Abstract
A total of 84 extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from cattle, farm workers, and the farm environment isolated from February to September 2008 in the Republic of Korea were investigated. All 84 ESBL-producing isolates carried blaCTX-M genes that belonged to the CTX-M-1 (n = 35) or CTX-M-9 (n = 49) family. The most predominant CTX-M type identified was CTX-M-14 (n = 49), followed by CTX-M-32 (n = 26). The blaCTX-M genes were identified most commonly in E. coli isolates from feces (n = 29), teats (n = 25), and milk (n = 14). A blaCTX-M-14 gene was also detected in an E. coli isolate from a farmer's hand. Transfer of the blaCTX-M gene from 60 blaCTX-M-positive E. coli isolates to the recipient E. coli J53 strain by conjugation was demonstrated. Plasmid isolation from blaCTX-M-positive transconjugants revealed a large (95- to 140-kb) conjugative plasmid. Almost all (82/84) blaCTX-M genes possessed an insertion sequence, ISEcp1, upstream of the blaCTX-M gene. Only in the case of the CTX-M-14 genes was IS903 downstream of the gene. The blaCTX-M genes were associated with seven kinds of addiction systems. Among them, pndAC, hok-sok, and srnBC were the most frequently identified addiction systems in both wild strains and transconjugants. The spread of blaCTX-M genes was attributed to both clonal expansion and horizontal dissemination. Our data suggest that a combination of multiple addiction systems in plasmids carrying blaCTX-M genes could contribute to their maintenance in the host cells. To our knowledge, the blaCTX-M-32 gene has not previously been reported in animal isolates from the Republic of Korea.
INTRODUCTION
The major mechanism of resistance to oxyimino-cephalosporins in Escherichia coli is the production of extended-spectrum β-lactamases (ESBLs) (1). The TEM- and SHV-type ESBLs were the most common ESBLs during the 1990s. However, CTX-M β-lactamases, a new group of plasmid-mediated ESBLs, have increased dramatically in the past decade and are now the dominant type of ESBL in most areas of the world (2). The global spread and high prevalence of the CTX-M type in E. coli is a matter of concern in both human and veterinary medicine.
The genetic basis contributing to the successful global dissemination of CTX-M β-lactamases remains poorly understood. Molecular epidemiological studies have shown a strong association of blaCTX-M genes with conjugative plasmids and successful bacterial clones (3). It has been suggested that introduction of IncFII plasmids encoding CTX-M-15 into the well-adapted ST131-O25:H4 E. coli clone and its subsequent spread could be involved in part in the successful dissemination of CTX-M-15-producing E. coli clones worldwide (4, 5). It has also been proposed that the plasmids carrying antimicrobial resistance or virulence determinants in bacteria exploit toxin-antitoxin gene pairs in order to maintain themselves during host replication. The toxin-antitoxin system, also known as the addiction system, eradicates plasmid-free cells and contributes to intra- and interspecies plasmid dissemination (6).
Recently, multiple addiction systems were reported in plasmids bearing blaCTX-M genes in E. coli, suggesting the contribution of multiple addiction systems to their maintenance in host strains (7). Similarly, the rapid spread of CTX-M-15-producing human clinical E. coli isolates in the Republic of Korea was attributed to clones with a high frequency of virulence determinants and addiction systems (8). More recently, we observed the predominance of CTX-M-14-producing E. coli isolates among healthy animals in the Republic of Korea (9). Addiction systems responsible for the maintenance and successful spread of CTX-M genes in E. coli among humans have been investigated, but their role in animal systems remains unknown. Therefore, the objective of this study was to characterize CTX-M β-lactamase and associated addiction systems in E. coli circulating among cattle, farm workers, and the farm environment in the Republic of Korea.
MATERIALS AND METHODS
Sampling and bacterial culture.
A total of 1,536 samples, including quarter milk samples (n = 559), cattle feces (n = 379), farm environmental samples (n = 512), and samples from farmers' hands (n = 43) and noses (n = 43), were collected from 22 different dairy cattle farms from February to September 2008. These farms were located in five different provinces (Jeonnam, n = 3; Jeonbuk, n = 8; Chungnam, n = 4; Gwangwon, n = 3; and Gyeonggi, n = 4) of the Republic of Korea and were chosen on the basis of likelihood of mastitis, farmers' interest, and compliance. Sample collection from each farm was done only once.
The quarter milk samples were taken aseptically from lactating cows directly after milking in sterile snap cap milk collection vials. The environmental samples were obtained from floors (n = 43), fences (n = 44), teats (n = 210), ventilation fans (n = 42), manure (n = 11), water (n = 30), feed (n = 30), and milking cups (n = 102). Fecal samples (20 to 50 g of fresh fecal mass) were collected in sterile stool collection containers. For farmers' nasal sampling, a cotton-tipped swab with Stuart's medium (Becton, Dickinson, Sparks, MD, USA), and for the farmers' hand sampling, four gauze pads (10 cm by 10 cm) wetted with 1% sterile skim milk were used. Drinking water and feed samples were collected in buckets and placed in 50-ml sterile containers. Other environmental samples were collected using four gauze pads wetted with 1% sterile skim milk and placed in sterile plastic bags. The samples were immediately transported to the laboratory in ice-cooled containers and processed within 24 h of collection.
The fecal samples were directly inoculated onto MacConkey agar plates supplemented with 2 μg/ml cefotaxime using cotton swabs and incubated aerobically overnight at 37°C. All gauze pad samples from environmental sources and farmers' hands were individually placed in a sterile stomacher bag containing 50 ml of 1% sterile skim milk and homogenized for 30 s. Nasal swabs, 1 ml of milk, 1 ml of water/feed, and 1 ml of homogenized gauze pad samples were inoculated into 9 ml tryptone soy broth containing 2 μg/ml cefotaxime and incubated for a further 16 to 20 h at 37°C. Then, one loopful (10 μl) of this broth was spread onto MacConkey agar plates supplemented with 2 μg/ml cefotaxime. Based on colony morphology and color, suspected large, smooth, pink colonies were subsequently subcultured on Chromogenic ESBL agar (bioMérieux, Marcy-l'Etoile, France). The identification of E. coli isolates and phenotypic confirmation of ESBL production were done as described previously (9). Altogether, 84 phenotypically ESBL-positive E. coli isolates were identified, which were further characterized in detail in the present study.
Antimicrobial susceptibility test.
