Abstract
Human cytomegalovirus (CMV) is a significant contributor to morbidity and mortality in immunocompromised patients, particularly in the transplant setting. The availability of anti-CMV drugs has improved treatment, but drug resistance is an emerging problem. Here, we describe an improved, rapid, sequencing-based assay for the two genes in CMV where drug resistance occurs, the UL97 and UL54 genes. This assay is performed in 96-well format with a single master mix and provides clinical results within 2 days. It sequences codons 440 to 645 in the UL97 gene and codons 255 to 1028 in the UL54 gene with a limit of detection of 240 IU/ml. With this assay, we tested 43 specimens that had previously been tested for UL97 drug resistance and identified 3 with UL54 mutations. One of these patients had no concurrent UL97 mutation, pointing toward the need for an assay that facilitates dual UL97/UL54 gene testing for complete resistance profiling.
INTRODUCTION
Human cytomegalovirus (CMV) causes morbidity and mortality in immunocompromised patients, including transplant and HIV-infected patients (1). The first-line drug therapy for CMV infection is ganciclovir (GCV) or its prodrug valganciclovir (VGCV). GCV or VGCV must be activated by phosphorylation before they act on human CMV. This phosphorylation is carried out by the viral kinase UL97, and activated GCV subsequently inhibits the viral DNA polymerase UL54. Clinically, GCV resistance usually arises first from a UL97 mutation resulting in decreased accumulation of the activated drug. Subsequent UL54 mutations can confer high levels of GCV resistance and various degrees of cross-resistance to the second-line drugs cidofovir (CDV) and foscarnet (FOS) (2).
Our group previously published a rapid PCR- and sequencing-based detection method for UL97 mutations conferring ganciclovir resistance (3). This test, currently used to detect mutations directly from clinical specimens, results in the sequence for UL97 gene codons 440 to 645. This region covers the known drug resistance mutation sites in the UL97 gene. However, because UL54 is the target of all currently marketed anti-CMV drugs, a detection method for UL54 mutations is also needed. Polymerase mutations are more likely to emerge after prolonged GCV treatment and typically add to the level of resistance conferred by UL97 mutations or introduce cross-resistance to CDV or FOS (4).
Despite its predicted utility in treatment management, clinical tests for UL54 drug resistance mutations have been slower to develop for a number of reasons. First, the coding sequence of UL54 is almost twice as long as that of UL97. In addition, the baseline sequences are more variable and the number and variety of resistance mutations are greater in the UL54 gene than in the UL97 gene. Polymerase genotypic testing therefore requires more-extensive sequencing, typically covering codons 300 to 1,000. To meet the clinical need for complete CMV drug resistance profiles, we adapted our previous UL97 sequencing method (3) to develop a single assay that amplifies six regions (2 in the UL97 gene and 4 in the UL54 gene) and is performed rapidly in a 96-well format with a single master mix. While other UL54 gene sequencing assays have been published (5–9), our method has the advantage of being performed in a 96-well format with a single master mix, without the need for multiple rounds of PCR, thus minimizing the potential for error when setting up 6 PCRs and 12 sequencing reactions per patient sample.
MATERIALS AND METHODS
PCR.
Four optimized primer sets were designed to amplify four regions of interest (ROI) within UL54 (Fig. 1 and Table 1). The forward primer for ROI 3 was adapted from Hantz et al. (5). The UL97 primer sets published by our laboratory for the previous UL97-only method were used in this protocol (3). The primer sets were aliquoted into separate wells for each region of interest and dried down into plates (10 min at 95°C) to facilitate the use of a single master mix for all regions.
Fig 1.
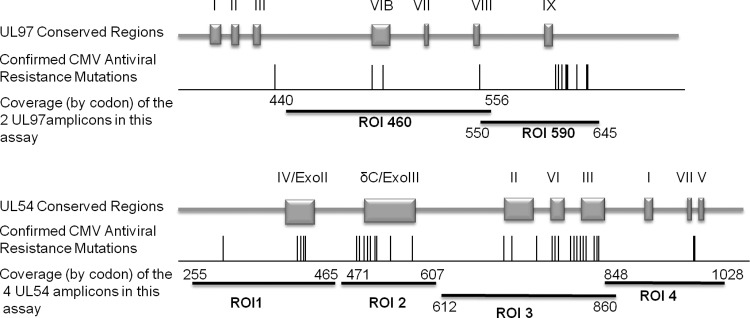
Schematic of CMV UL97 and UL54 and the regions of interest amplified by this assay. Adapted from reference 4 with permission.
