Abstract
Cryptococcus neoformans is an unconventional dimorphic fungus that can grow either as a yeast or in a filamentous form. To facilitate investigation of genetic factors important for its morphogenesis and pathogenicity, congenic a and α strains for a filamentous form were constructed. XL280 (α) was selected as the background strain because of its robust ability to undergo the morphological transition from yeast to the filamentous form. The MATa allele from a sequenced strain JEC20 was introgressed into the XL280 background to generate the congenic a and α pair strains. The resulting congenic strains were then used to test the impact of mating type on virulence. In both the inhalation and the intravenous infection models of murine cryptococcosis, the congenic a and α strains displayed comparable levels of high virulence. The a-α coinfections displayed equivalent virulence to the individual a or α infections in both animal models. Further analyses of the mating type distribution in a-α coinfected mice suggested no influence of a-α interactions on cryptococcal neurotropism, irrespective of the route of inoculation. Furthermore, deletion or overexpression of a known transcription factor, Znf2, in XL280 abolished or enhanced filamentation and biofilm formation, consistent with its established role. Overexpression of Znf2 in XL280 led to attenuation of virulence and a reduced abundance in the brain but not in other organs, suggesting that Znf2 might interfere with cryptococcal neurotropism upon extrapulmonary dissemination. In summary, the congenic strains provide a new resource for the exploration of the relationship in Cryptococcus between cellular morphology and pathogenesis.
INTRODUCTION
Cryptococcus neoformans is a human-pathogenic fungus that has a worldwide distribution in different environments. This fungus colonizes the lungs of susceptible individuals after being acquired from the environment through inhalation, and it can spread hematogenously to multiple organs. The predilection of Cryptococcus to infect the brain after extrapulmonary dissemination often leads to cryptococcal meningitis, a disease that claims more than half a million lives each year worldwide (1, 2). A monophyletic lineage, often referred to as the “Cryptococcus neoformans species complex,” comprises two species: C. neoformans and C. gattii. C. neoformans is responsible for the vast majority of cases of cryptococcosis and is further classified into var. grubii (serotype A), var. neoformans (serotype D), and their hybrid (serotype AD) (1, 3, 4). Both serotypes A and D have been extensively studied, in part due to the availability of robust molecular and genetic tools.
The Cryptococcus species are considered yeasts in their pathogenic form. However, other morphologies are reported within host tissues, including filamentous cells (pseudohyphae or hyphae) (5–10) and giant cells (11–13). This fungus also develops into a filamentous form during mating in vitro. Comparisons of strains reveal a wide range in their capabilities to undergo filamentation, both during conventional α-a mating and in a self-induced process called monokaryotic fruiting or unisexual mating. Research on different morphological forms has implicated positive roles for cell gigantism in disease (11–16) and negative roles for filaments in disease and conversely inducing a protective response from the host (17–22). Accordingly, the filamentous form is only occasionally observed in host tissues and is considered an atypical form for Cryptococcus in vivo (5–10). The association between morphogenesis and virulence in Cryptococcus is thus similar to that observed in other conventional dimorphic fungal pathogens, such as Histoplasma capsulatum and Blastomyces dermatitidis (23–26). However, strains of C. neoformans with a robust ability to undergo morphological transitions have been excluded from analyses, limiting our understanding of the relationship between morphology and developmental commitment as it relates to pathogenesis and sexual development.
An essential resource for any molecular biology research that utilizes genetics is congenic pairs, which are strains that are genetically identical, except for the mating type locus (MAT). The first and the widely used Cryptococcus congenic strains JEC21α and JEC20a, were constructed in the 1990s and sequenced in the early 2000s (27, 28). They were derived from two serotype D progenitor strains, NIH433 (a) and NIH12 (α) (27, 29) (Fig. 1). Both can cause fatalities in mice, with NIH12 displaying a higher level of virulence (30). However, JEC21α and JEC20a often fail to cause mortality in infected mice (31, 32), even with a high infection dose (1 × 107 cells/animal) and a long study period (32). Two other related serotype D congenic pairs, KN3501α/KN3501a and KN433α/KN433a (Fig. 1), also exhibit low levels of virulence in an intravenous infection model of murine cryptococcosis (33). Studying cryptococcal virulence with these serotype D congenic pairs often requires long study periods (40 to ∼>100 days), unnatural intravenous inoculation, and relatively high infection doses (1 × 106 to ∼1 × 107 cells/mouse) (27, 32–35).
Fig 1.
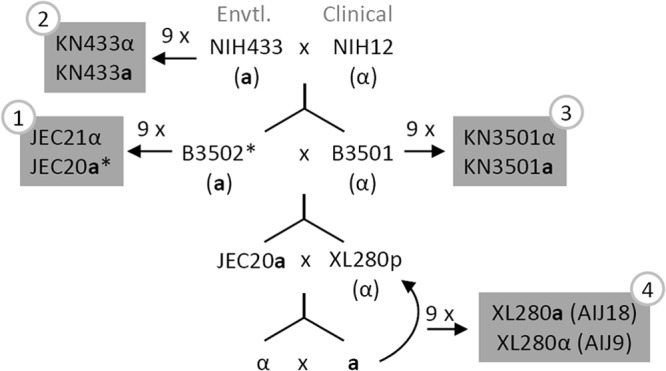
Crossing strategies used to generate the congenic strains XL280a and XL280α in this study and their relationship with other C. neoformans congenic pairs. NIH12 is a clinical isolate, and NIH433 is an environmental isolate (27, 29). B3501 and B3502 were generated from a cross between NIH12 and NIH433, and they were used to generate the first congenic pair, JEC21 and JEC20, in 1992 (27–29). Two other congenic pairs in the genetic background of NIH433 and B3501 were generated in 2005 (33). XL280p was generated previously by crossing B3501 and B3502 and was found to be hyperfilamentous on a variety of nutrient-limiting solid media (54). Based on next generation sequencing analysis, the genome sequence of XL280 is 81% identical to that of JEC21 (J. Heitman, personal communication). *, B3502 is nearly isogenic with JEC20 based on two previous studies (33, 58).
The construction of the H99 congenic pair strains (KN99α/KN99a) facilitated genetic analyses in the serotype A background. Examination of cryptococcal virulence with H99-derived strains permits lower inocula (1 × 103 to ∼1 × 105), shorter study periods (≤40 days), and the more natural inhalation route of infection (32, 36–38). However, rapid progression of the disease in H99-infected mice may present a challenge to detect a potential protective effect on animal survival of some drugs, including the commonly used antifungal fluconazole (39, 40). Thus, it would be valuable to have a well-characterized strain with a modestly lower virulence than H99.
One contentious topic in cryptococcal research is the impact of the mating type on fungal virulence. This question originated from the observations that the Cryptococcus population is almost exclusively composed of the α mating type (1, 41, 42). However, during the a-α bisexual matings, a and α yeast cells fuse, initiate filamentous growth, and generate equal numbers of a and α spores (29, 43–46). As there is no apparent growth advantage associated with the α mating type, several hypotheses have been proposed to explain the rise and the preservation of this largely unisexual α population that include (i) monokaryotic fruiting and (ii) mating type contribution to virulence.
Monokaryotic fruiting is a process to produce hyphae and spores by Cryptococcus cells of only one mating type (47–50). This is in contrast to bisexual mating, where α and a partners are required for filamentation and sporulation. Monokaryotic fruiting is observed under laboratory conditions and is also believed to occur in nature based on population genetics studies (31, 51, 52). Indeed, self-filamentous cryptococcal cells are observed occasionally even in hosts with cryptococcosis (5–10). Although isolates of both mating types possess the ability to undergo monokaryotic fruiting (49, 53), the mating type locus is a quantitative trait locus in which the α allele significantly enhances fruiting (54). It is proposed that the enhanced ability of α isolates to produce hyphae and spores might have tilted the cryptococcal population in favoring of the α mating type (51, 54). NIH12, one progenitor for the current serotype D congenic pairs (Fig. 1), is highly self-filamentous. However, none of the derived congenic pair strains undergoes fruiting, except for the stochastic fruiting observed in JEC21α. Nonetheless, B3502a derived from a cross between NIH433 and NIH12 is self-filamentous, and so are many progeny derived from a cross between B3501α and B3502a (54). However, a congenic pair in a self-filamentous genetic background has not been constructed because of the technical challenges of dissecting spores generated from the bisexual mating rather than from fruiting (29).