Antimicrobial susceptibility was tested by the disc diffusion method, in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (10) using commercial discs (BBL; Becton, Dickinson and Company, Cockeysville, MD). The antimicrobial drugs tested were selected based on their relevance in veterinary medicine and their importance in human medicine. They include ampicillin (10 μg), amoxicillin-clavulanic acid (20 and 10 μg), cephalothin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefoxitin (30 μg), imipenem (10 μg), chloramphenicol (30 μg), gentamicin (10 μg), neomycin (30 μg), streptomycin (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), and trimethoprim-sulfamethoxazole (1.25 to 23.75 μg). MICs of selected antimicrobials were tested using Etest strips (AB Biodisk). The diameters of inhibition zones surrounding the antimicrobial discs and Etest strips were interpreted according to the CLSI guidelines (10). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains for antimicrobial susceptibility tests.
β-Lactamase gene identification.
Screening of the blaCTX-M gene was done as described previously (11). To confirm blaCTX-M genes, group-specific primers for the CTX-M-1 and CTX-M-9 families were used as described previously (12). Finally, the combination of the CTX-M-1G FL-F (13) or CTX-M-9G FL-R (14) primer and the ISECP1U1 primer (15) was used to amplify and sequence the complete blaCTX-M gene. For the CTX-M-positive isolates, PCR amplification and sequencing of entire blaTEM and blaSHV genes was done as described previously (16). Sequence analyses and comparison with known sequences were performed with the BLAST programs at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST).
Conjugations.
The abilities of blaCTX-M-positive E. coli isolates to transfer blaCTX-M genes to sodium azide-resistant E. coli J53 by conjugation were determined by broth-mating experiments at 37°C, as described previously (17). Prior to the conjugation experiments, the blaCTX-M-positive E. coli isolates were tested for susceptibility to sodium azide. Transconjugants were selected on MacConkey agar plates supplemented with sodium azide (150 mg/liter; Sigma) and cefotaxime (2 μg/ml). Antimicrobial susceptibility and detection of β-lactamase genes were performed in the presumptive transconjugants as described above to confirm the transfer of β-lactamase genes.
Plasmid characterization.
Plasmid DNAs were extracted from the transconjugants using the QuickGene plasmid isolation system (Fujifilm Corporation, Tokyo, Japan) following the manufacturer's protocol. Plasmid size was estimated by comparison with the Bac-Tracker Supercoiled DNA Ladder (Epicentre Bitotechnologies, Madison, WI) after gel electrophoresis in 0.8% agarose gels. Replicon typing of the isolated plasmid DNA was done by a PCR-based replicon-typing (PBRT) method as described previously (18).
Exploration of the genetic environment of the blaCTX-M gene.
The genetic environment of the blaCTX-M gene identified in this study was investigated by PCR and by sequencing of the regions surrounding the gene. Forward primers of IS26 (19), ISCR1 (20), or ISEcp1 (15) and the CTX-M reverse consensus (MA2) primer (15) were used to examine regions upstream of the bla genes. Similarly, the MA1 primer (15) and reverse primers of IS903, orf477, and mucA genes (19) were used to investigate downstream of the bla genes.
Detection of addiction systems.
The presence of eight plasmid addiction systems, PemK-PemI (pemKI) (plasmid emergency maintenance), CcdA-CcdB (ccdAB) (coupled to cell division), RelB-RelE (relBE) (relaxed control of stable RNA synthesis), ParD-ParE (parDE) (DNA replication), VagC-VagD (vagCD) (virulence-associated protein), Hok-Sok (hok-sok) (host killing), PndA-PndC (pndAC) (promotion of nucleic acid degradation), and SrnB-SrnC (srnBC) (stable RNA), was determined by PCR using primer sets and conditions described previously (7).
PFGE.
Pulsed-field gel electrophoresis (PFGE) of XbaI (TaKaRa Bio Inc., Shiga, Japan)-digested genomic DNA of CTX-M β-lactamase-producing E. coli strains was carried out as described previously (21) using a Chef Mapper apparatus (Bio-Rad Laboratories, Hercules, CA). The PFGE conditions of XbaI macrorestriction analysis were 6 V/cm for 19 h with pulse times ranging from 2.16 to 54.17 s at a temperature of 14°C and an angle of 120°. The similarities of the restriction fragment length polymorphisms were analyzed using Bionumerics software, version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium) to produce a dendrogram. Clustering was carried out by the unweighted-pair group method with arithmetic averages (UPGMA), based on the Dice similarity index.
RESULTS
Antimicrobial susceptibility.
An antimicrobial susceptibility test showed that all 84 ESBL-producing isolates were resistant to ampicillin, cephalothin, and cefotaxime but that all of them were susceptible to imipenem and cefoxitin. Among the non-β-lactam drugs tested, resistance was most frequently observed to streptomycin (64/84; 76.5%) and tetracycline (64/84; 76.5%), followed by trimethoprim-sulfamethoxazole (55/84; 65.5%), nalidixic acid (50/84; 59.5%), neomycin (43/84; 51.2%), gentamicin (33/84; 39.3%), chloramphenicol (32/84; 38.1%), and ciprofloxacin (32/84; 38.1%). MIC ranges for cefotaxime, ceftazidime, cefepime, and imipenem were 4 to ≥16 μg/ml, ≤0.5 to 32 μg/ml, 0.5 to 16 μg/ml, and 0.19 to 0.25 μg/ml, respectively.
Characterization of β-lactamase genes.
All 84 ESBL-producing E. coli isolates carried blaCTX-M genes. The predominant CTX-M type identified was CTX-M-14 (n = 49), followed by CTX-M-32 (n = 26). Isolates producing CTX-M-15 (n = 9) were also identified. The distribution of 84 blaCTX-M genes according to sources of isolation is shown in Table 1. The blaCTX-M genes were identified most commonly in E. coli isolates from feces (n = 29), followed by teats (n = 25), milk (n = 14), and floor samples (n = 5). A blaCTX-M-14 gene was also detected in an E. coli isolate from a farmer's hand. Fifty-eight of these blaCTX-M-positive isolates also carried blaTEM-1, but none of them was positive for the blaSHV gene.
Table 1.
