OD600 again reached 0.1, and this cycle was repeated twice, resulting in three rounds in total (R0, R1, and R2). (G) At the end of each round, cells were sorted by FACS to determine the persister levels. No significant differences were observed between bench-top- and FACS-sorted samples (P = 0.37 for R0-ampicillin samples, 0.88 for R0-ofloxacin samples, 0.30 for R1-ampicillin samples, and 0.42 for R1-ofloxacin samples, using the t test). Note that P values for R2 samples could not be determined because the number of persisters was found to be under the limit of detection for these samples.
RESULTS
FACS methods to quantify persister metabolic and cell division distributions.
To determine whether persisters needed to be growth inhibited or metabolically inactive prior to antibiotic treatment, we required a method to quantify persister phenotype distributions. Previous studies have monitored persister growth rates in cultures of high-persistence (hipA7 and hipQ) mutants by using fluorescence microscopy (3); however, the low abundance of persisters in wild-type E. coli populations necessitated the use of a more rapid screening technique. FACS is a rapid, single-cell, quantitative screening method that has been used to study different aspects of the persistence phenotype (5, 7, 11, 25). Here we used FACS to quantify growth rate and metabolic activity distributions for persisters from exponential-phase E. coli cultures. In brief, FACS was used to segregate cultures into quantiles based on fluorescence signals (Fig. 1A to C), and each quantile was then treated with an antibiotic (ampicillin or ofloxacin) to enumerate the number of persisters present. To demonstrate that the FACS procedure did not alter persistence within the culture, we verified that fluorescence labeling (e.g., metabolic staining and fluorescent protein expression), flow cytometry, and segregation all did not change the level of persisters in the population (Fig. 1D to F).
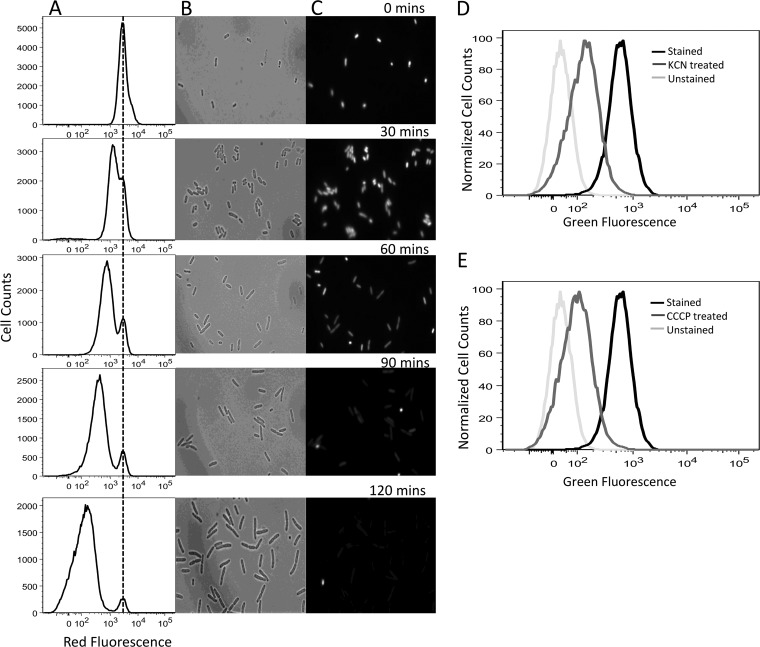
To monitor growth rate, we adapted the methodology of Roostalu and colleagues (7), which tracks cell division by dilution of a fluorescent protein. To generate a strain whose cell division could be monitored by red fluorescence, we knocked the mCherry gene, under the control of a strong, synthetic, IPTG-inducible promoter (T5p) (21), into the chromosome of an E. coli strain carrying a chromosomally integrated lacIq promoter mutation (MO001) (26). Background fluorescence from MO001 (without IPTG) was comparable to that of the parent strain without mCherry, whereas full induction produced fluorescence that was orders of magnitude above the background, providing us with a large dynamic range (see Fig. S1 in the supplemental material). To verify that this method monitored cell division, mCherry expression was induced during the overnight culture, and cells were washed to remove the inducer and inoculated into fresh medium without inducer. The dilution of mCherry protein within the cells due to cell proliferation was monitored using flow cytometry and fluorescence microscopy. As depicted in Fig. 2A to C, all cells started with high red fluorescence, and as cells divided, the red fluorescence of the population declined, except for a small subpopulation whose fluorescence remained constant due to a lack of division. To verify that mCherry was not degraded during the time frame of the experiments, cells were incubated in medium with chloramphenicol, an inhibitor of protein synthesis, and were demonstrated to retain their red fluorescence throughout the time course (see Fig. S2). These results demonstrate that this FACS method monitors cell division at the single-cell level.