The impact of mating type on cryptococcal virulence appears to be strain dependent (29, 32, 33). For instance, JEC21α is more virulent than its congenic pair strain JEC20a (27, 32), and KN433α is more virulent than KN433a (33), while there was no difference in virulence levels between the congenic KN3501α and KN3501a strains (33). KN99α and KN99a exhibit no difference in virulence when they infect mice individually in the inhalation model of cryptococcosis, but KN99α appears to be more efficient in colonizing the brain during mixed a-α coinfection (33, 37). In contrast, the recently constructed C. gattii congenic pair strains AIR265α and AIR265a are similarly virulent in mouse models, and neither of the mating types confers any competitive advantages during in vitro coculture or during a-α coinfection in mice, regardless of the route of inoculation (intranasal or intravenous), as noted in the accompanying article by Zhu et al. (55). One plausible reason to account for the observed differences between strains is that factors located outside the mating type locus are involved in controlling both virulence and mating behavior (27).
In this study, we used a series of backcrosses to generate a pair of congenic α and a strains in the highly self-filamentous XL280 background. We characterized the behaviors of the congenic strains in vitro and in two mouse models of cryptococcosis and further examined the impact of mating type on virulence and tissue tropism during a-α coinfection. Furthermore, we tested the function of a known regulator, Znf2, in the XL280 background in cellular and colony development and in mediating fungal virulence. This new strain set fills a missing resource for exploring morphology and pathogenesis in C. neoformans.
MATERIALS AND METHODS
Strains, crossing, and isolation of meiotic progeny.
Strains XL280p (α) and JEC20 (a) were used as the starting parental strains for crosses made in this study. XL280p was generated previously from a cross between two serotype D strains B3501 (α) and B3502 (a) (54). JEC20a is congenic with the sequenced strain JEC21α (27, 29, 56). Strains were maintained on yeast extract-peptone-dextrose (YPD) medium. For long-term storage, strains were saved in 15% glycerol stocks at −80°C. Crosses were set up by mixing yeast cells of α and a mating partners together on V8 agar plates (5% V8 juice, 0.5 g/liter KH2PO4, 4% agar, with pH adjusted to 7 with KOH). Because self-filamentation during the fruiting process on V8 media initiates later than the dikaryotic filamentation during a-α bisexual matings and because sporulation during fruiting is much less efficient than sporulation during bisexual matings, we chose to dissect the spores generated within 3 to 7 days to avoid complications due to sporulation generated from fruiting. Filaments formed on the edges and tops of the mated yeast colonies, and basidiospores were transferred to YPD plates. Spores were micromanipulated with a dissecting microscope, and their mating type was determined by successful mating of their derived colonies with either JEC20a or JEC21α. The mating type of a few randomly selected progeny was also tested by the presence or absence of the mating type-specific genes STE20α and STE20a as previously described (51, 57). The results obtained by both approaches were consistent (see Fig. S1 in the supplemental material).
Molecular markers.
The PCR-restriction fragment length polymorphism (RFLP) molecular markers used in this study were originally designed based on single nucleotide polymorphisms (SNPs) identified between B3501α and B3502a (nearly isogenic with JEC20a) (54, 58). Thirteen pairs of primers selected to include one on most chromosomes of C. neoformans were tested on strains JEC20a and XL280p (see Table S1 in the supplemental material). Eight of the combinations were different between these two parental strains, while five were identical, consistent with the close relationship between these two strains (Fig. 1). These eight markers were also used to test the final congenic pair, XL280α and XL280a. The PCR amplicons were digested with the appropriate restriction enzymes, and fragments were resolved on 1.4% agarose–1× Tris-acetate-EDTA gels.
Pulsed-field gel electrophoresis.
Chromosomal DNA was profiled by contour-clamped homogeneous electrical field (CHEF) electrophoresis. The conditions for preparation of chromosomal DNA and its resolution followed methods described previously (58). DNA was separated in a CHEF-DR III system (Bio-Rad) using the following conditions: block 1 was 75 to 150 s of switching with 4 V/cm at 12°C for 24 h, followed by block 2, which was 200 to 400 s of switching with 4 V/cm at 12°C for 24 h.
In vitro phenotypic assays of the congenic strains and the parental strains.
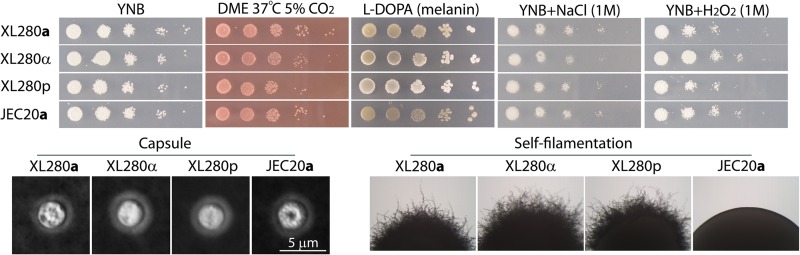
The in vitro phenotypic assays were performed as previously described (31, 35). Briefly, yeast cells were grown on YPD medium overnight and washed three times with water. Cells were suspended in distilled water, and the cell density was determined by measuring the optical density at 600 nm. Then cells were 10× serially diluted. To characterize capsule production, equal numbers of C. neoformans cells were transferred to Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, CA) and grown for 3 days at 37°C under 5% CO2. The appearance of a mucoid colony indicates capsule production. The capsule was visualized under light microscopy as a white halo surrounding the yeast cell in India ink due to the exclusion of ink particles. To examine melanin production, cells were spotted onto melanin-inducing medium containing l-dihydroxyphenylalanine (l-DOPA) (100 mg/liter) (59) and incubated at 22°C in the dark for 6 days. Melanization was observed as the colonies developed a brown color. To analyze growth on a minimal medium, cells were spotted onto yeast nitrogen base (YNB) medium. To analyze resistance to osmotic and oxidative stresses, cells were grown on YNB medium supplemented with 1 M of NaCl or 1 mM H2O2, respectively.
The segregation of znf2 mutations in the XL280 strain background.
The ZNF2 gene, encoding a zinc finger transcription factor that controls filamentation and biofilm formation, was replaced by the nourseothricin acetyltransferase (NAT) marker gene (60). The ZNF2oe strain was generated by expressing ZNF2 under the control of the promoter of the GPD1 gene linked with the G418 resistance marker in the znf2Δ mutant background (17). These mutants in the XL280p (α) background were crossed with XL280a to isolate the mutations in the MATa background. Dissected progeny were examined for their mating type by crossing with JEC21α or JEC20a on V8 medium, their resistance to the drug NAT and/or G418 by growing them on plates containing YPD plus NAT plus G418 (YPD+NAT/G418 plates), their ability to undergo self-filamentation on V8 medium, and their ability to form a complex colony morphology (which correlates with biofilm formation) (19, 61–64) on YNB medium. The ratios of these phenotypes in the progeny indicated Mendelian segregation of the mating type and each drug resistance marker. It also indicated the linkage between Znf2 mutations and their corresponding phenotypes.
Virulence assays in two mouse models of cryptococcosis.
Mouse studies were performed basically as described previously (19, 40). The animal models of systemic cryptococcosis were induced by two routes: intravenous and intranasal infection. In each infection model, 9 or 10 8- to 10-week-old female A/J mice (Jackson Laboratory) were infected by each Cryptococcus strain and were considered one group. All animals per group were used for the survival study, and five or six animals per group were used to examine the fungal burden. For the intravenous infection, each mouse was challenged with 1.0 × 106 fungal cells suspended in 50 μl of saline. For the intranasal infection, animals were first sedated with ketamine and xylazine, and then 1.0 × 106 fungal cells (or 5 × 105 and 5 × 106 cells for the pilot experiment) suspended in 50 μl of saline were slowly inoculated into the left nostril of sedated animals. After infection, animals were weighed daily and monitored twice a day for disease progression, including weight loss, gait changes, labored breathing, or fur ruffling. Moribund mice were sacrificed, and their organs were dissected. For the intranasal infection model, the lungs, brain, spleen, and the left kidney from terminated mice were harvested. For the intravenous infection model, the brain, spleen, and the left kidney were harvested. The dissected organs were homogenized in 2 ml of cold phosphate-buffered saline (PBS) buffer using an IKA* Ultra-Turrax T18 homogenizer with the same setting for each type of organ. The tissue suspensions were serially diluted (10×), plated onto YNB agar medium, and incubated at 30°C for 2 days such that the colonies became visible to count CFU.
For the examination of the impact of znf2 mutations in the XL280 background on cryptococcal virulence, animals were infected by the wild type, the znf2Δ mutant, and the ZNF2oe strain intranasally with the inoculum of 1.0 × 106 fungal cells per animal as described above. Because the PGPD1-ZNF2 strain exhibits heterogeneity in cell morphology and its population is a mixture of yeast cells and filamentous cells, only cells in the yeast form were used for animal inoculation to obtain an accurate inoculation and to avoid potential problems caused by differences in cell types at initial infection as we described previously (19). The PGPD1-ZNF2 culture with a mixed morphotype was centrifuged briefly at a low speed to allow the enrichment of yeast cells on the top. The top layer was then centrifuged again at a higher speed, and yeast cells were collected for infection.