Distribution of blaCTX-M genes identified in this study according to sources of isolation
| Source | No. of samples investigated | No. of isolates producing: |
Total no. (%) | ||
|---|---|---|---|---|---|
| CTX-M-14 | CTX-M-15 | CTX-M-32 | |||
| Feces | 379 | 16 | 7 | 6 | 29 (34.5) |
| Teat | 210 | 18 | 7 | 25 (29.7) | |
| Milk | 559 | 6 | 1 | 7 | 14 (16.7) |
| Floor | 43 | 3 | 2 | 5 (5.9) | |
| Fence | 44 | 2 | 2 | 4 (4.8) | |
| Ventilation fan | 42 | 1 | 1 | 2 (2.4) | |
| Feed | 30 | 1 | 1 (1.2) | ||
| Water | 30 | 1 | 1 (1.2) | ||
| Manure | 11 | 1 | 1 (1.2) | ||
| Farmers' hands | 43 | 1 | 1 (1.2) | ||
| Farmers' noses | 43 | ||||
| Milking cups | 102 | 1 | 1 (1.2) | ||
| Total | 1,536 | 49 | 9 | 26 | 84 (100) |
Transferability of blaCTX-M genes.
Transfer of the cefotaxime resistance phenotype and blaCTX-M gene from 60 blaCTX-M-positive E. coli isolates to the recipient E. coli strain J53 by conjugation was successful. The characteristics of conjugation-positive E. coli isolates carrying blaCTX-M-14 and blaCTX-M-32 genes are shown in Table 2 and Table 3, respectively. Conjugative-transfer frequencies varied from 5.0 × 10−4 to 4.9 × 10−2 transconjugants per recipient. PCR analysis showed the presence of the respective blaCTX-M genes from all the transconjugants. However, the blaTEM-1 gene from only 14 out of the 58 E. coli isolates that carried both the blaCTX-M and blaTEM-1 genes was transferred to the recipient E. coli strain J53. In addition to the cephalosporin resistance phenotype, resistance traits to non-β-lactam antibiotics also transferred along with blaCTX-M genes.
Table 2.
Characteristics of 34 conjugation-positive E. coli isolates carrying blaCTX-M-14 genes described in the studya
| Isolate | Farm | Origin | Etest MIC (mg/liter) |
Other β-lactamase | Addiction system(s)b | Plasmid Inc type | Non-β-lactam resistance pattern | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CT | CTL | TZ | TZL | |||||||
| M9-7 | L | Cattle feces | >16 | 0.023 | ≤0.5 | ≤0.064 | TEM-1 | ccdAB, pndCA, srnBC | I1-Iγ | GEN STR |
| M9-8 | L | Calf feces | >16 | 0.032 | ≤0.5 | 0.064 | TEM-1 | ccdAB | NT | GEN STR |
| M9-9 | L | Calf feces | 16 | 0.032 | ≤0.5 | 0.064 | TEM-1 | srnBC | NT | GEN STR |
| M9-10 | L | Calf feces | >16 | 0.023 | ≤0.5 | 0.064 | TEM-1 | pndCA, srnBC | NT | GEN STR SXT TET |
| M9-13 | M | Calf feces | >16 | 0.023 | ≤0.5 | 0.094 | hok-sok, pndAC, srnBC | F | NEO STR TET | |
| M9-14 | M | Calf feces | >16 | 0.023 | ≤0.5 | 0.064 | TEM-1 | ccdAB, vagCD, srnBC | F | NEO NAL SXT TET |
| M9-15 | N | Calf feces | 16 | 0.047 | ≤0.5 | 0.125 | pndCA | I1-Iγ | APR CIP NEO NAL STR SXT TET | |
| M9-16 | N | Calf feces | >16 | 0.023 | ≤0.5 | 0.094 | TEM-1 | ccdAB, vagCD, srnBC | NT | CHL CIP GEN NEO NAL STR SXT TET |
| M9-17 | L | Feed | >16 | 0.032 | ≤0.5 | 0.064 | TEM-1 | pndCA, srnBC | NT | NEO STR TET |
| M9-18 | L | Ground floor | 12 | 0.032 | 0.5 | 0.094 | TEM-1 | ccdAB, srnBC | NT | NEO NAL STR SXT TET |
| M9-19 | L | Ground fence | 12 | 0.032 | ≤0.5 | 0.064 | TEM-1 | pndCA, srnBC | NT | APR CIP NEO NAL STR SXT TET |
| M9-20 | L | Parlor fence | 16 | 0.023 | ≤0.5 | 0.064 | TEM-1 | pndCA, srnBC | NT | NEO STR TET |
| M9-21 | L | Parlor floor | >16 | 0.032 | ≤0.5 | 0.064 | pndAC, srnBC | NT | NEO NAL STR SXT TET | |
| M9-22 | L | Milking cup | >16 | 0.023 | ≤0.5 | 0.064 | TEM-1 | ccdAB, pndAC, srnBC | NT | APR CIP NEO NAL STR SXT TET |
| M9-23 | L | Teat | 16 | 0.032 | 0.5 | 0.064 | TEM-1 | pndAC, srnBC | NT | NEO STR TET |
| M9-24 | L | Teat | >16 | 0.023 | ≤0.5 | 0.064 | TEM-1 | ccdAB, pndAC, srnBC | NT | NEO NAL STR SXT TET |
| M9-25 | L | Teat | 8 | 0.023 | ≤0.5 | 0.064 | TEM-1 | pndAC, srnBC | NT | APR CIP NEO NAL STR SXT TET |
| M9-26 | L | Teat | 16 | 0.032 | ≤0.5 | 0.064 | TEM-1 | pndAC, srnBC | NT | NEO NAL STR SXT TET |
| M9-27 | L | Teat | 16 | 0.016 | ≤0.5 | ≤0.064 | TEM-1 | ccdAB, srnBC | NT | NEO NAL STR SXT TET |
| M9-28 | L | Teat | 16 | 0.032 | ≤0.5 | 0.094 | TEM-1 | ccdAB, srnBC | NT | APR CIP NEO NAL STR SXT TET |
| M9-29 | L | Teat | 8 | 0.032 | 0.5 | 0.094 | TEM-1 | ccdAB, srnBC | NT | NEO NAL STR TET |
| M9-30 | L | Teat | 16 | 0.023 | ≤0.5 | 0.064 | TEM-1 | pndAC, srnBC | NT | NEO NAL STR SXT TET |
| M9-31 | L | Teat | 12 | 0.023 | ≤0.5 | 0.064 | TEM-1 | ccdAB, srnBC | NT | APR CIP, NEO NAL STR SXT TET |
| M9-32 | L | Farmer's hand | 12 | 0.023 | ≤0.5 | 0.064 | TEM-1 | pndAC, srnBC | NT | NEO STR TET |
| M9-33 | M | Ventilation | >16 | 0.032 | 0.5 | 0.094 | TEM-1 | ccdAB, vagCD, srnBC | F | NEO NAL STR SXT TET |
| M9-34 | M | Teat | >16 | 0.047 | ≤0.5 | 0.094 | TEM-1 | F | APR CIP, NEO NAL STR SXT TET | |
| M9-35 | M | Milk | >16 | 0.032 | ≤0.5 | 0.064 | TEM-1 | ccdAB, vagCD, srnBC | F | NAL STR TET |
| M9-36 | P | Cattle feces | 8 | 0.032 | ≤0.5 | 0.064 | TEM-1 | hok-sok, pndAC, srnBC | F | NEO NAL STR SXT TET |
| M9-37 | P | Cattle feces | 8 | 0.032 | ≤0.5 | 0.064 | TEM-1 | hok-sok, pndAC, srnBC | F | APR CIP NEO NAL STR SXT TET |
| M9-41 | P | Teat | 4 | 0.023 | ≤0.5 | 0.064 | hok-sok, pndAC, srnBC | F, I1-Iγ | TET | |
| M9-42 | P | Teat | >16 | 0.023 | ≤0.5 | 0.064 | hok-sok, srnBC | F | ||
| M9-43 | P | Teat | 16 | 0.023 | ≤0.5 | 0.064 | hok-sok, srnBC | F | ||
| M9-44 | P | Teat | 16 | 0.032 | ≤0.5 | 0.064 | hok-sok, srnBC | F | ||
| M9-45 | P | Teat | 12 | 0.023 | ≤0.5 | 0.064 | hok-sok, srnBC | F | ||
Etest, Epsilometer test; NT, not typeable by PCR-based replicon typing; CT, cefotaxime; CTL, cefotaxime-clavulanic acid; TZ, ceftazidime; TZL, ceftazidime-clavulanic acid; APR, apramycin; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; NEO, neomycin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline.