Fig 2.
Cell division and metabolic activity reporters. Division at the single-cell level was monitored by flow cytometry and microscopy. After accumulation of mCherry protein during overnight culture, cells were washed to remove the inducer and inoculated into fresh medium without inducer. When the cells started to divide, the red fluorescence of the population decreased, except for a small subpopulation whose fluorescence remained constant (nongrowing cells). Panel A shows the flow cytometry data, whereas panels B and C show phase-contrast and fluorescence images taken by microscopy, respectively, at the indicated time points during exponential-phase growth. (D and E) Cellular metabolic activity was characterized by RSG staining. RSG produces a stable green fluorescence signal when reduced by bacterial reductases. The fluorescence signals were reduced when the cells were treated with KCN and CCCP, which block respiration and deplete the proton motive force, respectively.
To study single-cell metabolic activity, we employed RSG, a fluorogenic redox indicator that yields green fluorescence when reduced by bacterial reductases (19, 27, 28). To ensure that the measured fluorescence reported on metabolic activity, we used the chemical inhibitor KCN to block respiration and CCCP to deplete the proton motive force. As depicted in Fig. 2D and E, both CCCP and KCN significantly reduced staining with RSG, demonstrating that RSG is a robust indicator of bacterial metabolism (P = 0.024 and 0.030 for mean fluorescence values of CCCP- and KCN-treated samples, respectively, compared with that of RSG-only-stained samples, using the t test).
Persisters can originate from the fastest-growing subpopulation of cells.
To separate the nongrowing cells from the growing subpopulation in exponential-phase cultures, mCherry expression was induced from MO001 during overnight culture, and cells were washed to remove the inducer and inoculated into fresh medium without inducer. At 2.5 h postinoculation (OD600 = 0.1), the nongrowing population constituted approximately 4% of all cells (Fig. 3A). To enumerate persisters, FACS-sorted samples were treated with ampicillin or ofloxacin and CFU were monitored as a function of time. As depicted in Fig. 3B and C, a rapid killing regimen, representing the death of normal cells, was followed by a slower killing regimen, indicating the presence of persisters. This biphasic kill curve verified that 5 h of antibiotic treatment under the conditions described here was sufficient to quantify persister levels (15). Figure 3B and C also demonstrate that MO001 had persister levels indistinguishable from those of the wild type. To determine whether the nongrowing subpopulation was enriched with persisters, we sorted the population into three regions based on red fluorescence and treated the samples with ampicillin or ofloxacin for 5 h. While the majority of persisters were found to be within the nongrowing subpopulation (77.26% ± 6.11% of ampicillin persisters and 78.55% ± 3.61% of ofloxacin persisters), we also measured persisters within the growing subpopulation (Fig. 3D and E). Surprisingly, 22.73% ± 6.42% of ampicillin persisters and 21.44% ± 3.79% of ofloxacin persisters were within the growing subpopulation. We note that a previous study has shown that persisters can replicate at rates that are 10-fold lower than those of normal cells (3) but that this is the first report of persisters being found in the fastest-dividing subpopulation (A*) of an exponentially growing culture (doubling time of 25.41 ± 0.38 min) (see Fig. S4 in the supplemental material).