Examination of cellular morphology in vitro and during infection.
To examine cell morphology under culture conditions that are relevant to host physiology, strains XL280α, XL280a, and the mixture of equal numbers of XL280α and XL280a cells were inoculated into fetal bovine serum and RPMI liquid medium with a final cell density of 1 × 105 cells/ml. Cells cultured in rich YPD liquid medium under the same condition were used as controls. The cells were incubated at 37°C under 5% CO2. Photographs of the cells were taken after 3 days of incubation.
To examine cellular morphology of XL280 during infection, animals were infected with XL280 at the dose of 5 × 105 cells per mouse intranasally as described above. Moribund mice were sacrificed, and lungs, brains, and kidneys dissected from the sacrificed animals were fixed in 10% formalin, embedded in paraffin, sectioned at a thickness of 5 μm, and stained with Gomori methenamine silver (GMS) as previously described (19). Fungal cell morphology was then examined microscopically.
In vitro growth competition assay between the congenic a and α strains.
The congenic strains XL280a and XL280α were grown in YPD liquid medium at 30°C overnight and then washed three times with sterile water. The cell density of the suspension of each strain was determined based on hemocytometer counting. XL280a and XL280α cells of equal numbers were then mixed and inoculated into liquid DMEM to the final concentration of 1 × 105 cells/ml. The initial inocula were confirmed by plating the XL280a and XL280α cell suspension prior to the mixing onto YNB medium, and their CFU were counted after colonies became visible. The a-α coculture in liquid DMEM was incubated at 37°C under 5% CO2. Aliquots of the in vitro coculture were removed at days 3 and 9 of incubation and plated onto YNB plates with serial dilutions. Single colonies were randomly picked and examined for their mating type through crossing with reference strains JEC20a and JEC21α separately on V8 juice agar medium at 22°C in the dark. These single colonies by themselves were included as the negative control. The crosses were examined after 2 to 3 days. Crosses with abundant hyphal formation and sporulation compared to the tested colony alone were scored as successfully mating with the reference strain.
Congenic a and α coinfection in two mouse models of cryptococcosis.
For a-α coinfection experiments, mice were challenged with a 1:1 ratio of XL280a and XL280α cells (1 × 106 total fungal cells per animal) intranasally or intravenously. The a and α cell numbers of the initial mixed inocula were confirmed by measuring the CFU of serial dilutions. The ratio of a to α cells in the original inocula was confirmed based on the CFU of a and α cell suspensions prior to the mixing. The organs of sacrificed mice were dissected and processed as described earlier. To determine the ratio of the a to α cells in various organs, single colonies recovered from each organ dissected from terminated animals were scored for their mating type by crossing with the reference strains JEC20a and JEC21α as described above. In total, more than 9,000 matings were performed and screened to examine the mating type distribution in different organs.
Statistical analysis.
Statistical significance of the survival data between different groups was assessed by the Gehan-Breslow Wilcoxon test. The one-way analysis of variance (ANOVA) tests were used in the fungal burden studies. Fisher's exact test was used to analyze the distribution of the a and α isolates during the in vitro coculture competition. All statistical analyses were performed using the Graphpad Prism 5 program, with P values lower than 0.05 considered statistically significant.
Animal ethics statement.
The mouse experiments were performed according to the guidelines of the National Institutes of Health and the Texas A&M University Institutional Animal Care and Use Committee (IACUC). The protocol number is 2011-22.
RESULTS
Generation of a congenic pair of strains in a C. neoformans filamentous background by a series of backcrosses.
We chose to use XL280 as the genetic background to generate a pair of congenic α and a strains that can undergo fruiting for the following three reasons. (i) XL280 was an α progeny isolated from a cross between the nonfilamentous B3501 strain and a filamentous B3502 strain (50, 54) (Fig. 1). It is one of the most robust fruiting strains examined, and it can self-filament on a variety of nutrient-poor media (54). (ii) A genetic map and the genome sequences for XL280's parental strains B3501 and B3502 (near isogenic with JEC20a) are available (48, 53). The genome of XL280 itself has been sequenced (J. Heitman, personal communication). (iii) In the past few years since its creation, XL280 has already been used by several laboratories in a variety of studies (19, 50, 54, 60, 65–67). Here we refer to this strain as XL280p to distinguish it from the congenic pair strains that will be described later.
We selected JEC20a as the donor for the a mating type locus for the congenic a strain because sequences of its mating type locus are available. Furthermore, JEC20a is closely related to XL280p, and the contribution of this particularly mating type allele in virulence has been tested in three other strain backgrounds (Fig. 1). A progeny with an a mating type from a cross between XL280p and JEC20a was selected at random and then backcrossed to XL280p. After an additional 9 rounds of backcrosses between a randomly selected a progeny with XL280p, we obtained the strain pair XL280a (AIJ18) and XL280α (AIJ9) (Fig. 1; see Fig.S2 in the supplemental material). During backcrossing, we noticed that in multiple generations α progeny in general underwent relatively more robust fruiting than a progeny (see Fig. S3 in the supplemental material). This observation is consistent with the previous study showing that the α mating type allele is one of the most significant quantitative trait loci associated with enhanced self-filamentation (54).
XL280p and JEC20a are siblings from crosses between B3501 and B3502. Hence, XL280p and JEC20a already share a large proportion of their genetic material based on their pedigrees (27, 29, 56). A comparison of the genome sequences of XL280p and JEC21α indicates that their genomes are 81% identical (J. Heitman, personal communication). Thus, after the cross between XL280p and JEC20a and 9 backcrosses to XL280p, in theory both XL280a and XL280α should be 99.98% identical to the parental strain XL280p, with the exception of the mating type locus in XL280a. Consistently, when the 8 polymorphic markers between XL280p and JEC20a were tested on XL280α and XL280a, all of their alleles were identical to those of XL280p (Fig. 2A), indicating that the genetic material from XL280p has been successfully incorporated into the congenic pair strains during the bisexual a-α backcrosses (a × XL280p). A comparison of the chromosomal profiles of these strains using CHEF gel analysis indicates identical chromosomal profiles between XL280p and the congenic pair of strains (Fig. 2B). These results suggest no gross chromosomal rearrangement or insertions or deletions of large regions during the backcrosses. The results are consistent with the stable phenotypes and genotypes observed in the related serotype D strains previously generated (Fig. 1). Thus, XL280α and XL280a are considered congenic, differing at the mating type locus.
Fig 2.
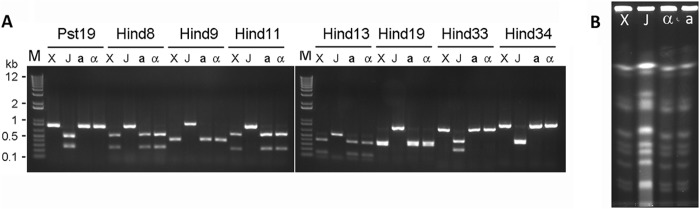
Genetic typing of the XL280 congenic pair strains. (A) After 10 backcrosses, both congenic strains XL280α (α) and XL280a (a) contain all of the alleles from XL280p (X) rather than JEC20a (J). The molecular marker segregation indicates that genetic materials from XL280p were transferred to the congenic pairs during backcrosses. DNA regions of the four strains were amplified by PCR, the products were digested with the diagnostic restriction enzyme, and the fragments were resolved on agarose gels. M, Invitrogen 1 Kb+ markers, with a subset of sizes indicated in kilobase pairs. (B) CHEF analyses of chromosomal DNA from the congenic pair strains XL280α (α) and XL280a (a), XL280p (X), and JEC20a (J) indicate identical chromosomal profiles between the congenic pair and XL280p. The MATa donor JEC20a shows a different chromosomal profile.
We further examined the phenotypes of the congenic pair strains XL280α and XL280a in vitro. As shown in Fig. 3, the congenic pair strains and their parental strains, XL280p and JEC20a, are all prototrophic, capable of growing on minimum YNB media. All of the strains grew well and produced capsule visible under a light microscope when incubated on DMEM at 37°C under 5% CO2, a condition that is relevant to the host physiology. All strains produced melanin on l-DOPA medium and showed a similar level of resistance toward NaCl and H2O2. Thus, no drastic difference was detected among the strains with respect to these in vitro phenotypes. One apparent exception is their ability to undergo fruiting: JEC20a is nonfilamentous, XL280a produces filaments profusely, and XL280α and XL280p are much more robust in self-filamentation (Fig. 3).
Fig 3.