Underlined bla genes, addiction system and resistance markers transferred to the recipient E. coli J53 strain by conjugation.
Table 3.
Characteristics of 26 conjugation-positive E. coli isolates carrying blaCTX-M-32 genes described in this studya
| Isolateb | Origin | Etest MIC (mg/liter) |
Other β-lactamase | Addiction system(s)c | Transconjugant plasmid Inc type(s) | Non-β-lactam resistance pattern | |||
|---|---|---|---|---|---|---|---|---|---|
| CT | CTL | TZ | TZL | ||||||
| M1-4 | Cattle feces | >16 | 0.023 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, N, P, I1-Iγ | CHL CIP GEN NEO NAL STR SXT TET |
| M1-5 | Cattle feces | >16 | 0.023 | 32 | 0.125 | TEM-1 | hok-sok, pndAC, srnBC | N | CHL CIP GEN NEO NAL STR SXT TET |
| M1-6 | Cattle feces | >16 | 0.023 | 16 | 0.094 | TEM-1 | hok-sok, srnBC | N | CHL CIP NEO NAL STR SXT TET |
| M1-7 | Cattle feces | >16 | 0.023 | 32 | 0.023 | relBE | N | CHL CIP NEO NAL STR SXT TET | |
| M1-8 | Cattle feces | >16 | 0.023 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | N | CHL CIP GEN NEO NAL STR SXT TET |
| M1-9 | Calf feces | >16 | 0.032 | 16 | 0.125 | TEM-1 | hok-sok, pndAC, srnBC | P, I1-Iγ | AMC CIP NEO NAL STR SXT TET |
| M1-11 | Manure | >16 | 0.023 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-12 | Ventilation | >16 | 0.023 | 12 | 0.125 | TEM-1 | hok-sok, srnBC | F, FIB, N | CHL CIP NEO NAL STR SXT TET |
| M1-13 | Ground floor | >16 | 0.023 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-14 | Ground fence | >16 | 0.016 | 8 | 0.094 | TEM-1 | hok-sok, srnBC | N | CHL CIP NEO NAL STR SXT TET |
| M1-15 | Parlor floor | >16 | 0.016 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-16 | Parlor fence | >16 | 0.016 | 12 | 0.094 | TEM-1 | pndAC | N, P, I1-Iγ | CHL CIP GEN NEO NAL STR SXT TET |
| M1-17 | Teat | >16 | 0.023 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | N | CHL CIP NEO NAL STR SXT TET |
| M1-18 | Teat | >16 | 0.023 | 8 | 0.125 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-19 | Teat | >16 | 0.023 | 12 | 0.125 | TEM-1 | hok-sok, pndAC, srnBC | N | CHL CIP GEN NEO NAL STR SXT TET |
| M1-20 | Teat | >16 | 0.023 | 8 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | NT | CHL CIP NEO NAL STR SXT TET |
| M1-21 | Teat | >16 | 0.023 | 12 | 0.125 | TEM-1 | hok-sok, srnBC | F, FIB, N | AMC CHL CIP NEO NAL STR SXT TE |
| M1-22 | Teat | >16 | 0.023 | 8 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-23 | Teat | >16 | 0.016 | 12 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-24 | Milk | >16 | 0.032 | 16 | 0.032 | TEM-1 | hok-sok, pndAC, srnBC | FIB, N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-25 | Milk | >16 | 0.023 | 32 | 0.125 | TEM-1 | hok-sok, pndAC, srnBC | FIB, N, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-26 | Milk | >16 | 0.032 | 8 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | N | CHL CIP NEO NAL STR SXT TET |
| M1-27 | Milk | >16 | 0.023 | 8 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, P, I1-Iγ | CHL CIP GEN NEO NAL STR SXT TET |
| M1-28 | Milk | >16 | 0.023 | 12 | 0.125 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, N, P, I1-Iγ | APR CHL CIP NEO NAL STR SXT TET |
| M1-29 | Milk | >16 | 0.023 | 12 | 0.125 | TEM-1 | hok-sok, srnBC | F, FIB, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
| M1-30 | Milk | >16 | 0.023 | 8 | 0.094 | TEM-1 | hok-sok, pndAC, srnBC | F, FIB, P, I1-Iγ | CHL CIP NEO NAL STR SXT TET |
Etest, Epsilometer test; NT, not typeable by PCR-based replicon typing; CT, cefotaxime; CTL, cefotaxime-clavulanic acid; TZ, ceftazidime; TZL, ceftazidime-clavulanic acid; AMC, ampicillin-clavulanic acid; APR, apramycin; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; NEO, neomycin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline.
All E. coli strains carrying blaCTX-M-32 genes were isolated from farm N.