Table 1.
UL54 primers used in this study
| UL54 ROI | Direction | Sequence | Sequence coverage (codons) |
|---|---|---|---|
| 1 | Forward | GGT GCT CCG TGA ATC GTT AC | 255–465 |
| Reverse | GTG AGA AGC CGA GGG AAA GG | ||
| 2 | Forward | CGG CCG CCA CCA AGG TGT ATA TTG | 471–607 |
| Reverse | GCA CCG TCG TAC CTT TGC TGT AG | ||
| 3 | Forward | CGT TGC TGT GTC ACC TAA CG | 612–860 |
| Reverse | AAC ACG GCT CTG AAA A G/A TTG | ||
| 4 | Forward | CGC GGT TCA TCA AAG ACA AC | 848–1028 |
| Reverse | CAC GCC GTA TTT CTT GAC TT |
Amplification by these primers was performed on a StepOne plus instrument (Life Technologies, Carlsbad, CA), using the Roche FastStart high-fidelity PCR system, with the inclusion of SYBR green to allow real-time monitoring of the accumulation of the PCR product. The UL97 and UL54 ROI 1, 2, and 4 primers were used at a final concentration of 400 nM. The ROI 3 primers were used at a concentration of 600 nM. The ROI 3 reverse primer was created by combining two primers to achieve a G/A mix at the 17th base of the primer. The program for cycling was as follows: denaturation at 95°C for 2 min, followed by 45 cycles of 95°C for 20 s, 55°C for 30 s, and 72°C for 30 s. The final elongation was done at 72°C for 4 min, with cooling at 37°C for 1 min prior to a melt curve stage of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The melt curve was included in the cycling conditions as a quality control measure. If a single peak was observed at the appropriate temperature, the amplification was considered successful and the PCR product was sequenced. The melting temperatures (°C) for each region are as follows: UL97 460, 86.2; UL97 590, 87.1; ROI 1, 84.2, ROI 2, 83.8; ROI 3, 85.4; and ROI 4, 86.2. Negative reactions resulted in melting temperatures between 63 and 78°C. The resulting amplicons were sequenced in both the forward and reverse direction, using the same primers used for amplification, utilizing the ABI Prism BigDye version 3.1 terminator cycle sequencing kit (0.5 μl BigDye, 0.75 μl reaction buffer, 0.12 μl 0.4 μM primer, 2.38 μl distilled H2O, 1.25 μl PCR product cleaned with ExoSap-It enzyme [Life Technologies, Carlsbad, CA]) according to the manufacturer's directions on an Applied Biosystems 3730XL DNA analyzer, and the sequence was analyzed using Applied Biosystems SeqScape software.
Sensitivity and extraction.
The sensitivity of the assay was determined by analyzing an Accrometrix CMVtc panel (Life Technologies, Carlsbad, CA) containing intact, encapsulated viral particles of CMV strain AD169. The panel consisted of a normal human plasma (tested nonreactive for CMV DNA) and four viral dilutions over a 10-fold concentration gradient from 240 to 240,000 IU/ml. For each concentration, 1 ml was extracted and eluted to 100 μl on a Roche MagnaPure LC using the large-volume total nucleic acid extraction kit (Roche Diagnostics, Basel, Switzerland). PCR was performed as described above, and a 1.5% Tris-borate-EDTA (TBE) agarose gel stained with ethidium bromide was loaded with 10 μl PCR product and 4 μl loading dye and run at 120 V to assess product formation at different template concentrations. The PCR products from the lowest dilution were also analyzed by sequencing. Additionally, two serially diluted patient samples were analyzed to assess the sensitivity on clinical specimens.
Frequency of UL54 mutations in clinical specimens.