In vitro phenotypic assays of the parental strains XL280p and JEC20a and the two congenic strains XL280α and XL280a. Yeast cells of each strain were grown in liquid YPD medium overnight, and serial dilutions (10-fold) of cells were spotted onto different media. Cells were grown on minimal YNB medium at 22°C for 3 days as a control for growth (first column from the left); cells were grown on DMEM (DME) at 37°C under 5% CO2 to assay growth under host-relevant conditions (2nd column) and capsule production (images below); cells were grown on the l-DOPA medium at 22°C for 6 days (3rd column) to assay melanin production (brown pigment); cells were grown on YNB supplemented with 1 M NaCl or with 1 mM H2O2 to assay resistance to osmotic stress (4th column) or oxidative stress (last column). Self-filamentation of these strains on the L-DOPA medium is shown below.
XL280p is virulent to mice and it grows as yeast during infection.
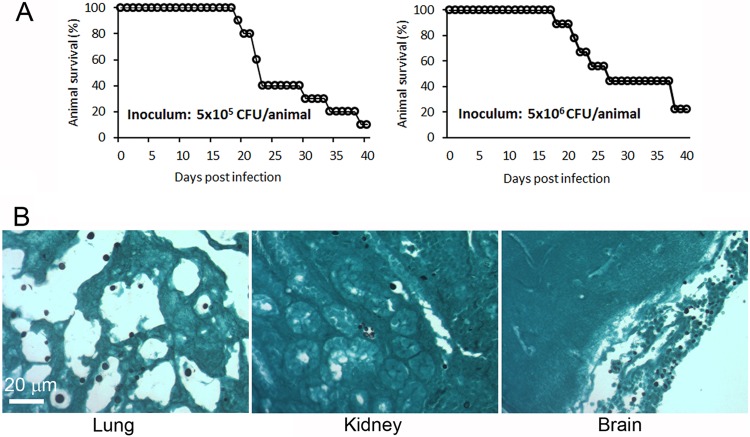
We performed pilot experiments to determine if XL280p is virulent to mice. We tested XL280p in the inhalation model with two infection doses (5 × 105 and 5 × 106 CFU per animal) in two independent experiments and monitored animal survival for 40 days after the infections. As shown in Fig. 4A, most animals succumbed to XL280p infection within 40 days postinfection in both pilot experiments and the 10× difference in inoculum did not appear to have any drastic effect on animal survival. This is similar to what has been observed in the serotype A strain H99, where lowering the infection inoculum by 100-fold results in only a short delay in causing mortality in mice both in the inhalation infection model and in the intravenous infection model (37, 40).
Fig 4.
XL280p is virulent in the inhalation infection model of murine cryptococcosis, and it grows as yeast during infection. (A) Each mouse was inoculated intranasally with 5 × 105 or 5 × 106 fungal cells of XL280p, and animal survival was monitored for 40 days. (B) Mice were inoculated with 5 × 105 fungal cells intranasally. Lungs, brains, and kidneys were dissected from sacrificed moribund mice, fixed, and processed for GMS staining to reveal fungal cell morphology in the organs (shown in black).
Although XL280p and its congenic pair strains have been observed to grow as yeast in various liquid media and in rich solid media (Fig. 3; see Fig. S4 in the supplemental material), these strains can grow in the filamentous form on solid substrates in vitro under a variety of nutrient-limiting conditions (Fig. 3) (54). To ascertain the cell morphology of XL280p during infection, we performed histological examination of the lungs (the initial site of infection), kidneys (a dissemination site), and brains (the site of fatal infection) of the XL280p-infected mice. As shown in Fig. 4B, XL280p was in the yeast form in all of the organs examined. Thus, like most clinical isolates, XL280p does not generate hyphae in vivo, where it grows as yeast.
The congenic pair strains show similar levels of virulence in two mouse models of cryptococcosis.
To examine the virulence of the congenic strains, we used the well-established inhalation and intravenous infection models of murine cryptococcosis. We chose to use the intermediate inoculum of 1 × 106 fungal cells per animal for both models and included XL280p in the comparative virulence assays to determine if the virulence potential of the congenic pair differs from that of their background parental strain. The congenic pair strains and XL280p caused 100% fatalities in infected mice within 6 days after being introduced into animals intravenously (Fig. 5B) and ∼90 to 100% animal mortality rates within 40 days postinfection after being introduced into animals via the respiratory route (Fig. 5A). The XL280 congenic strains are thus much more virulent than other serotype D reference strains, which typically require 30 to 100 days at a similar or higher dose to cause significant mortality in mice, even when these strains are introduced to animals intravenously (11, 27, 32, 39).
Fig 5.
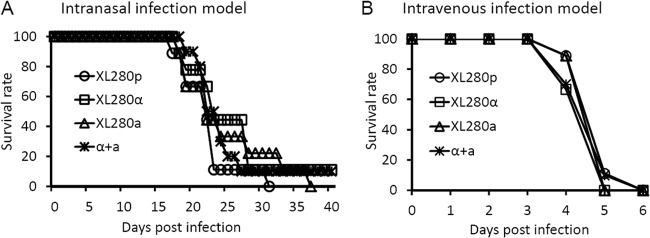
The congenic strains XL280α and XL280a have equal virulence to each other and the parental XL280p strain individually or coinfected. (A) Mice were inoculated with 1 × 106 fungal cells intranasally, and their survival was monitored for 40 days. There was no difference observed in survival rates among the strains. The P values were 0.833 (XL280p/XL280α), 0.252 (XL280p/XL280a), 0.984 (XL280p/a + α), 0.075 (XL280α/XL280a), 0.754 (XL280α/a + α), and 0.053 (XL280a/a + α). (B) Mice were inoculated with 1 × 106 fungal cells intravenously, and there was no difference among these strains. The P values were 0.317 (XL280p/XL280α), 0.317 (XL280p/XL280a), 1 (XL280p/a + α), 1 (XL280α/XL280a), 0.317 (XL280α/a + α), and 0.317 (XL280a/a + α).
At the inoculum used in these experiments, we did not observe any difference in rates of animal mortality caused by the parental strain XL280p and the congenic strains XL280α and XL280a in either the inhalation infection model (Fig. 5A) or the intravenous infection model (Fig. 5B). The comparable levels of virulence exhibited by the congenic XL280α and XL280a strains in these two animal models exclude the mating type locus as one major factor influencing cryptococcal overall virulence in this genetic background under the tested conditions. The high similarity between XL280p and XL280α both in vitro and in vivo is consistent with the predicted isogenicity between these two strains.
The a-α coinfection in the inhalation infection model of murine cryptococcosis.
One interesting phenomenon of the C. neoformans serotype A congenic pair strains KN99a and KN99α is that the α strain tends to dominate in cerebrospinal fluid (CSF) during mixed a-α coinfection in the inhalation infection model, even though KN99a and KN99α are equivalent in virulence when used in infection individually (37, 68). No a-α coinfection experiment has been performed for other congenic pairs, except the recently constructed C. gattii AIR265α and AIR265a strains. AIR265α and AIR265a are similar in virulence when tested individually in both the inhalation and the intravenous infection models of murine cryptococcosis (55). Furthermore, neither the α nor the a mating type allele confers any apparent advantages in terms of mortality or neurotropism in the AIR265 background during a-α coinfection in either model of murine cryptococcosis (55).
Here, we tested the impact of potential interactions between a and α cells on virulence in the inhalation infection model using the coinfection of the congenic pair strains. As shown in Fig. 5A, the XL280a and XL280α coinfection exhibited similar dynamics in causing animal fatality as either a or α strain infection alone in the inhalation infection model. Next, we examined the distributions of the a and α mating types in various organs during a-α mixed coinfection. We chose the unmarked XL280a and XL280α strains for this experiment to preclude the addition of any potential variation by the introduction of genetic markers to the mating type locus, as was employed in the previous study (68). We first performed an in vitro experiment to determine if there is any proliferation advantage conferred by one mating type when the a and α cells are cocultured together under a condition that is host physiologically relevant. We inoculated a mixture of a and α cells into the mammalian cell culture medium DMEM at 37°C under 5% CO2 at a 1:1 ratio (51.5% a versus 48.5% α; n = 822). The a/α ratio in the coculture was measured again after 3 and 9 days of incubation. At day 3 postinoculation, there were 44.2% a cells and 55.8% α cells (n = 95); at day 9 postinoculation, there were 46.5% a cells and 53.5% α cells (n = 127). Thus, the a/α ratio maintained a 1:1 level in the coculture and there was no statistically significant difference among the time points examined (P = 0.273). Therefore, neither the a nor the α mating type allele appears to confer any apparent growth advantage in the coculture under this in vitro condition.