Underlined bla genes, addiction systems and resistance markers transferred to the recipient E. coli J53 strain by conjugation.
Analysis of blaCTX-M plasmids.
Plasmid DNA preparation from blaCTX-M-positive E. coli J53 transconjugants revealed a large conjugative plasmid, ranging in size from 95 to 140 kb. Various replicon types, including IncF, IncI1-Iγ, IncN, IncFIB, and IncP, either alone or in combination, were identified. All but one plasmid harboring blaCTX-M-14 genes were positive for a single replicon or were nontypeable (Table 2), whereas plasmids harboring blaCTX-M-32 genes were multireplicon plasmids, with the exception of eight IncN plasmids (Table 3). Twenty-one of 60 conjugative plasmids were nontypeable for the 18 incompatibility groups sought by the PBRT method. All of them encoded CTX-M-14, except one that encoded CTX-M-32.
Genetic environment of the blaCTX-M gene.
PCR identified ISEcp1 upstream of blaCTX-M genes in 82 out of 84 E. coli isolates and in their 60 corresponding transconjugants. Similarly, IS903 was identified downstream of blaCTX-M genes in 48 E. coli isolates. These 82 isolates bearing ISEcp1 upstream of the bla gene were classified into two different types based on their genetic environments. Type I (n = 47) was characterized by the presence of ISEcp1 and IS903 elements upstream and downstream of the blaCTX-M gene, respectively, whereas type II (n = 35) showed a genetic environment identical to that of type I in the upstream region of the blaCTX-M gene, but the downstream IS903 was absent.
Prevalence of addiction systems.
Altogether, 188 plasmid addiction systems were detected among the CTX-M-producing E. coli strains. Of the eight addiction systems sought, seven different kinds, namely, pemKI, ccdAB, relBE, vagCD, pndAC, hok-sok, and srnBC, were identified (Table 4). None of these strains harbored the parDE system. The mean number of addiction systems detected was highest (3.3) among the CTX-M-15-producing E. coli isolates, followed by CTX-M-32-producing (2.7) and CTX-M-14-producing (1.8) isolates. Overall, 46 addiction systems were detected in the transconjugants. However, only the pndAC, hok-sok, and srnBC systems were identified among the transconjugants. The mean numbers of addiction systems detected among blaCTX-M-32- and blaCTX-M-14-positive transconjugants were 1.1 and 0.5, respectively.
Table 4.
Nature and number of addiction systems according to blaCTX-M type identified in E. coli strains and their transconjugants
| Origin | CTX-M type | n | No. of strains with the indicated addiction system |
Total | Mean | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pemKI | ccdAB | relBE | parDE | vagCD | hok-sok | pndAC | srnBC | |||||
| Wild strains | CTX-M-14 | 49 | 1 | 15 | 0 | 0 | 6 | 8 | 20 | 33 | 87 | 1.8 |
| CTX-M-15 | 9 | 4 | 4 | 0 | 0 | 4 | 6 | 3 | 9 | 30 | 3.3 | |
| CTX-M-32 | 26 | 0 | 0 | 1 | 0 | 0 | 25 | 20 | 25 | 71 | 2.7 | |
| Any | 84 | 5 | 19 | 1 | 0 | 10 | 39 | 43 | 67 | 188 | 2.2 | |
| Transconjugants | CTX-M-14 | 34 | 0 | 0 | 0 | 0 | 0 | 7 | 3 | 7 | 17 | 0.5 |
| CTX-M-32 | 26 | 0 | 0 | 0 | 0 | 0 | 11 | 14 | 4 | 29 | 1.1 | |
| Any | 60 | 0 | 0 | 0 | 0 | 0 | 18 | 17 | 11 | 46 | 0.8 | |
PFGE analysis.
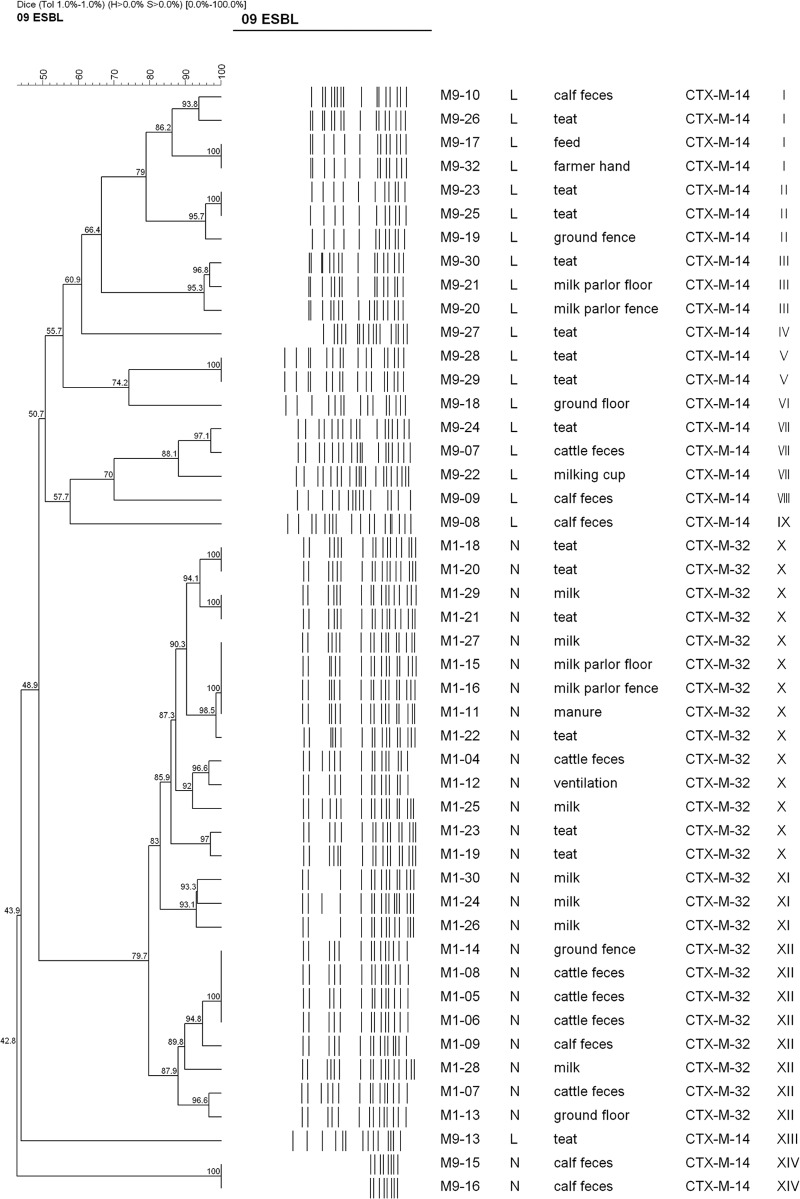
XbaI PFGE analysis of a subset of 47 E. coli strains carrying blaCTX-M-14 (n = 22) and blaCTX-M-32 (n = 25) genes from two different farms (farm L and farm N) in which the isolation rate of E. coli strains producing CTX-M β-lactamase was highest demonstrated at least 13 arbitrary (designated I through XIII) pulsotypes (Fig. 1), based on the similarity value of 0.85. The 22 CTX-M-14-producing strains represented 10 pulsotypes, including five singleton types. In contrast, 25 CTX-M-32-producing strains represented three predominant pulsotypes, X (n = 14), XII (n = 8), and XI (n = 3). DNA from two strains consistently autodigested. Thus, a cluster formed by these strains is ignored throughout this paper.