The combined UL97/UL54 method was validated against the previous UL97-only method by testing 43 retrospective patient plasma samples from 41 patients. The UL54 results were compared to previous reference laboratory test results when available. Mutations were also confirmed by performing sequencing in both the forward and reverse directions. A sample exchange with an outside reference laboratory (ARUP Laboratories, Salt Lake City, UT) was also performed to validate the results for the eight specimens with leftover sample available. These samples were residual samples after routine clinical testing for UL97 resistance from 2005 to 2012, stored frozen at −20°C. Clinical data were collected for each of the 41 patients, with approval from the University of Washington Internal Review Board.
RESULTS
Sensitivity.
The sensitivity of the combined UL97/UL54 method, amplifying 2 regions of UL97 (3) and 4 regions of UL54 (Fig. 1 and Table 1), was assessed with an Accrometrix four-member, 10-fold CMV dilution panel. After PCR amplification, gel electrophoresis showed that the assay successfully amplified the lowest dilution of CMV (240 IU/ml) in all UL97 and UL54 regions (Fig. 2). Furthermore, the PCR products from the lowest dilution were successfully sequenced. Our laboratory's conversion factor for IU/ml to copies/ml puts the limit of detection of this assay around 1,000 copies/ml or better, similar to the sensitivity of the previous, UL97-only method (3). The assay was repeated on 2-fold serial dilutions of two patient samples, and sequencing was successfully performed on the PCR product from the 780- or 390-copies/ml dilutions for all 6 amplicon regions on both patient samples (see Fig. S1 in the supplemental material).
Fig 2.
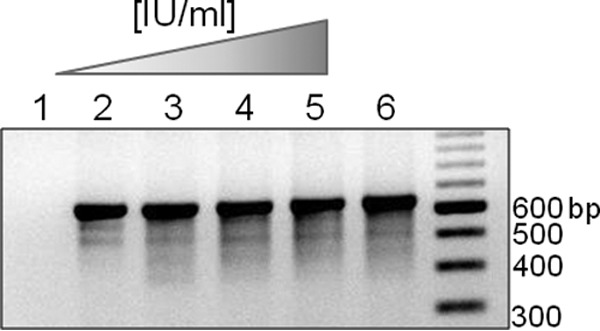
Assay sensitivity was assessed by amplification of an Accrometrix CMVtc panel. Gel electrophoresis (1.5% TBE gel) of ROI 1 PCR, representative of all regions, using an Accrometrix CMVtc panel as the template. Lanes 1 to 5 represent panel components as follows: 1, negative control; 2, 240 IU/ml; 3, 2,400 IU/ml; 4, 24,000 IU/ml; 5, 240,000 IU/ml. Lane 6 is a CMV-positive control. Equivalent results were obtained for all UL97 and UL54 ROIs.
UL54 mutations in patient specimens.
We evaluated residual samples from patients previously tested for mutations in the UL97 region to determine the frequency of UL54 mutations in that population. A total of 43 such residual patient plasma samples from 41 patients were available for testing by our combined UL97/UL54 assay. All six amplicon regions sequenced well for all but three samples, where ROI 3 or 4 failed to amplify due to low viral template. There was good agreement between the previous UL97-only method and the new combined UL97/UL54 method for all patients. Twenty-four patients with no UL97 mutations by the UL97-only method were confirmed to have the wild-type UL97 sequence with the combined UL97/UL54 method. Six of these samples were confirmed as wild type by ARUP Laboratories. Sixteen patients with UL97 mutations detected by the UL97-only method were confirmed to have the same UL97 mutations by the combined UL97/UL54 method (Table 2). Two of these samples, including one with both UL97 and UL54 mutations (patient 14), were confirmed by ARUP Laboratories. Four additional patients had predominantly wild-type UL97 sequences with minor drug-resistant mutant variants (A594V and A594S) detected by the original UL97-only method. Subsequent testing with the UL97/UL54 method identified only the majority sequence. This discrepancy is likely due to the well-known, inherent limitations of detecting minor variants by Sanger sequencing (10, 11) combined with the long-term storage of these samples.
Table 2.