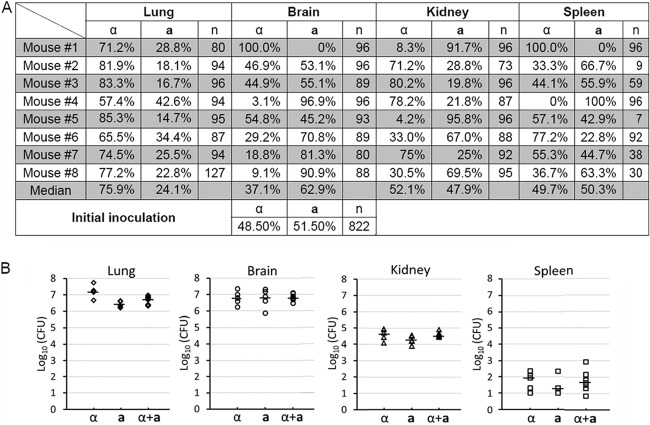
We then examined the mating type distribution in eight terminated animals infected by the a-α mixture (Fig. 6A). We randomly picked ∼96 colonies from each of the 32 organs (lungs, brains, kidneys, and spleens of the 8 animals) and determined their mating type. We could not perform as many tests for some spleens due to the low numbers of fungal cells recovered (Fig. 6B). As shown in Fig. 6A, there were uniformly more α cells than a cells recovered from lungs of all eight animals examined. The median percentile for α cells in the lungs was ∼76%, and that for a cells was 24%. This uniform overrepresentation of α cells in the lungs during a-α coinfection likely reflects the better proliferation of α cells in the lungs, as there was higher lung fungal burden in animals infected by α cells alone than in those infected by a cells alone at the time of termination (Fig. 6B). In contrast to the uniform overrepresentation of α cells in the lungs during the coinfection, there was no consistent pattern of mating type distribution in other organs. In the brain, numbers of a and α cells were roughly equal in 3 out of 8 mice, with a cells dominating in 4 mice and α cells dominating in 1 mouse. In the kidney, there was an even split between α and a cells, with a cells dominating in 4 mice and α cells dominating in the remaining 4 mice. A similar even split between a and α cells was also observed in the spleen. The different pattern of the a and α cell distributions in these organs during a-α coinfection is consistent with the organ fungal burden. Here, animals infected by the α strain showed a higher fungal burden in the lungs, but their fungal burdens in the brain, kidney, and spleen showed no statistically significant difference from those infected by a cells or the a-α mixture (Fig. 6B). It is not clear why higher proliferation of α cells in the lungs did not cause higher fungal burden in other organs and why overrepresentation of the α mating type in the lungs during the coinfection did not generate any obvious ripple effect in other organs since these organs were infected after extrapulmonary dissemination.
Fig 6.
The mating type distribution during a-α coinfection in the inhalation infection model of cryptococcosis. (A) Animals were challenged with the mixture of a-α fungal cells (1 × 106/animal) by inhalation. At the time of termination, lungs, brains, kidneys, and spleens from eight animals per group were dissected and homogenized. The mating type of randomly picked Cryptococcus cells recovered from each organ was determined. (B) Animals were challenged with 1 × 106 fungal cells (α, a, or the a-α mixture) intranasally. At the time of termination, lungs, brains, kidneys, and spleens from five animals per group were dissected and homogenized. Serial dilutions of the homogenized tissue were plated, and CFU were used to determine the organ fungal burden. The short, bold, horizontal lines indicate the median. The fungal burden of these groups was statistically different in the lungs (P = 0.001). No statistically significant difference was found among the groups in the brain (P = 0.973), the kidney (P = 0.110), or the spleen (P = 0.575).
The a-α coinfection in the intravenous infection model of murine cryptococcosis.
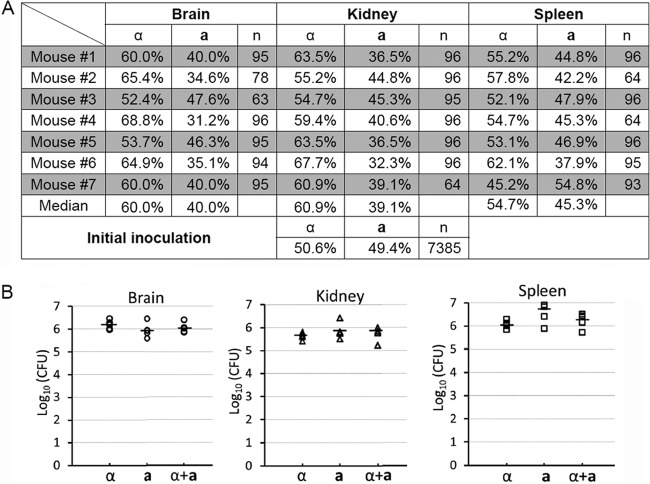
We decided to examine further the outcome of a-α coinfection in the intravenous infection model, whereby the pulmonary infection is bypassed. As shown in Fig. 5B, the a-α coinfection exhibited similar dynamics in causing animal fatality as the infection by either the a or the α strain alone in this intravenous infection model. Here we tested the mating type of fungal cells recovered from brains, kidneys, and spleens of seven mice infected by the a-α mixture (21 organs in total). As shown in Fig. 7A, there were slightly more α cells than a cells in the lungs in all seven mice examined. A similar slightly higher representation of α was also uniformly observed in other two organs in all seven mice. The median α/a ratios were 6:4 in the brain, 6:4 in the kidney, and 5.5:4.5 in the spleen. It is not clear why the minor advantages in amplification of α cells during coinfection in this intravenous model did not cause any apparent differences in terms of virulence among the α, a, or α-a infections (Fig. 5B). We speculate that rapid progression of the disease caused by these intravenous infections (death within 6 days after infection) could potentially render such modest advantages not important for the animal's survival. Consistent with this speculation, the fungal burdens in all organs examined at 5 days postinoculation were all high (typically ∼106 CFU/organ in the brain, kidney, and spleen), and there were no statistically significant differences among the α, a, or a-α mixture groups in this intravenous model used in this study (Fig. 7B). Neither of the mating type alleles appeared to confer any apparent advantage in neurotropism during individual infection or coinfection under the tested conditions (Fig. 7B).
Fig 7.
The mating type distribution during a-α coinfection in the intravenous infection model of cryptococcosis. (A) Animals were challenged with the mixture of a-α fungal cells (1 × 106) intravenously. At the time of termination, brains, kidneys, and spleens from seven animals per group were dissected and homogenized. The mating type of randomly picked Cryptococcus cells recovered from each organ was determined. (B) Animals were challenged with 1 × 106 fungal cells (α, a, or the a-α mixture) intravenously. At day postinfection 5 and also the time of termination, brains, kidneys, and spleens from five animals per group were harvested. Serial dilutions of the homogenized tissue were plated, and CFU were used to determine the organ fungal burden. The short, bold, horizontal lines indicate the median. No statistically significant difference was found among the groups in the brain (P = 0.370), the kidney (P = 0.410), or the spleen (P = 0.070).
Znf2 is a common factor that governs morphogenesis and mediates fungal ability to cause diseases in the varieties of Cryptococcus neoformans.
As XL280 can easily undergo the yeast-filament morphological transition on its own under appropriate culture conditions, XL280 could be particularly useful in facilitating the research into the factors that control morphogenesis and virulence in Cryptococcus. The association between morphological transition and fungal virulence is well known in several major fungal pathogens, such as Candida albicans, B. dermatitidis, and H. capsulatum (23–26, 69, 70). The existence of morphotype-associated pathogenicity is also observed in C. neoformans (18, 19, 60, 71). Although the physical aspects of morphogenesis may themselves be one important factor controlling virulence, alterations in cell surface molecules during morphogenesis could change the interaction between the pathogen and the host and consequently shape the outcome of fungal infections (17, 72–75).
We decided to examine a known factor that controls morphogenesis to assess its impact on virulence in the XL280 background. We chose to investigate the zinc finger transcription factor Znf2, as it was previously demonstrated to govern the yeast-filament transition in C. neoformans (18, 19, 60). In addition, this regulator governs the formation of complex colony morphology (biofilm) through its control of the expression of multiple cell surface proteins, including the adhesion protein Cfl1 (19).