To confirm that persisters can arise from the growing subpopulation, we sought to provide further evidence in support of our FACS results. Specifically, we sought to (i) quantify the precision of our FACS experiments by measuring the number of nongrowing cells that were improperly sorted into the fastest-growing quantile and (ii) demonstrate further that our FACS technique does not generate persisters (in addition to the controls depicted in Fig. 1D to F). Measuring the precision of our FACS procedure allowed calculation of the proportion of persisters in the fastest-growing subpopulation that may have arisen from nongrowing cells and was an important control given the ∼100-fold difference in persister frequencies between the nongrowing and growing subpopulations (Fig. 3D). To generate these data, we performed a tandem FACS experiment where cells were sorted and then immediately reanalyzed on the same cell sorter (see Materials and Methods). We determined that <0.2% of cells in the fastest-growing subpopulation (A*) were subsequently sorted as nongrowing cells (C*) (see Fig. S3 in the supplemental material). Given the frequencies of persisters in A* and C*, we calculated that >80% of persisters measured in A* were normally replicating cells. Although the cells were replicating normally prior to sorting, the possibility remained that the FACS procedure stimulated some cells to become persisters. As depicted in Fig. 1D to F, we performed a control to test this for the entire population and showed that the persister levels were indistinguishable with and without sorting (as analyzed by ANOVA). However, to lend further experimental support for our FACS procedure, we sought to execute the same protocol on a population of exponential-phase cells devoid of persisters and to show that persisters were unequivocally not generated by our method. To do this, we sought to demonstrate that our results were in agreement with those of Keren and colleagues (8), who found that continued culturing by dilution in fresh medium eliminates persisters from a culture. We repeated this experiment under normal assay conditions (bench top) as well as with our FACS method (see Materials and Methods) and found the results between the two to be indistinguishable (Fig. 3G). Repeated inoculation into fresh medium eliminated the nongrowing subpopulation (Fig. 3F), as well as all persisters from the growing subpopulation (Fig. 3G). These results are consistent with those of Keren and colleagues and provide further evidence that our FACS method does not alter persister levels. Interestingly, normally dividing cells lose the ability to become persisters after continued culturing, suggesting that aspects of their physiology that confer persistence are lost by continued rounds of replication. We note that in order for cells to be within the A* region of Fig. 3A, they had to divide, on average, >3 times after inoculation from stationary phase, whereas cells present in the R2 culture, which did not contain persisters, divided, on average, >14 times. Altogether, these data demonstrate that while nongrowing cells are more likely to be persisters than growing cells, persisters can be found in the normally dividing subpopulation, and that a lack of growth does not confer persistence, since 99.57% ± 0.048% of nongrowing cells are not ampicillin persisters, and similarly, 99.26% ± 0.064% of nongrowing cells are not ofloxacin persisters.
Persisters exhibit heterogeneous metabolic activity.
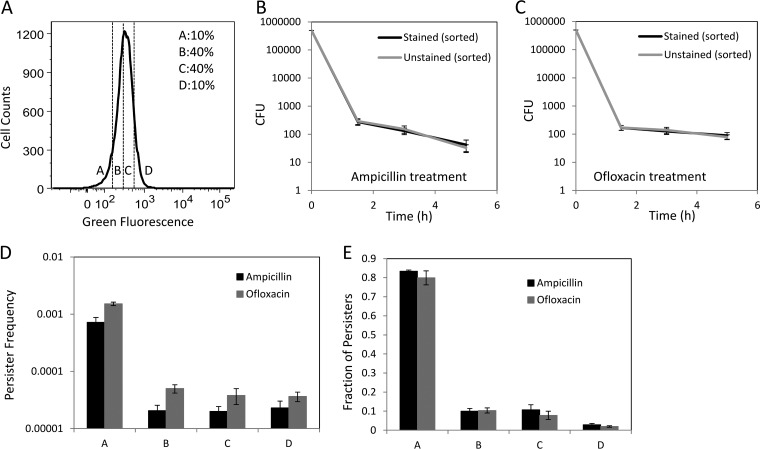
Cells from early exponential phase (OD600 = 0.1) were incubated with RSG for 30 min and analyzed by flow cytometry (see Materials and Methods). As expected, staining of cells with RSG exhibited increased green fluorescence compared to that of the unstained, CCCP-treated, and KCN-treated controls (Fig. 2D and E). To determine where persisters existed within the phenotypic distribution of reductase activity, we sorted cells from four different regions of the distribution, as indicated in Fig. 4A. The A region contained cells with metabolic activity in the lowest ∼10% of the population, the B region contained cells with low-intermediate metabolic activity (∼40% of the population), the C region contained cells with intermediate-high metabolic activity (∼40% of the population), and the D region contained cells with metabolic activity in the highest ∼10% of the population. To enumerate persisters, FACS-sorted samples were treated with ampicillin or ofloxacin and CFU were monitored. The biphasic kill curves verified that 5 h of antibiotic treatment under the conditions described here was sufficient to quantify persister levels (Fig. 4B and C). Furthermore, Fig. 4B and C demonstrate that RSG staining had no effect on the persistence of the population. The frequencies of persisters within these populations are presented in Fig. 4D, whereas the fraction of persisters from the total population in each quantile is provided in Fig. 4E. We found that 83.33% ± 0.67% and 79.92% ± 3.62% of all ampicillin and ofloxacin persisters, respectively, were present in region A of the reductase distribution, demonstrating that 4 of 5 persisters contained low reductase activity (Fig. 4E). This is the first direct measurement of metabolic activity in persisters, and while the data are supportive of the notion that persisters are largely metabolically dormant, the regions with higher reductase activity (B, C, and D) did contain a nonnegligible portion of persisters: ∼20%. These data demonstrate that persisters can be metabolically active and retain their antibiotic tolerances, suggesting that metabolic dormancy prior to antibiotic treatment is not a requirement of persistence. In addition, the frequency of persisters in the region of low reductase activity (A) was only 0.07% ± 0.01% for ampicillin and 0.15% ± 0.01% for ofloxacin, suggesting that metabolic dormancy is not sufficient to confer persistence.