Retrospective patient samples tested for UL54 mutations and concordance of UL97 results between the combined UL97/UL54 method and UL97-only methoda
| Patient | Mutation found in indicated gene using: |
Viral quantity (copies/ml)b | ||
|---|---|---|---|---|
| Combined method |
UL97-only method (UL97 gene) | |||
| UL54 gene | UL97 gene | |||
| 3 | N408D | — | — | 2,200 |
| 14 | V715 M | L595S | L595S | 6,800 |
| 38 | N408D | H520Q | H520Q | 1,200 |
| 11 | — | M460V | M460V | 12,000 |
| 12 | — | A594V | A594V | 140,000,000 |
| 13 | — | L595W | L595W | 14,000 |
| 17 | — | L595S | L595S | 300,000 |
| 18 | — | Del600-603 | Del600-603 | 200,000 |
| 19 | — | Del596-603 | Del596-603 | 470,000 |
| 21 | — | M460V | M460V | 4,600 |
| 27 | — | H520Q | H520Q | 20,000 |
| 32 | — | H520Q | H520Q | 12,000 |
| 40 | — | A594V | A594V | 1,400 |
| 41 | — | L595F | L595F | 91,000 |
| 42 | — | L595F | L595F | 11,000 |
| 43 | — | L595S | L595S | NA |
—, no mutation present; NA, no result available.
Viral quantity is from original clinical viral load testing prior to sample storage.
Importantly, previously unidentified UL54 mutations were revealed by the new combined UL97/UL54 method for three patients. For two of these (patients 14 and 38), the UL54 mutations occurred in the setting of previously selected UL97 mutations (Table 2). For the final patient (patient 3), a UL54 mutation was present in the absence of a UL97 mutation.
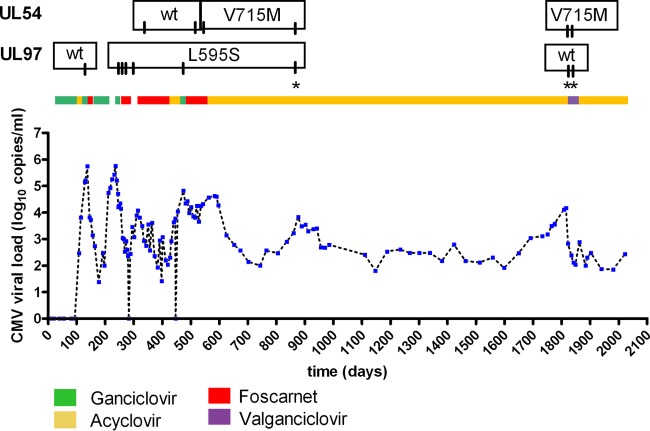
For these three patients, we reviewed their clinical histories pertaining to CMV status for the period around the sample draw date to investigate how UL97/UL54 mutations affect the course of CMV disease and to check for concordance of our UL54 results with previous reference laboratory results. Patient 14 is a 66-year-old male first diagnosed with chronic obstructive pulmonary disease (COPD) at age 45. He received a bilateral lung transplant in February 2007 and was given ganciclovir prophylactically to prevent CMV infection (CMV serostatus, donor positive/recipient negative [D+/R−]). Upon completion of his treatment, his viral loads began increasing, as shown by the data in Figure 3. During an initial increase in CMV viral load, UL97 resistance testing indicated a wild-type genotype. However, a subsequent increase in viral load was presumed to result from the L595S UL97 resistance mutation, which confers ganciclovir resistance (3, 12, 13). Once resistance was identified, drug treatment was switched from ganciclovir to foscarnet for an extended period, during which UL54 resistance testing (performed by an outside reference laboratory) initially revealed a wild-type UL54 genotype. However, the V715M UL54 mutation, conferring foscarnet resistance (14–17), was identified during subsequent testing by the reference laboratory. These mutations were confirmed with our laboratory's combined UL97/UL54 sequencing method.
Fig 3.
CMV viral load, treatment, and mutation time course for patient 14. Blue dots indicate quantitation by quantitative PCR. Tick marks in mutation labels indicate dates of testing. Asterisk indicates resistance testing performed for this study. UL97 results without an asterisk were from clinical testing using the UL97-only method. UL54 results without an asterisk were done by a reference laboratory.