Fig 1.
Dendrogram showing the cluster analysis of XbaI PFGE patterns of 47 CTX-M-producing E. coli strains isolated from cattle, farm workers, and the farm environment in the Republic of Korea. The cluster analysis was performed by using the Dice coefficient and the unweighted-pair group method with arithmetic averages. Details given in the first two columns are % similarity and PFGE banding pattern, respectively. The numbers at the nodes indicate % similarity. Details given in the third through seventh columns from the left are the strain number, farm investigated, source of isolation of the blaCTX-M gene, CTX-M type, and pulsotype for each strain. XbaI macrorestriction analysis yielded few or no DNA banding patterns in the two E. coli strains due to constant autodigestion of the genomic DNA during agarose plug preparation, and thus, a cluster formed by these strains is ignored throughout this paper.
DISCUSSION
The phenotypic and genotypic characteristics of ESBL-producing E. coli strains isolated from cattle, farm workers, and the farm environment from February to September 2008 in the Republic of Korea were investigated in this study. Overall, E. coli isolates from 84 (5.5%) of the 1,536 samples examined demonstrated CTX-M-type ESBL production. This percentage is higher than that recently found in fecal samples from healthy cattle (1/654; 0.2%) in the Republic of Korea (9). Of these, 35 (41.7%) belonged to the CTX-M-1 family and 49 (58.3%) to the CTX-M-9 family. Our results are consistent with previous molecular epidemiological studies of ESBL-producing E. coli strains reporting dissemination of E. coli isolates harboring blaCTX-M genes of the CTX-M-1 or CTX-M-9 family among food animals from the Republic of Korea (9), China (22), Hong Kong SAR (23, 24), and Europe (25, 26, 27). Similarly, these findings mirror the trend observed in humans in the Republic of Korea, where the significant increase in the incidence of ESBLs in human clinical isolates of E. coli was attributed to the dissemination of the CTX-M-1 and CTX-M-9 families of β-lactamases (28).
In this study, a total of three blaCTX-M alleles (CTX-M-14, -15, and -32) were detected. Except for blaCTX-M-32, the blaCTX-M genes identified in this work were previously observed in E. coli isolates from animals in the Republic of Korea (9, 29, 30). Recently, we also reported blaCTX-M-14 and blaCTX-M-15 in nontyphoid Salmonella isolates among food animals and humans (31). However, CTX-M-32-producing organisms are relatively infrequent on a worldwide basis. CTX-M-32 was first reported in 2004 from the northern part of Spain and has mostly been reported from Spain (32, 33). The amino acid sequence of CTX-M-32 differs from that of CTX-M-1 by a single Asp240-Gly substitution, which is responsible for its enhanced level of resistance to ceftazidime (32). The results of our study, which showed a higher MIC value for cefotaxime, as well as ceftazidime, among isolates producing CTX-M-32, support this fact. Although a single clinical E. coli isolate producing CTX-M-32 was previously observed in human medicine (34), to our knowledge, the blaCTX-M-32 gene has not previously been reported in animal isolates from the Republic of Korea.
In the present study, all blaCTX-M-32 genes and a majority of blaCTX-M-14 genes (34/49; 69.4%) transferred to a recipient E. coli strain, along with the ESBL phenotype, by conjugation. However, none of the blaCTX-M-15 genes were transferred to the recipient strain, which indicates that this genetic determinant may be located only on a nonconjugative plasmid or in the chromosome. Moreover, each transconjugant apparently carried a single large conjugative plasmid. The replicon typing of these transconjugant plasmids demonstrated significant plasmid diversity, which is consistent with that of blaCTX-M plasmids previously reported from the Republic of Korea (9) and the United Kingdom (35). In general, IncN and IncF were the predominant replicon types among the plasmids carrying blaCTX-M-32 and blaCTX-M-14 genes, respectively, suggesting their roles in the horizontal dissemination of these genes. Consistent with our findings, Novais et al. previously reported IncN plasmids bearing blaCTX-M-32 from various human clinical E. coli strains from Spain (33). As in this study, blaCTX-M-14 genes were mostly carried on IncF plasmids in the previous reports from the Republic of Korea (9, 36). In contrast, the spread of blaCTX-M-14 among E. coli isolates in Spain has been reported to be mainly mediated by IncK plasmids (37). Nevertheless, dissemination of IncN plasmids, which are typically conjugative and broad-host-range plasmids, may contribute to the further spread of isolates producing CTX-M-32 enzymes among other members of the Enterobacteriaceae (33).
ISEcp1 is frequently found in the upstream region of blaCTX-M-type genes and plays a critical role in the efficient capture, expression, and mobilization of blaCTX-M genes (15, 38). In agreement with previous studies (9, 31, 32), ISEcp1 was detected in 97.6% of wild strains and in all the corresponding transconjugants. In addition, the absence of IS26 or ISCR1 upstream of the blaCTX-M genes indicates that these elements were not involved in the mobilization of CTX-M genes, as has been previously reported (26). IS903 was detected downstream of the blaCTX-M-14 gene in all but one isolate carrying the gene. However, orf477 and mucA, sought downstream of the blaCTX-M gene, were not detected in any isolate in this study. In contrast, IS903 and orf477 have previously been reported downstream of most CTX-M-9 and CTX-M-1 family genes, respectively (19, 36, 39).