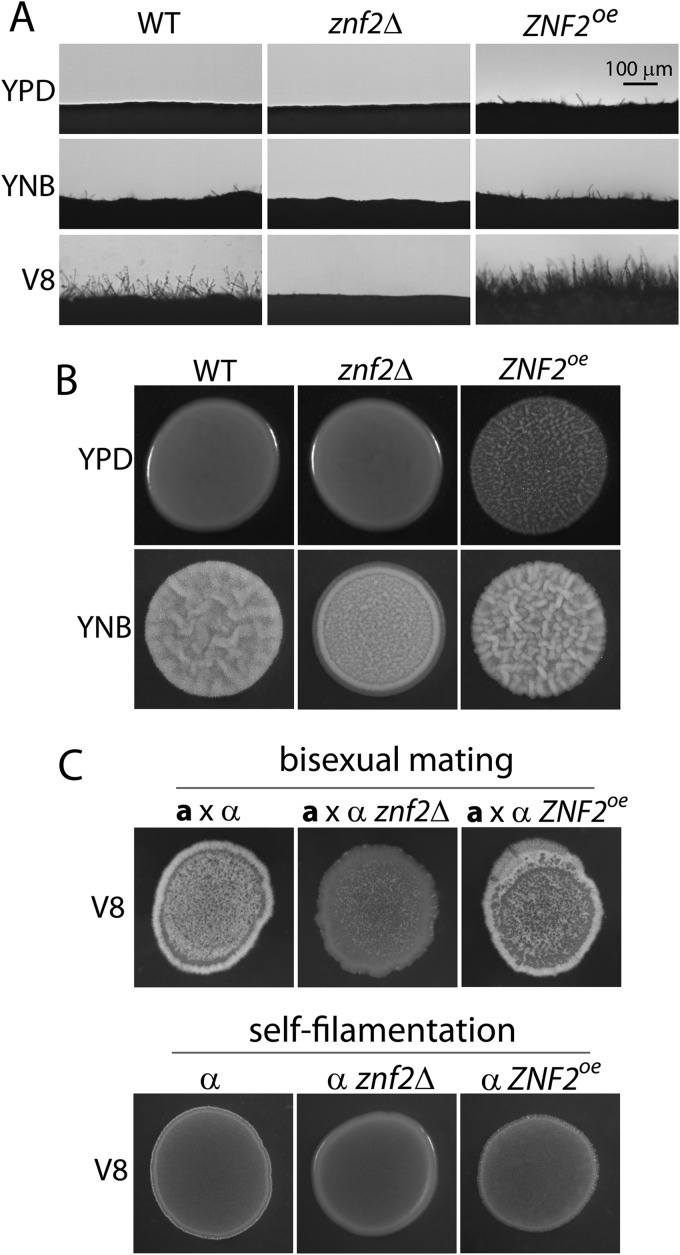
To examine the role of Znf2 in the morphotype transition in the XL280 background, we inoculated yeast cells of the wild-type strain, the znf2Δ mutant, and the ZNF2oe strain (PGPD1-ZNF2 in the znf2Δ mutant background) on YPD, YNB, and V8 media and examined their self-filamentation. As expected, the wild-type strain did not filament on the rich YPD medium, produced rudimentary filaments on the YNB medium, and generated robust filamentation on the V8 juice medium (Fig. 8A). The deletion of ZNF2 abolished the ability of XL280 to produce any filaments under all conditions, while the overexpression of ZNF2 conferred more robust filamentation on both YNB and V8 media and enabled XL280 to produce some filaments under the otherwise suppressive conditions of the YPD medium (Fig. 8A). Thus, Znf2 directs self-filamentation in the XL280 background. It has been shown previously that filamentation during bisexual matings between a znf2Δ mutant and a wild-type partner is significantly reduced due to a dose effect, and mating hypha production is completely abolished in crosses involving both znf2Δ mutant partners in the JEC21α/JEC20a or KN99α/KN99a background (60). A similar phenotype was also observed when ZNF2 was disrupted in XL280 (Fig. 8C). Disruption of ZNF2 in both partners in the XL280 background also completely eliminated production of mating hyphae (data not shown). Taken together, Znf2 is required for filamentation derived from either monokaryotic fruiting (self-filamentation) or a-α bisexual matings. In addition to its fundamental role in filamentation, Znf2 also controls biofilm formation in XL280 as indicated by the increasingly complex colony morphology with increased ZNF2 expression (znf2Δ mutant < wild type < ZNF2oe strain) (Fig. 8B). This phenotype is consistent with previous observations made in the serotype A H99 background (19).
Fig 8.
Znf2 controls filamentation and the formation of complex colony morphology in XL280 background. (A) The wild type, the znf2Δ mutant, and the ZNF2oe strain with the inoculum were cultured on complete YPD medium, minimum YNB medium, and V8 juice medium at 22°C in the dark for 2 days. Photographs of the colony edge were shown to display filamentation (fluffy edge) or yeast cells (smooth edge). (B) The wild type, the znf2Δ mutant, and the ZNF2oe strain with the inoculum were cultured on YPD and YNB media at 22°C in the dark for 4 days. The wrinkled colony indicates the complex colony morphology. (C) The nonfilamentous strain JEC20a was mated with the wild type, the znf2Δ mutant, and the ZNF2oe α strains on V8 juice agar medium. The mating mixtures were cultured at 22°C in the dark for 9 days before the photographs were taken. The white fluffy phenotype reflects aerial hyphal production in the mating colony. The cultures of the wild type, the znf2Δ mutant, and the ZNF2oe α strains without a mating partner are shown below for comparison.
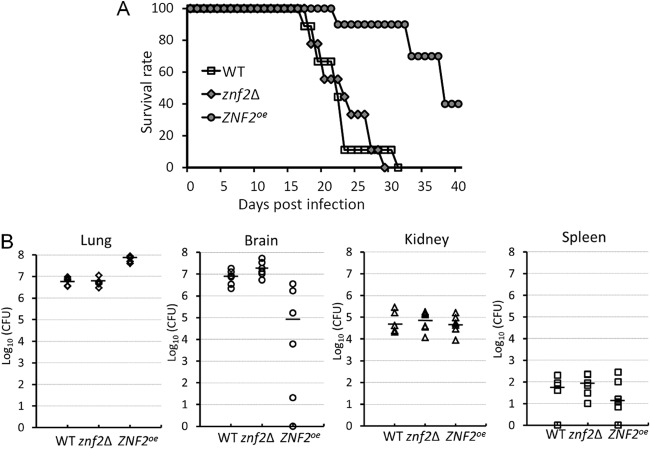
Previously we showed that the expression level of Znf2 is inversely linked to the ability of strain H99 to cause fatal diseases in the inhalation model of murine cryptococcosis. That is, the ZNF2oe strain in an H99 background was completely avirulent, while the znf2Δ mutant was slightly more virulent than the wild-type H99 strain (19, 60). Here we tested the wild type, the znf2Δ mutant, and the ZNF2oe strain in the XL280 background in the inhalation model of murine cryptococcosis to assess the role of this regulator in virulence. As shown in Fig. 9A, the overexpression of ZNF2 resulted in a significant attenuation in virulence. However, the reduction in virulence of the ZNF2oe strain is not as dramatic in XL280 as that in H99 (19, 60). We could not detect any statistically significant difference between the wild type and the znf2Δ mutant. This result is not unexpected given that the znf2Δ mutant is only slightly more virulent than the wild type in the H99 background, and the effect of Znf2 mutations on virulence in XL280 might be less drastic.
Fig 9.
Znf2 mediates the fungal ability to cause fatal diseases in the murine inhalation model of cryptococcosis. (A) Mice were inoculated with 1 × 106 fungal cells intranasally, and survival was monitored for 40 days. There was no statistically significant difference between the wild type and the znf2Δ mutant. The P values were 0.07 for WT/znf2Δ, and <0.001 for WT/ZNF2oe and ZNF2oe/znf2Δ. (B) Mice were inoculated with 1 × 106 fungal cells intranasally. At the time of termination, lungs, brains, kidneys, and spleens from six animals per group were dissected and homogenized. Serial dilutions of the homogenized tissue were plated, and CFU were used to determine the organ fungal burden. The short, bold, horizontal lines indicate the medians. No colonies were recovered from a few dissected organs even with all the suspended tissue plated. Because the y axis is log10 of the CFU, “n/a” (not applicable) was used to show them as zero on the bottom of the y axis. The fungal burdens among the wild type, the znf2Δ mutant, and the ZNF2oe strain were statistically different in the lungs (P < 0.001) and the brain (P = 0.004). There was no statistically significant difference in the kidney (P = 0.874) or in the spleen (P = 0.442).
The ZNF2oe strain in the H99 background is avirulent, and it did not disseminate to the brain tissue in the inhalation infection model of murine cryptococcosis (19). However, it was not clear if the absence of brain fungal burden was an indirect effect due to the low fungal burden in the lungs or reflects defective extrapulmonary dissemination of the ZNF2oe strain to the brain (19). Here we examined the fungal burden of the lungs, brains, kidneys, and spleens of the mice infected by the wild-type strain, the znf2Δ mutant, and the ZNF2oe strain in the XL280 background at the time of their termination. As shown in Fig. 9B, the fungal burden in the lungs was high in mice infected by all of the strains, with the ZNF2oe strain showing a fungal burden more than 10-fold higher than that of the wild-type strain or the znf2Δ mutant. In contrast, the presence of the ZNF2oe strain in the brain was about 100-fold lower with larger variations (median, ∼105 CFU/organ) than the wild type, while the znf2Δ mutant showed a slightly higher fungal burden in the brain (>107 CFU/organ) (Fig. 9B). The lower burden of the ZNF2oe strain in the brain was unlikely a result of poor dissemination from the lungs, as there was no difference observed among all of the strains in the kidney and the spleen (Fig. 9B). The results suggest that Znf2 might have a role in interfering with extrapulmonary dissemination to the brain specifically. Further investigation is needed to examine the role of Znf2 and its downstream factors in mediating cryptococcal neurotropism during infection.