During the past nearly 4 years, the patient was on a herpes simplex virus prophylactic dose of acyclovir, with a brief treatment of valganciclovir during a CMV viral load increase around 1,800 days. Our laboratory obtained a residual specimen from quantitation assays performed during this increase in viral load and tested for UL97 and UL54 resistance mutations. These specimens revealed that the UL97 mutation, L595S, was lost and the UL54 mutation, V715M, was maintained years after exposure to foscarnet. Detailed analysis of the sequencing performed around 870 days revealed that the L595S UL97 mutation was present as a mix of the variant and the wild type in one sequencing direction and only the variant in the other direction, indicating a small proportion of UL97 wild-type virus present in a background of the L595S variant. The UL54 sequencing at this time indicated 100% V715M variant in both sequencing directions.
Patient 38 did not receive direct clinical care from our institution, so only viral load and resistance profile data were available for analysis. After multiple instances of UL97 resistance testing that identified wild-type CMV during an acute viral load increase, UL97 mutation H520Q, conferring ganciclovir resistance (15, 16), was identified (Fig. 4). Our laboratory's validation testing of the combined UL97/UL54 method identified that this UL97 mutation was accompanied by a UL54 mutation, N408D, conferring cidofovir and ganciclovir resistance (14, 16–18).
Fig 4.
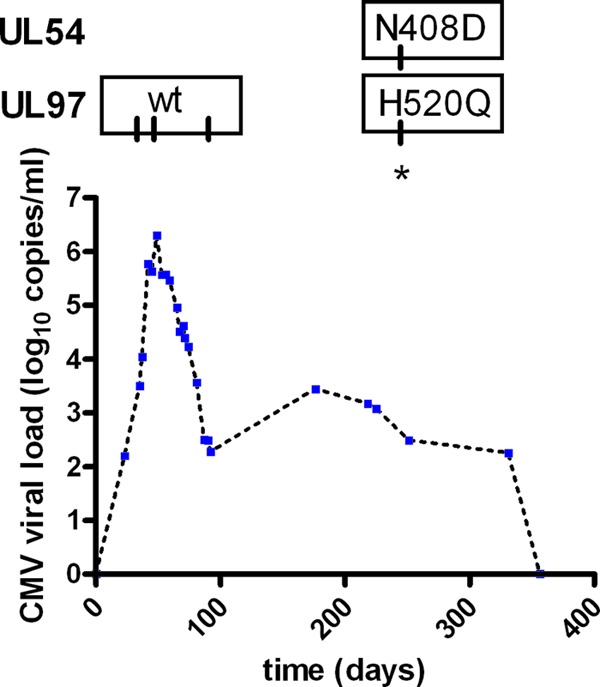
CMV viral load and mutation profile for patient 38. Blue dots indicate quantitation by quantitative PCR. Tick marks in mutation labels indicate dates of testing. Asterisk indicates resistance testing performed for this study. UL97 results without an asterisk were from clinical testing using the UL97-only method.
Patient 3 also did not receive direct clinical care from our institution and had only one viral load quantitation and one drug resistance testing performed at our institution. This patient was clinically tested for UL97 resistance mutations and found to have a wild-type genotype. Subsequent testing for validation of the combined UL97/UL54 assay confirmed a wild-type UL97 genotype and identified a UL54 mutation, N408D.
In addition to the three patients with UL54 mutations, we retrieved viral load data for 33 of the 43 specimens sequenced. Analysis of viral load profiles for these patients failed to reveal any consistent patterns in viral load in patients with or without drug resistance mutations (see Fig. S2 in the supplemental material), possibly due to the heterogeneous nature of our population and the complicated and unpredictable nature of the host immune response to viral reactivation and infection in immunosuppressed individuals. These data point to the difficulty of predicting resistance during CMV infection and account for our laboratory's typical yield on resistance testing: from 2005 to 2012, only 31% of specimens ordered for UL97 sequencing carried drug resistance mutations.
DISCUSSION
Human cytomegalovirus (CMV) results in serious complications in immunocompromised patients, particularly in a bone marrow or solid-organ transplantation setting. Tissue-invasive CMV disease, such as pneumonia and hepatitis, is responsible for high morbidity and mortality rates in transplant patients. The use of prophylactic anti-CMV therapy beginning early posttransplant has reduced the incidence of CMV-associated morbidity and mortality. However, with the increased administration of anti-CMV drugs, there has been an increased emergence of drug-resistant CMV. The incidence of drug resistance varies depending on the organ transplanted and the immunosuppressive regimen, but it is commonly seen in 5 to 10% of transplant patients who are CMV seronegative receiving a CMV-seropositive organ (D+/R−) (4).