Fig 4.
Metabolic activity of persisters. (B and C) CFU were determined at the indicated time points during the antibiotic treatment. RSG staining did not affect the persister levels (P = 0.58 for ampicillin persisters at the 5-h time point and 0.52 for ofloxacin persisters at the 5-h time point, using the t test). Persister frequencies (D) as well as fractions (E) were quantified after 5 h of antibiotic treatment of FACS-sorted cells from regions A, B, C, and D, based on RSG staining (A).
Most persisters exhibit low metabolic activity due to their nonreplicating state.
Since low metabolic activity and lack of replication are predictive of persister enrichment, we sought to determine whether these results were coupled or could be used in conjunction to further increase the persister frequency in a sample. To do this, we performed a 2D FACS experiment in which growing and nongrowing cells could be discerned using mCherry (Fig. 5A), and metabolic activity was measured using RSG. We observed that the nongrowing subpopulation fluoresced far less on the green channel (RSG) than the growing subpopulation (P = 0.022 by the t test), suggesting that nongrowing cells generally harbored lower reductase activity than growing cells (Fig. 5B). After verifying that RSG-stained, mCherry-expressing cells did not have a changed persistence phenotype (Fig. 5C and D), we found that the persister enrichment associated with low metabolic activity was dependent on replication (Fig. 5E and F). When the growing and nongrowing subpopulations were split in half based on metabolic activity (GI and GII populations and NGI and NGII populations), indistinguishable persister frequencies were obtained (Fig. 5E). These data demonstrate that persistence depends more on replication status than on metabolic activity, and thus 2D sorting based on metabolic status and growth rate is unlikely to yield persister enrichment beyond that which is attained based simply on growth rate.
Fig 5.
2D FACS sorting. (A) Unstained exponential-phase MO001 cells. (B) RSG-stained MO001 cells. (C and D) CFU were determined at the indicated time points during the antibiotic treatment. Persister frequencies (E) as well as fractions (F) were quantified after 5 h of antibiotic treatment of FACS-sorted cells from the NGI, NGII, GI, and GII regions (B). There was no significant difference in persister frequencies between NGI and NGII (P = 0.77 for ampicillin samples and P = 0.09 for ofloxacin samples, using the t test) or between GI and GII (P = 0.45 for ampicillin samples and P = 0.86 for ofloxacin samples, using the t test).
DISCUSSION
The dominant model of persistence posits that persisters are growth-inhibited cells with depressed cellular functions whose corruption by bactericidal antibiotics fails to produce cellular death (3). This model has stimulated a significant amount of research into different mechanisms of growth inhibition and their association with persistence (5, 9, 10, 14–16, 18, 29–31) and has facilitated the extrapolation of dormancy to additional cellular processes, such as metabolism, for which supporting evidence remains indirect (4–6). However, it has remained unclear to what extent growth inhibition prior to antibiotic treatment characterizes persistence. Several studies have shown that specific antibiotics can kill bacteria in nongrowing cultures (8, 17, 32, 33), and data from Roostalu and colleagues suggest that a lack of growth under growth-promoting conditions is insufficient to confer persistence in response to ampicillin (7). The results presented here agree with and expand upon these findings, as ampicillin and ofloxacin persisters were found to be rare (<1%) within the nongrowing subpopulation of an exponential-phase culture. These data suggest that factors beyond lack of growth define which nongrowing cells become persisters and which do not. Possible reasons for why nongrowing cells might not be persisters include the following: the cells may have been dead prior to antibiotic treatment or may have resumed replication during antibiotic treatment (3, 15, 23). We note that under the experimental conditions investigated here, 2.93% ± 0.68% of the nongrowing subpopulation could have been dead prior to antibiotic treatment, 29.17% ± 14.94% of the nongrowing subpopulation began replicating after 5 h of incubation in the absence of antibiotic, and 24.08% ± 11.92% and 19.83% ± 9.28% of the nongrowing subpopulation experienced a reduction in mCherry content after 5 h of incubation in the presence of ampicillin and ofloxacin, respectively. The remaining nongrowing subpopulation can be classified as viable but nonculturable (VBNC), since its members stained as live cells, did not resume replication during antibiotic treatment, and failed to form a colony afterward. These data, in conjunction with the previous investigations discussed above, demonstrate that growth inhibition is insufficient to confer persistence.