Plasmid addiction systems presumably contribute to the stability and maintenance of plasmids in colonized hosts and facilitate further dissemination even in the absence of antibiotic selection (6, 35, 40). Although seven different addiction systems were detected associated with blaCTX-M genes in this work, pndAc, hok-sok, and srnBC were the most frequently identified addiction systems in both the CTX-M-producing wild E. coli strains and their transconjugants. Interestingly, there was complete absence of certain addiction systems in the transconjugants compared to wild strains in this study. This might, in part, be explained by the failure of transfer of CTX-M-15-positive plasmids bearing some of these addiction systems. Furthermore, it implies that some types of addiction systems may be located in the chromosome or nonconjugative plasmids, in addition to addiction systems present in the conjugative plasmids bearing resistance markers. Mnif et al. reported multiple addiction systems in plasmids bearing the blaCTX-M gene among E. coli isolates producing CTX-M-15 or CTX-M-9 from France (7), which is similar to our results. Moreover, multiple addiction systems were identified in multiresistant IncF plasmids bearing blaCTX-M genes among clinical E. coli isolates in the United Kingdom (35). However, the prevalence and nature of addiction systems have never been reported before among CTX-M-producing isolates from animals across the globe. Further analysis of addiction systems among the transconjugants showed that hok-sok and srnBC systems were more commonly associated with the IncF replicon, whereas the pndAC system was associated with the Inc I1-Iγ replicon (Table 5). Mnif et al. also reported an association between the vagCD and hok-sok systems and FIA and FIB replicons (7). Thus, these findings suggest linkage between addiction systems and specific plasmid backbones.
Table 5.
Distribution of addiction systems according to plasmid replicon type among transconjugants
| Inc/replicon type | n | No. of isolates with the indicated addiction system |
Total | Mean | ||
|---|---|---|---|---|---|---|
| hok-sok | pndAC | srnBC | ||||
| CTX-M-14 plasmids | ||||||
| F | 11 | 6 | 0 | 6 | 12 | 1.1 |
| I1-Iγ | 2 | 0 | 2 | 0 | 2 | 1 |
| F, I1-Iγ | 1 | 1 | 1 | 1 | 3 | 3 |
| Unidentified | 20 | 0 | 0 | 0 | 0 | |
| CTX-M-32 plasmids | ||||||
| N | 8 | 0 | 0 | 0 | 0 | |
| P, I1-Iγ | 1 | 0 | 1 | 0 | 1 | 1 |
| F, FIB, I1-Iγ | 1 | 1 | 0 | 0 | 1 | 1 |
| F, FIB, N | 2 | 2 | 0 | 1 | 3 | 1.5 |
| N, P, I1-Iγ | 4 | 0 | 4 | 0 | 4 | 1 |
| N, P, FIB, I1-Iγ | 2 | 1 | 2 | 0 | 3 | 1.5 |
| F, FIB, P, I1-Iγ | 2 | 2 | 2 | 0 | 4 | 2 |
| F, N, P, FIB, I1-Iγ | 5 | 5 | 5 | 3 | 13 | 2.6 |
| Unidentified | 1 | 0 | 0 | 0 | 0 | |
| Total | 60 | 18 | 17 | 11 | 46 | 0.77 |
PFGE analysis showed that the blaCTX-M-32-positive strains were genetically homogeneous, suggesting clonal dissemination of CTX-M-32-producing E. coli strains within the farm. In contrast, CTX-M-14-producing E. coli strains were genetically more heterogeneous, indicating that the spread of these strains may be attributed to a combination of both clonal expansion and horizontal transmission. Furthermore, a CTX-M-14-producing E. coli isolate from a farmer's hand was indistinguishable from an isolate from a feed sample, which was, in turn, very closely related to E. coli isolates from teats and calf feces (Fig. 1), suggesting clonal dissemination of specific clones of CTX-M-producing E. coli isolates among animals, humans, and the farm environment, which constitutes a potential public health threat, especially to farmers, ranchers, and animal handlers.
In conclusion, we identified and characterized CTX-M β-lactamase and associated addiction systems among E. coli isolates from animals, farmers, and the farm environment. Our data suggest that a combination of multiple addiction systems in plasmids carrying blaCTX-M genes could contribute to maintenance of these plasmids in their host cells.
ACKNOWLEDGMENT
This work was supported by a grant from the Animal, Plant, and Fisheries Quarantine and Inspection Agency, Ministry of Food, Agriculture, Forestry, and Fisheries, Republic of Korea.
Footnotes
Published ahead of print 12 April 2013
REFERENCES
- 1. Batchelor M, Threlfall EJ, Liebana E. 2005. Cephalosporin resistance among animal-associated enterobacteria: a current perspective. Expert Rev. Anti Infect. Ther. 3:403–417 [DOI] [PubMed] [Google Scholar]
- 2. Rossolini GM, D'Andrea MM, Mugnaioli C. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14:33–41 [DOI] [PubMed] [Google Scholar]
- 3. Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17:83–97 [DOI] [PubMed] [Google Scholar]
- 4. Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321 [DOI] [PubMed] [Google Scholar]
- 5. Marcade G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 63:67–71 [DOI] [PubMed] [Google Scholar]
- 6. Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499 [DOI] [PubMed] [Google Scholar]
- 7. Mnif B, Vimont S, Boyd A, Bourit E, Picard B, Branger C, Denamur E, Arlet G. 2010. Molecular characterization of addiction systems of plasmids encoding extended-spectrum β-lactamases in Escherichia coli. J. Antimicrob. Chemother. 65:1599–1603 [DOI] [PubMed] [Google Scholar]
- 8. Shin J, Kim DH, Ko KS. 2011. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 63:39–47 [DOI] [PubMed] [Google Scholar]
- 9. Tamang MD, Nam HM, Kim SR, Chae MH, Jang GC, Jung SC, Lim SK. 2013. Prevalence and molecular characterization of CTX-M β-lactamase-producing Escherichia coli isolated from healthy swine and cattle. Foodborne Pathog. Dis. 10:13–20 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100-S20-U Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Batchelor M, Hopkins K, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH, Liebana E. 2005. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 49:1319–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Branger C, Zamfir O, Geoffroy S, Laurans G, Arlet G, Thien HV, Gouriou S, Picard B, Denamur E. 2005. Genetic background of Escherichia coli and extended-spectrum β-lactamase type. Emerg. Infect. Dis. 11:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu Y, Ji S, Chen Y, Zhou W, Wei Z, Li L, Ma Y. 2007. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect. 54:53–57 [DOI] [PubMed] [Google Scholar]