DISCUSSION
In this study, we used a series of backcrosses to generate a pair of congenic α and a strains in the highly self-filamentous XL280 background. We characterized the behaviors of the congenic strains in vitro and in two mouse models of cryptococcosis and found that they showed similar phenotypes both in vitro and in vivo. The one obvious exception was better self-filamentation observed in the α strain, which is consistent with previous research showing that the mating type is a quantitative trait locus controlling self-filamentation (54). Furthermore, we tested the function of a known master regulator of morphogenesis, Znf2, in the XL280 background and found that Znf2 plays predicted roles in filamentation, biofilm formation, and mediation of fungal virulence. As XL280 has been used in a variety of studies to investigate morphogenesis, sporulation, and cryptococcal interaction with various hosts in the past few years (19, 50, 54, 60, 65–67), the congenic pair strains generated from the present study, in combination with its recently sequenced genome (J. Heitman, personal communication), will enable XL280 to be a useful model for future analysis of morphogenesis and virulence in C. neoformans.
XL280 was derived from the clinical isolate NIH12, which can also undergo robust monokaryotic fruiting. The related congenic pairs constructed previously were purposely made in genetic backgrounds that do not self-filament or do so poorly (Fig. 1) (27). Among these congenic pairs, the pair JEC21α/JEC20a has been used most widely. However, all of the serotype D congenic strains, including JEC21α/JEC20a, display low levels of virulence in mouse models. Investigation of fungal virulence in these backgrounds requires high inocula and often intravenous inoculation (29). Here we found that the animal survival curves produced by XL280 and its congenic pair strains with an inoculum of 1 × 106 in both the inhalation and the intravenous infection models are similar to what we and others observe for the serotype A strain H99 with an inoculum of 1 × 105 (19, 37, 40). Although there is no direct comparison to other serotype D strains, XL280 congenic pair strains are likely much more virulent than their related serotype D strains in murine models (Fig. 1) (32–35). Based on the pedigree of these serotype D congenic pairs (Fig. 1), JEC21α/JEC20a, KN3501α/KN3501a, and XL280α/XL280a in theory should all harbor approximately 50% of the overall genetic background derived from each of the NIH12 and NIH433 genomes. The differences between these pairs in terms of morphogenesis and virulence likely derive from allelic combinations generated by recombination events during meiosis.
We noticed a different pattern of fungal proliferation in various organs in the inhalation model and the intravenous model of murine cryptococcosis at the time of animal termination. The high fungal burden observed in the lungs in the inhalation model is expected, as is the preferential proliferation in the brain over other organs (e.g., kidney or spleen) in this model, given the well-known neurotropism of cryptococcal infections (Fig. 6B). In sharp contrast, the fungal presence in all organs examined was uniformly high in the intravenous infection model and there was no apparent tissue tropism (Fig. 7B). These observations indicate that the neurotropism of cryptococcal infections is apparent upon extrapulmonary dissemination, but not with direct infusion of fungal cells into the blood. Further supporting evidence of the difference of these two models comes from the study of the mating type distribution in various organs during a-α coinfection. When mixed a and α cells were introduced into animals intravenously, there was nondiscriminating slightly higher representation of α in all organs and in all animals examined, likely reflecting slightly better proliferation of the α cells in vivo. In contrast, although α cells uniformly dominated the lungs in all animals when mixed a and α cells were introduced into animals through the respiratory tract, a and α showed showed equal predominance in the kidney and the spleen, whereas a cells might even have a slightly better chance than α cells in dominating the brain. Thus, extrapulmonary dissemination appears to be a critical step in determining cryptococcal tissue tropism.
It is possible that for Cryptococcus cells to disseminate successfully from lungs, specific alterations in their cells, such as cell morphology (size and shape) and cell surface composition, might have to happen (14–17, 19). These alterations might render a higher affinity of these Cryptococcus cells to the brain. For instance, it is known that capsule structure differs at different stages of infection and in different organs, and capsule and cell size change during organ invasion (76–78). It is conceivable then that certain changes associated with pulmonary extrusion may better assist cryptococcal invasion of the brain. Alternatively, Cryptococcus can disseminate through a Trojan horse mechanism, and its neurotropism could be derived indirectly from the affinity between the host cells hijacked by Cryptococcus and the brain. For instance, monocytes have been shown to assist in cryptococcal dissemination and brain invasion (79). The identification of cryptococcal factors and host factors that affect cryptococcal neurotropism could help in testing these hypotheses.
XL280 congenic pair strains might be adept in regulating cell size in response to host factors. We noticed, for example, that their cell sizes increased when cells were incubated in serum at 37°C under 5% CO2 (see Fig. S4 in the supplemental material). It is known that cell size regulation is important for cryptococcal virulence (14–16), and it is not clear if this or other traits of XL280 contribute to its heightened virulence compared to those of other related serotype D strains. We also noticed that the XL280-infected mice more often showed apparent neurological disorders when they became moribund, while the H99-infected mice more often displayed severe pulmonary stress. In accordance with this observation, the fungal burden in lungs is typically 1 order higher than that in the brain in H99-infected mice (68, 80). In contrast, the fungal burden in the brain is at the same order as that in the lungs in XL280-infected mice (Fig. 6B and 9B). For comparison, the Cryptococcus gattii reference strain R265 predominantly amplifies in the lungs and very little in the brain (55, 80). Further investigation into the molecular mechanisms underlying the strain or serotype-specific differences in neurotropism is warranted. It is also interesting to note that overexpression of Znf2 in XL280 led to drastically lower fungal burden in the brain, even though it was present at higher numbers in the lungs and comparable levels in the kidney and spleen to those in the wild type (Fig. 9B). Thus, Znf2 may play a role in interfering with cryptococcal dissemination specifically to the brain after pulmonary extrusion. Given that Znf2 is a transcription factor and its regulon is highly enriched with cell surface proteins (75), it would be interesting to examine surface molecules altered by Znf2 and to investigate their roles in cryptococcal tissue tropism in the future.
In conclusion, the congenic α and a strains constructed in the genetic background of the highly filamentous strain XL280 are highly virulent, making them a preferred system for both cryptococcal morphogenesis and virulence studies and enabling the uses of this species complex to understand the genetic basis for dimorphic transitions in pathogenic fungi.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Chinese Scholarship Council (to P.Z.), the American Heart Association (grant 0BGIA3740040 to X.L.), the Norman Hackerman Advanced Research Program (grant 01957 to X.L.), and National Institutes of Health NIAID grants (AI097599 to X.L. and AI094364 to A.I.).
Footnotes
Published ahead of print 13 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00259-13.