Though insufficient, growth inhibition prior to antibiotic exposure has commonly been considered a necessary condition of persistence (34). Several recent studies have investigated stress-induced persistence, a phenomenon in which a nonpersister cell is transformed into a persister cell through the action of signal transduction pathways (9, 10). However, it is important that none of these studies determined the growth state of the persisters formed prior to antibiotic treatment. Indeed, even the study of Dorr and colleagues, in which ciprofloxacin itself stimulated persister formation through the SOS response, did not determine whether the persisters formed were growing normally prior to antibiotic exposure (9, 10). Most recently, a study by Wakamoto and colleagues established that persisters observed in response to a prodrug antibiotic can be replicating normally prior to and during exposure (35). Cell death was found to be associated with the presence of catalase, the prodrug-activating enzyme, and independent of growth. While Wakamoto and colleagues established that persistence in response to prodrugs does not require growth inhibition, the same phenomenon for antibiotics that do not require activation (e.g., β-lactams and fluoroquinolones) had not been established. Here we discovered that growth inhibition prior to antibiotic challenge is not necessary for a cell to be a persister in response to ampicillin and ofloxacin, both of which are antibiotics that do not require activation by an enzyme to kill bacteria. We demonstrated that persisters can arise from normally replicating bacteria, though at significantly lower frequencies than those observed for nongrowing bacteria, and that the proportion of persisters from normally replicating bacteria is significant compared to that originating from nongrowing bacteria, amounting to approximately 20% of all persisters present. This observation challenges what most investigations had assumed was a requirement of the persistence phenotype (2, 34) and highlights the diversity of persisters present in bacterial populations (14).
Interestingly, we observed that the ability of a normally replicating cell to form a persister is lost after continuous exponential-phase propagation, a result that is consistent with observations from a previous study (8). Though we did not identify the cause of this phenotypic change, we postulate that this phenomenon is associated with dilution of a specific component, perhaps a protein, that is present in stationary phase and requires a >512-fold dilution to drop to levels that no longer confer persistence. From Fig. 3, it is obvious that the R0 population retained stationary-phase characteristics, as the mCherry concentration remained largely above background. The fluorescence of the R1 population resembles the background fluorescence and that of R2, but cells in R1 had undergone 5 or 6 fewer divisions than those in R2. Since the number of persisters in R1 exceeded that which could have originated from the nongrowing subpopulation of R0 (at most, 1 to 3 in 500,000 cells could have been persisters from the R0 nongrowing subpopulation), the ability to form persisters was still retained by the growing subpopulation. This ability was lost after 5 or 6 subsequent divisions, which characterizes R2. Specifically, in the absence of degradation or export, stationary-phase components would be diluted 512-fold, on average, in R1 and 16,384-fold in R2. Since specific protein concentrations can exceed 200,000 molecules per cell in stationary-phase cells (36), there are stationary-phase characteristics that are more likely to be lost in cells of R2 than in cells of R1. To determine whether this is the case is an interesting topic for a future investigation.