- 14. Xiong Z, Li T, Xu Y, Li J. 2007. Detection of CTX-M-14 extended-spectrum beta-lactamase in Shigella sonnei isolates from China. J. Infect. 55:e125–e128. 10.1016/j.jinf.2007.07.017 [DOI] [PubMed] [Google Scholar]
- 15. Saladin M, Cao VT, Lambert T, Donay JL, Herrmann JL, Ould-Hocine Z, Verdet C, Delisle F, Philippon A, Arlet G. 2002. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161–168 [DOI] [PubMed] [Google Scholar]
- 16. Rayamajhi N, Kang SG, Lee DY, Kang ML, Lee SI, Park KY, Lee HS, Yoo HS. 2008. Characterization of TEM-, SHV- and AmpC-type beta-lactamases from cephalosporin-resistant Enterobacteriaceae isolated from swine. Int. J. Food Microbiol. 124:183–187 [DOI] [PubMed] [Google Scholar]
- 17. Tamang MD, Oh JY, Seol SY, Kang HY, Lee JC, Lee YC, Cho DT, Kim J. 2007. Emergence of multidrug-resistant Salmonella enterica serovar Typhi associated with a class 1 integron carrying the dfrA7 gene cassette in Nepal. Int. J. Antimicrob. Agents 30:330–335 [DOI] [PubMed] [Google Scholar]
- 18. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 19. Eckert C, Gautier V, Arlet G. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14–23 [DOI] [PubMed] [Google Scholar]
- 20. Brizio A, Vasco S, Goncalves AR, Lito LM, Cristino JM, Salgado MJ, Duarte A. 2006. Survey of extended-spectrum β-lactamases in Escherichia coli isolates from a Portuguese hospital and characterisation of a novel class 1 integron (In60A) carrying the blaCTX-M-9 gene. Int. J. Antimicrob. Agents 28:320–324 [DOI] [PubMed] [Google Scholar]
- 21. Gautom RK. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tian GB, Wang HN, Zou LK, Tang JN, Zhao YW, Ye MY, Tang JY, Zhang Y, Zhang AY, Yang X, Xu CW, Fu YJ. 2009. Detection of CTX-M-15, CTX-M-22, and SHV-2 extended-spectrum beta-lactamases (ESBLs) in Escherichia coli fecal-sample isolates from pig farms in China. Foodborne Pathog. Dis. 6:297–304 [DOI] [PubMed] [Google Scholar]
- 23. Ho PL, Chow KH, Lai EL, Lo WU, Yeung MK, Chan J, Chan PY, Yuen KY. 2011. Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to ‘critically important’ antibiotics among food animals in Hong Kong, 2008–10. J. Antimicrob. Chemother. 66:765–768 [DOI] [PubMed] [Google Scholar]
- 24. Duan RS, Sit THC, Wong SSY, Wong RCW, Chow KH, Mak GC, Ng LT, Yam WC, Yuen KY, Ho PL. 2006. Escherichia coli producing CTX-M β-lactamases in food animals in Hong Kong. Microb. Drug Resist. 12:145–148 [DOI] [PubMed] [Google Scholar]
- 25. Blanc V, Mesa R, Saco M, Lavilla S, Prats G, Miro E, Navarro F, Cortés P, Llagostera M. 2006. ESBL- and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet. Microbiol. 118:299–304 [DOI] [PubMed] [Google Scholar]
- 26. Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec JY. 2006. CTX-M-1- and CTX-M-15 type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int. J. Antimicrob. Agents 28:402–407 [DOI] [PubMed] [Google Scholar]
- 27. Madec JY, Lazizzera C, Châtre P, Meunier D, Martin S, Lepage G, Ménard MF, Lebreton P, Rambaud T. 2008. Prevalence of fecal carriage of acquired expanded-spectrum cephalosporin resistance in Enterobacteriaceae strains from cattle in France. J. Clin. Microbiol. 46:1566–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song W, Lee H, Lee K, Jeong SH, Bae IK, Kim JS, Kwak HS. 2009. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum β-lactamase in clinical isolates of Escherichia coli from Korea. J. Med. Microbiol. 58:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim SK, Lee HS, Nam HM, Jung SC, Bae YC. 2009. CTX-M-type beta-lactamase in Escherichia coli isolated from sick animals in Korea. Microb. Drug Resist. 15:139–142 [DOI] [PubMed] [Google Scholar]
- 30. Tamang MD, Nam HM, Jang GC, Kim SR, Chae MH, Jung SC, Byun JW, Park YH, Lim SK. 2012. Molecular characterization of extended-spectrum-β-lactamase-producing and plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob. Agents Chemother. 56:2705–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamang MD, Nam HM, Kim TS, Jang GC, Jung SC, Lim SK. 2011. Emergence of extended-spectrum β-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J. Clin. Microbiol. 49:2671–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cartelle M, Del Mar TM, Molina F, Moure R, Villanueva R, Bou G. 2004. High-level resistance to ceftazidime conferred by a novel enzyme, CTM-M-32, derived from CTX-M-1 through a single Asp240-Gly substitution. Antimicrob. Agents Chemother. 48:2308–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novais A, Canton R, Moreira R, Peixe L, Baquero F, Coque TM. 2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51:796–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee SG, Jeong SH, Lee H, Kim CK, Lee Y, Koh E, Chong Y, Lee K. 2009. Spread of CTX-M-type extended-spectrum β-lactamases among bloodstream isolates of Escherichia coli and Klebsiella pneumoniae from a Korean hospital. Diagn. Microbiol. Infect. Dis. 63:76–80 [DOI] [PubMed] [Google Scholar]
- 35. Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. 2012. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 67:878–885 [DOI] [PubMed] [Google Scholar]
- 36. Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. 2011. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J. Antimicrob. Chemother. 66:1263–1268 [DOI] [PubMed] [Google Scholar]
- 37. Valverde A, Canton R, Garcillan-Barcia MP, Novais A, Galan JC, Alvarado A, de la Cruz F, Baquero F, Coque TM. 2009. Spread of blaCTX-M-14 is mainly driven by IncK plasmids disseminated among A (ST10), B1 (ST155/ST359) and D Escherichia coli phylogroups in Spain. Antimicrob. Agents Chemother. 53:5204–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun Y, Zeng Z, Chen S, Ma J, He L, Liu Y, Deng Y, Lei T, Zhao J, Liu JH. 2010. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 16:1475–1481 [DOI] [PubMed] [Google Scholar]
- 40. Perichon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M. 2008. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob. Agents Chemother. 52:2581–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]