CMV drug resistance arises from mutations in the viral kinase UL97 or the DNA polymerase UL54. Clinical genotypic testing of the UL97 gene and, less frequently, the UL54 gene, leads to diagnosis of CMV drug resistance. The current CMV treatment guidelines call for initial genotyping of the UL97 gene when drug resistance is suspected. If a drug-resistant variant is identified in the UL97 gene, subsequent UL54 gene sequencing is recommended (19). Many cases of resistance have been documented, both as case reports (6, 20–26) and cohort studies (5, 15, 27). Most of these cases report resistance arising from canonical UL97 ganciclovir resistance mutations. Some also examine the incidence of UL54 mutations providing cross-resistance to cidofovir, foscarnet, and ganciclovir.
Here, we present a rapid method to sequence the conserved regions of the UL97 and UL54 genes where most resistance mutations occur. As evidenced by the multitude of case studies presenting UL97 and UL54 genotyping data, PCR methods for identifying resistance have been available for quite some time. However, most of these methods rely on nested PCRs (5, 23), which require multiple rounds of PCR before sequencing, increasing the time to result and risk of contamination. Methods that do not rely on nested PCR utilize a LightCycler instrument (6), requiring PCR setup in capillaries with subsequent PCR product selection and clean up via agarose gel electrophoresis, all of which can become cumbersome for high-throughput clinical analysis. Our method utilizes dried-down primer sets in a 96-well format, so a single master mix can be multichannel pipetted for PCR amplification and sequencing reactions. Confirmation of PCR product formation is performed with melt curve analysis, abrogating the need for gel electrophoresis, a step that adds time and the potential for sample contamination or mix-up. This rapid, combined UL97/UL54 method is highly scalable and results in a 2-day turnaround time from specimen extraction to results.
Using this combined UL97/UL54 sequencing method, we identified 3 patients out of 41 tested who had UL54 resistance mutations. The mutations we identified, V715M (patient 14) and N408D (patients 3 and 38), are well-characterized resistance mutations (2, 10, 12, 13, 18). Patient 14 is a lung transplant recipient who developed multiple-drug-resistant CMV. Cases of patients with dual UL97 and UL54 mutations are particularly prevalent in lung transplant recipients. A recent 39-patient study showed that lung transplant recipients accounted for 57% of patients with dual UL97 and UL54 mutations, even though that transplant procedure accounted for only 22% of CMV-positive patients (27). Several cases of multidrug-resistant, dual UL97/UL54 mutant CMV viruses have been reported in solid organ and stem cell transplant recipients (21, 24, 25). A 2002 cohort study identified a lung transplant recipient with a UL54 V715M mutation associated with the UL97 mutation A594V (15).
Interestingly, in the absence of ganciclovir treatment, patient 14 lost the UL97 L595S variant but maintained the UL54 V715M mutant over years of treatment with just acyclovir and reduced immunosuppression. It is likely that the L595S mutation causes a fitness disadvantage to CMV such that once the selective pressure of ganciclovir was removed, the small population of virus carrying wild-type UL97 (evident in the sequencing performed around 870 days) rebounded. This phenomenon has been previously documented in patients with CMV retinitis when therapy was shifted from ganciclovir to cidofovir (26, 28).
Identification of the UL54 mutant in the absence of a UL97 mutation points to the question of whether UL54 mutations can exist by themselves. The current paradigm of treatment calls for the identification of a UL97 mutation before testing for a UL54 mutation. However, we have identified two patients, 14 and 3, that at some point during treatment harbored a UL54 mutation but not a UL97 mutation. In patient 14, the presence of just a UL54 mutation is likely the result of a fitness cost of the prior UL97 mutation. For patient 3, the clinical history is unknown. It is possible that the UL54 mutation N408D arose without a prior UL97 mutation, although no previous samples are available. In a 2010 study of transplant recipients, a kidney transplant recipient was identified as having CMV with UL54 mutation A834P, conferring resistance to ganciclovir, cidofovir, and foscarnet, without a concomitant UL97 mutation 158 days after antiviral exposure (5).