REFERENCES
- 1. Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 2. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 3. Kwon-Chung KJ, Varma A. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6:574–587 [DOI] [PubMed] [Google Scholar]
- 4. Lin X, Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60:69–105 [DOI] [PubMed] [Google Scholar]
- 5. Gazzoni AF, Severo CB, Barra MB, Severo LC. 2009. Atypical micromorphology and uncommon location of cryptococcosis: a histopathologic study using special histochemical techniques (one case report). Mycopathologia 167:197–202 [DOI] [PubMed] [Google Scholar]
- 6. Freed ER, Duma RJ, Shadomy HJ, Utz JP. 1971. Meningoencephalitis due to hyphae-forming Cryptococcus neoformans. Am. J. Clin. Pathol. 55:30–33 [DOI] [PubMed] [Google Scholar]
- 7. Lurie HI, Shadomy HJ. 1971. Morphological variations of a hypha-forming strain of Cryptococcus neoformans (Coward strain) in tissues of mice. Sabouraudia 9:10–14 [DOI] [PubMed] [Google Scholar]
- 8. Staib F, Mishra SK, Grosse G. 1977. Further observations about selective involvement of the brain in experimental cryptococcosis, p 169–176 In Iwata K. (ed), Recent advances in medical and veterinary mycology. University Park Press, Baltimore, MD [Google Scholar]
- 9. Shadomy HJ, Utz JP. 1966. Preliminary studies on a hypha-forming mutant of Cryptococcus neoformans. Mycologia 58:383–390 [PubMed] [Google Scholar]
- 10. Williamson JD, Silverman JF, Mallak CT, Christie JD. 1996. Atypical cytomorphologic appearance of Cryptococcus neoformans: a report of five cases. Acta Cytol. 40:363–370 [DOI] [PubMed] [Google Scholar]
- 11. Neill JM, Abrahams I, Kapros CE. 1950. A comparison of the immunogenicity of weakly encapsulated and of strongly encapsulated strains of Cryptococcus neoformans (Torula histolytica). J. Bacteriol. 59:263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cruickshank JG, Cavill R, Jelbert M. 1973. Cryptococcus neoformans of unusual morphology. Appl. Microbiol. 25:309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Love GL, Boyd GD, Greer DL. 1985. Large Cryptococcus neoformans isolated from brain abscess. J. Clin. Microbiol. 22:1068–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. 10.1371/journal.ppat.1000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953. 10.1371/journal.ppat.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, Nielsen K. 2012. Titan cell production enhances the virulence of Cryptococcus neoformans. Infect. Immun. 80:3776–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L, Lin X. 2012. Morphogenesis in fungal pathogenicity: shape, size, and surface. PLoS Pathog. 8:e1003027. 10.1371/journal.ppat.1003027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect. Genet. Evol. 9:401–416 [DOI] [PubMed] [Google Scholar]
- 19. Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 8:e1002765. 10.1371/journal.ppat.1002765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fromtling RA, Blackstock R, Hall NK, Bulmer GS. 1979. Kinetics of lymphocyte transformation in mice immunized with viable avirulent forms of Cryptococcus neoformans. Infect. Immun. 24:449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fromtling RA, Blackstock R, Hall NK, Bulmer GS. 1979. Immunization of mice with an avirulent pseudohyphal form of Cryptococcus neoformans. Mycopathologia 68:179–181 [DOI] [PubMed] [Google Scholar]
- 22. Fromtling RA, Blackstock R, Bulmer GS. 1980. Immunization and passive transfer in immunity in murine cryptococcosis, p 122–124 In Kuttin ES, Baum GL. (ed), Human and animal mycology: Proceedings of the VII Congress of ISHAM Excerpta Medica, New York, NY [Google Scholar]
- 23. Klein BS, Tebbets B. 2007. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 10:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nemecek JC, Wuthrich M, Klein BS. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588 [DOI] [PubMed] [Google Scholar]
- 25. Nguyen VQ, Sil A. 2008. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc. Natl. Acad. Sci. U. S. A. 105:4880–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Webster RH, Sil A. 2008. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc. Natl. Acad. Sci. U. S. A. 105:14573–14578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwon-Chung KJ, Edman JC, Wickes BL. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D'Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJ, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler KB, Maiti R, Marra MA, Marra RE, Mathewson CA, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heitman J, Allen B, Alspaugh JA, Kwon-Chung KJ. 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28:1–5 [DOI] [PubMed] [Google Scholar]
- 30. Kwon-Chung KJ, Hill WB. 1981. Sexuality and pathogenicity of Filobasidiella neoformans (Cryptococcus neoformans), p 243–250 In Vanbreusighem R, DeVroy C. (ed), Sexuality and pathogenicity of fungi. Masson, New York, NY [Google Scholar]
- 31. Lin X, Litvintseva A, Nielsen K, Patel S, Kapadia Z, Floyd A, Mitchell TG, Heitman J. 2007. αADα hybrids of Cryptococcus neoformans: evidence of same sex mating in nature and hybrid fitness. PLoS Genet. 3:e186. 10.1371/journal.pgen.0030186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barchiesi F, Cogliati M, Esposto MC, Spreghini E, Schimizzi AM, Wickes BL, Scalise G, Viviani MA. 2005. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J. Infect. 51:10–16 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen K, Marra RE, Hagen F, Boekhout T, Mitchell TG, Cox GM, Heitman J. 2005. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics 171:975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469–485 [DOI] [PubMed] [Google Scholar]
- 35. Lin X, Nielsen K, Patel S, Heitman J. 2008. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect. Immun. 76:2923–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177–194 [PMC free article] [PubMed] [Google Scholar]
- 37. Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Litvintseva AP, Mitchell TG. 2009. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect. Immun. 77:3188–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhai B, Lin X. 2013. Evaluation of the anticryptococcal activity of the antibiotic polymyxin B in vitro and in vivo. Int. J. Antimicrob. Agents 41:250–254 [DOI] [PubMed] [Google Scholar]
- 40. Zhai B, Wu C, Wang L, Sachs MS, Lin X. 2012. The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob. Agents Chemother. 56:3758–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwon-Chung KJ, Bennett JE. 1978. Distribution of alpha and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337–340 [DOI] [PubMed] [Google Scholar]
- 42. Hull CM, Heitman J. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557–615 [DOI] [PubMed] [Google Scholar]
- 43. Kwon-Chung KJ. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197–1200 [PubMed] [Google Scholar]
- 44. McClelland CM, Chang YC, Varma A, Kwon-Chung KJ. 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 12:208–212 [DOI] [PubMed] [Google Scholar]
- 45. Alspaugh JA, Perfect JR, Heitman J. 1998. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet. Biol. 25:1–14 [DOI] [PubMed] [Google Scholar]
- 46. Kwon-Chung KJ. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821–833 [PubMed] [Google Scholar]
- 47. Todd RL, Herrmann WW. 1936. The life cycle of the organism causing yeast meningitis. J. Bacteriol. 32:89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erke KH. 1976. Light microscopy of basidia, basidiospores, and nuclei in spores and hyphae of Filobasidiella neoformans (Cryptococcus neoformans). J. Bacteriol. 128:445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wickes BL, Mayorga ME, Edman U, Edman JC. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc. Natl. Acad. Sci. U. S. A. 93:7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021 [DOI] [PubMed] [Google Scholar]
- 51. Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 5:e1000283. 10.1371/journal.ppat.1000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bui T, Lin X, Malik R, Heitman J, Carter D. 2008. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, α mating type populations. Eukaryot. Cell 7:1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tscharke RL, Lazera M, Chang YC, Wickes BL, Kwon-Chung KJ. 2003. Haploid fruiting in Cryptococcus neoformans is not mating type alpha-specific. Fungal Genet. Biol. 39:230–237 [DOI] [PubMed] [Google Scholar]
- 54. Lin X, Huang JC, Mitchell TG, Heitman J. 2006. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genet. 2:e187. 10.1371/journal.pgen.0020187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu P, Zhai B, Lin X, Idnurm A. 2013. Congenic strains for the genetic analysis of virulence traits in Cryptococcus gattii. Infect. Immun. 81:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heitman J, Casadevall A, Lodge JK, Perfect JR. 1999. The Cryptococcus neoformans genome sequencing project. Mycopathologia 148:1–7 [DOI] [PubMed] [Google Scholar]
- 57. Lengeler KB, Cox GM, Heitman J. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marra RE, Huang JC, Fung E, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. 2004. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167:619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chaskes S, Tyndall RL. 1978. Pigment production by Cryptococcus neoformans and other Cryptococcus species from aminophenols and diaminobenzenes. J. Clin. Microbiol. 7:146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 6:e1000953. 10.1371/journal.pgen.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stovicek V, Vachova L, Palkova Z. 2012. Yeast biofilm colony as an orchestrated multicellular organism. Commun. Integr. Biol. 5:203–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zara S, Bakalinsky AT, Zara G, Pirino G, Demontis MA, Budroni M. 2005. FLO11-based model for air-liquid interfacial biofilm formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schembri MA, Dalsgaard D, Klemm P. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186:1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Voordeckers K, De Maeyer D, van der Zande E, Vinces MD, Meert W, Cloots L, Ryan O, Marchal K, Verstrepen KJ. 2012. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 86:225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hsueh YP, Xue C, Heitman J. 2009. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J. 28:1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee SC, Heitman J. 2012. Function of Cryptococcus neoformans KAR7 (SEC66) in karyogamy during unisexual and opposite-sex mating. Eukaryot. Cell 11:783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77:4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK, Jr, Giles SS, Mitchell TG, Casadevall A, Perfect JR, Heitman J. 2005. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 73:4922–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu H, Kohler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 70. Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 71. Magditch DA, Liu TB, Xue C, Idnurm A. 2012. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog. 8:e1002936. 10.1371/journal.ppat.1002936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seider K, Heyken A, Luttich A, Miramon P, Hube B. 2010. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr. Opin. Microbiol. 13:392–400 [DOI] [PubMed] [Google Scholar]
- 73. Kanetsuna F, Carbonell LM. 1971. Cell wall composition of the yeastlike and mycelial forms of Blastomyces dermatitidis. J. Bacteriol. 106:946–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rappleye CA, Eissenberg LG, Goldman WE. 2007. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. U. S. A. 104:1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gantner BN, Simmons RM, Underhill DM. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Garcia-Hermoso D, Dromer F, Janbon G. 2004. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect. Immun. 72:3359–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chretien F, Lortholary O, Kansau I, Neuville S, Gray F, Dromer F. 2002. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J. Infect. Dis. 186:522–530 [DOI] [PubMed] [Google Scholar]
- 78. Charlier C, Chretien F, Baudrimont M, Mordelet E, Lortholary O, Dromer F. 2005. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am. J. Pathol. 166:421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. 2009. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect. Immun. 77:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ. 2012. The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3(3):e00103–12. 10.1128/mBio.00103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.