In association with growth inhibition, metabolic dormancy has been a commonly cited characteristic of persisters, despite the existence of only indirect evidence (17, 34). Therefore, in addition to cell division, we monitored the metabolic state of cells and found that a lower reductase activity prior to antibiotic treatment increased the likelihood that a cell would be a persister. Interestingly, lower reductase activity was also insufficient to define persistence, and we further found that sorting based on cell division and reductase activity did not provide improved persister enrichment beyond that attained with cell division alone. Though persister metabolism could not be leveraged in conjunction with cell division to improve persister enrichment, the metabolic measurements presented here suggest that persisters can contain a wide range of metabolic activities and thereby support the idea of persister diversity (14). Inherently, persister diversity will complicate efforts to eradicate these cells as sources of chronic and recurrent infections; however, charting persister diversity, as was done in this study, will place the field in the best position possible to develop broad-spectrum antipersister therapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina J. DeCoste and Christi A. O'Donnell for their technical support with flow cytometry and Zemer Gitai and his lab for providing pAS08.3 mCherry and for technical support with fluorescence microscopy. We also thank A. James Link and Ned Wingreen for their thoughtful suggestions.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI105342, by the Department of the Army under award number W81XWH-12-2-0138, and with start-up funds from Princeton University.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Published ahead of print 29 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00243-13.
REFERENCES
- 1. Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372 [DOI] [PubMed] [Google Scholar]
- 3. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 4. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shah D, Zhang ZG, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. 10.1186/1471-2180-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaldalu N, Mei R, Lewis K. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roostalu J, Joers A, Luidalepp H, Kaldalu N, Tenson T. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8:68. 10.1186/1471-2180-8-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keren I, Kaldalu N, Spoering A, Wang YP, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18 [DOI] [PubMed] [Google Scholar]
- 9. Dorr T, Lewis K, Vulic M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5:e1000760. 10.1371/journal.pgen.1000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. 10.1371/journal.pbio.1000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8:431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu YX, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 56:4922–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moker N, Dean CR, Tao JS. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192:1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allison KR, Brynildsen MP, Collins JJ. 2011. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 14:593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gefen O, Balaban NQ. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33:704–717 [DOI] [PubMed] [Google Scholar]
- 16. Kint CI, Verstraeten N, Fauvart M, Michiels J. 2012. New-found fundamentals of bacterial persistence. Trends Microbiol. 20:577–585 [DOI] [PubMed] [Google Scholar]
- 17. Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 57:1468–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2008. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2:696–706 [DOI] [PubMed] [Google Scholar]
- 20. Gray DR, Yue S, Chueng CY, Godfrey W. 2005. Bacterial vitality detected by a novel fluorogenic redox dye using flow cytometry, p 331 Abstr. 105th Gen. Meet. Am. Soc. Microbiol [Google Scholar]
- 21. Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle MT, Dobberstein B. 1987. A T5 promoter-based transcription translation system for the analysis of proteins in-vitro and in-vivo. Methods Enzymol. 155:416–433 [DOI] [PubMed] [Google Scholar]
- 22. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jõers A, Kaldalu N, Tenson T. 2010. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J. Bacteriol. 192:3379–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma C, Sim SZ, Shi WL, Du LJ, Xing DM, Zhang Y. 2010. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 303:33–40 [DOI] [PubMed] [Google Scholar]
- 25. Kim J-S, Heo P, Yang T-J, Lee K-S, Jin Y-S, Kim S-K, Shin D, Kweon D-H. 2011. Bacterial persisters tolerate antibiotics by not producing hydroxyl radicals. Biochem. Biophys. Res. Commun. 413:105–110 [DOI] [PubMed] [Google Scholar]
- 26. Calos MP. 1978. DNA sequence for a low-level promoter of the lac repressor gene and an ‘up’ promoter mutation. Nature 274:762–765 [DOI] [PubMed] [Google Scholar]
- 27. Hyser JM, Utama B, Crawford SE, Estes MK. 2012. Genetic divergence of rotavirus nonstructural protein 4 results in distinct serogroup-specific viroporin activity and intracellular punctate structure morphologies. J. Virol. 86:4921–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lidstrom ME, Konopka MC. 2010. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 6:705–712 [DOI] [PubMed] [Google Scholar]
- 29. Vázquez-Laslop N, Lee H, Neyfakh AA. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim Y, Wang X, Zhang X-S, Grigoriu S, Page R, Peti W, Wood TK. 2010. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 12:1105–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Falla TJ, Chopra I. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fung DKC, Chan EWC, Chin ML, Chan RCY. 2010. Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob. Agents Chemother. 54:1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 35. Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95 [DOI] [PubMed] [Google Scholar]
- 36. Nair S, Finkel SE. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Amato SM, Orman MA, Brynildsen MP. 9 May 2013, posting date Metabolic control of persister formation in Escherichia coli. Mol. Cell 10.1016/j.molcel.2013.04.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.