This new rapid method for combined UL97/UL54 drug resistance testing contributes significantly to higher-throughput clinical testing for CMV drug resistance diagnosis. It simplifies UL97 and UL54 analysis, allowing for more-comprehensive resistance genotyping that is as sensitive as the previous UL97-only method. Although the current consensus recommends testing only UL97 initially, we highlight the possibility of UL54 mutations occurring without UL97 mutations. With this new, more-efficient assay, routine combined testing can be prospectively studied to define its clinical utility.
Supplementary Material
ACKNOWLEDGMENTS
We thank Melanie Mallory, ARUP Laboratories, Salt Lake City, UT, for her assistance with the sample exchange. We also thank Tanis Dingle, University of Washington, Seattle, WA, for assistance with patient chart review.
Published ahead of print 15 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00611-13.
REFERENCES
- 1. Scott GM, Isaacs MA, Zeng F, Kesson AM, Rawlinson WD. 2004. Cytomegalovirus antiviral resistance associated with treatment induced UL97 (protein kinase) and UL54 (DNA polymerase) mutations. J. Med. Virol. 74:85–93 [DOI] [PubMed] [Google Scholar]
- 2. Gilbert C, Azzi A, Goyette N, Lin SX, Boivin G. 2011. Recombinant phenotyping of cytomegalovirus UL54 mutations that emerged during cell passages in the presence of either ganciclovir or foscarnet. Antimicrob. Agents Chemother. 55:4019–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castor J, Cook L, Corey L, Jerome KR. 2007. Rapid detection directly from patient serum samples of human cytomegalovirus UL97 mutations conferring ganciclovir resistance. J. Clin. Microbiol. 45:2681–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hantz S, Garnier-Geoffroy F, Mazeron MC, Garrigue I, Merville P, Mengelle C, Rostaing L, Saint Marcoux F, Essing M, Rerolle JP, Cotin S, Germi R, Pillet S, Lebranchu Y, Turlure P, Alain S, French CMV Resistance Survey Study Group 2010. Drug-resistant cytomegalovirus in transplant recipients: a French cohort study. J. Antimicrob. Chemother. 65:2628–2640 [DOI] [PubMed] [Google Scholar]
- 6. Yeh S, Fahle G, Forooghian F, Faia LJ, Weichel ED, Stout JT, Flaxel CJ, Lauer AK, Sen HN, Nussenblatt RB. 2012. Polymerase chain reaction-based ganciclovir resistance testing of ocular fluids for cytomegalovirus retinitis. Arch. Ophthalmol. 130:113–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu H, Jabs DA, Forman MS, Martin BK, Dunn JP, Weinberg DV, Davis JL, Cytomegalovirus Retinitis and Viral Resistance Study Group 2002. Comparison of cytomegalovirus (CMV) UL97 gene sequences in the blood and vitreous of patients with acquired immunodeficiency syndrome and CMV retinitis. J. Infect. Dis. 185:861–867 [DOI] [PubMed] [Google Scholar]
- 8. Lurain NS, Weinberg A, Crumpacker CS, Chou S, Adult AIDS Clinical Trials Group-CMV Laboratories 2001. Sequencing of cytomegalovirus UL97 gene for genotypic antiviral resistance testing. Antimicrob. Agents Chemother. 45:2775–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spector SA, Hsia K, Wolf D, Shinkai M, Smith I. 1995. Molecular detection of human cytomegalovirus and determination of genotypic ganciclovir resistance in clinical specimens. Clin. Infect. Dis. 21(Suppl 2):S170–S173 [DOI] [PubMed] [Google Scholar]
- 10. Vandenbroucke I, Van Marck H, Verhasselt P, Thys K, Mostmans W, Dumont S, Van Eygen V, Coen K, Tuefferd M, Aerssens J. 2011. Minor variant detection in amplicons using 454 massive parallel pyrosequencing: experiences and considerations for successful applications. Biotechniques 51:167–177 [DOI] [PubMed] [Google Scholar]
- 11. Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baldanti F, Underwood MR, Stanat SC, Biron KK, Chou S, Sarasini A, Silini E, Gerna G. 1996. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J. Virol. 70:1390–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chou S, Van Wechel LC, Lichy HM, Marousek GI. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cihlar T, Fuller MD, Cherrington JM. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lurain NS, Bhorade SM, Pursell KJ, Avery RK, Yeldandi VV, Isada CM, Robert ES, Kohn DJ, Arens MQ, Garrity ER, Taege AJ, Mullen MG, Todd KM, Bremer JW, Yen-Lieberman B. 2002. Analysis and characterization of antiviral drug-resistant cytomegalovirus isolates from solid organ transplant recipients. J. Infect. Dis. 186:760–768 [DOI] [PubMed] [Google Scholar]
- 16. Baldanti F, Lurain N, Gerna G. 2004. Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs. Hum. Immunol. 65:403–409 [DOI] [PubMed] [Google Scholar]
- 17. Gilbert C, Bestman-Smith J, Boivin G. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updat. 5:88–114 [DOI] [PubMed] [Google Scholar]
- 18. Gilbert C, Boivin G. 2005. New reporter cell line to evaluate the sequential emergence of multiple human cytomegalovirus mutations during in vitro drug exposure. Antimicrob. Agents Chemother. 49:4860–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Snydman DR, Limaye AP, Potena L, Zamora MR. 2011. Update and review: state-of-the-art management of cytomegalovirus infection and disease following thoracic organ transplantation. Transplant. Proc. 43:S1–S17 [DOI] [PubMed] [Google Scholar]
- 20. Limaye AP, Corey L, Koelle DM, Davis CL, Boeckh M. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645–649 [DOI] [PubMed] [Google Scholar]
- 21. Springer KL, Chou S, Li S, Giller RH, Quinones R, Shira JE, Weinberg A. 2005. How evolution of mutations conferring drug resistance affects viral dynamics and clinical outcomes of cytomegalovirus-infected hematopoietic cell transplant recipients. J. Clin. Microbiol. 43:208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boutolleau D, Deback C, Bressollette-Bodin C, Varnous S, Dhedin N, Barrou B, Vernant JP, Gandjbakhch I, Imbert-Marcille BM, Agut H. 2009. Resistance pattern of cytomegalovirus (CMV) after oral valganciclovir therapy in transplant recipients at high-risk for CMV infection. Antiviral Res. 81:174–179 [DOI] [PubMed] [Google Scholar]
- 23. Boivin G, Goyette N, Gilbert C, Covington E. 2005. Analysis of cytomegalovirus DNA polymerase (UL54) mutations in solid organ transplant patients receiving valganciclovir or ganciclovir prophylaxis. J. Med. Virol. 77:425–429 [DOI] [PubMed] [Google Scholar]
- 24. Baldanti F, Lilleri D, Campanini G, Comolli G, Ridolfo AL, Rusconi S, Gerna G. 2004. Human cytomegalovirus double resistance in a donor-positive/recipient-negative lung transplant patient with an impaired CD4-mediated specific immune response. J. Antimicrob. Chemother. 53:536–539 [DOI] [PubMed] [Google Scholar]
- 25. Goldsmith PM, Husain MM, Carmichael A, Zhang H, Middleton SJ. 2012. Case report: multidrug-resistant cytomegalovirus in a modified multivisceral transplant recipient. Transplantation 93:e30–e32 [DOI] [PubMed] [Google Scholar]
- 26. Bowen EF, Johnson MA, Griffiths PD, Emery VC. 1997. Development of a point mutation assay for the detection of human cytomegalovirus UL97 mutations associated with ganciclovir resistance. J. Virol. Methods 68:225–234 [DOI] [PubMed] [Google Scholar]
- 27. Iwasenko JM, Scott GM, Naing Z, Glanville AR, Rawlinson WD. 2011. Diversity of antiviral-resistant human cytomegalovirus in heart and lung transplant recipients. Transplant Infect. Dis. 13:145–153 [DOI] [PubMed] [Google Scholar]
- 28. Bowen EF, Cherrington JM, Lamy PD, Griffiths PD, Johnson MA, Emery VC. 1999. Quantitative changes in cytomegalovirus DNAemia and genetic analysis of the UL97 and UL54 genes in AIDS patients receiving cidofovir following ganciclovir therapy. J. Med. Virol. 58:402–407 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.