Abstract
We have developed a novel blood lysis-centrifugation approach for highly sensitive Mycobacterium tuberculosis detection in large volumes of blood with the Xpert MTB/RIF assay. One through 20 ml of blood was spiked with 0.25 to 10 CFU/ml of the M. tuberculosis surrogate M. bovis BCG. Multiple replicates of each sample were processed by a new lysis-centrifugation method and tested with the Xpert MTB/RIF assay. The assay was very sensitive with increased blood volumes. In the 20-ml samples, BCG was detected in blood spiked with 10, 5, 1, and 0.25 CFU/ml 100, 100, 83, and 57% of the time, respectively, compared to 100, 66, 18, and 18%, of the time, respectively, in 1-ml blood samples. Assay sensitivity was influenced by the type of anticoagulant used, with acid-citrate-dextrose solution B (ACD-B) providing the best results. A limit of detection of 10 CFU/ml was established with BCG spiked into ACD-B-treated blood, and 92, 36, and 33% of the samples with 5, 1, and 0.5 CFU/ml, respectively, were assay positive. The lysis buffer was stable both at room temperature and at 4°C for 2 months. The assay was tested with blood stored for 8 days without a change in sensitivity as measured by cycle threshold. This new assay format extends the capability of the Xpert MTB/RIF test, enabling up to 20 ml of blood to be tested rapidly for the presence of M. tuberculosis. This approach may be a useful method to detect extrapulmonary tuberculosis and the risk of death in immunocompromised patients.
INTRODUCTION
Tuberculosis (TB) is one of the leading causes of death from an infectious disease worldwide (1). Diagnostic delays contribute significantly to death. In the case of pulmonary TB (PTB), a diagnosis can often be made by testing expectorated sputum samples. However, some patients are not able to expectorate sputum, especially if they are very sick or young. Extrapulmonary TB (EPTB) is common (2, 3), especially in patients with human immunodeficiency virus (HIV) infection. The death rate among subjects with dual HIV and Mycobacterium tuberculosis infections can be very high (4, 5). EPTB is usually diagnosed by performing microscopic, culture-based, or nucleic acid amplification tests (NATs) of aspirated fluid or tissue biopsy material, depending on the site that is infected (6, 7). Microscopy has a low sensitivity for the diagnosis of EPTB, and reliance on culture-based detection methods can lead to substantial delays in diagnosis (8, 9). Rapid TB NATs have not been optimized for nonpulmonary samples and have had variable performance in detecting TB in tissue or fluid samples (9–12). Thus, there is a real need for improved methods to test extrapulmonary samples.
Blood is an attractive option for the detection of TB, especially in HIV-infected patients. Blood is easier and more reliable to sample than sputum, especially in patients who are confused or obtunded or who do not have a productive cough. M. tuberculosis can be cultured from 2 to 64% of the blood samples collected, depending on the study population (13). In sub-Saharan Africa, M. tuberculosis is a common cause of bacteremia, accounting for 54% of all bloodstream infections (BSI), especially among HIV-positive patients (14). Furthermore, disseminated M. tuberculosis infection resulting in a BSI may be an important predictor of death (4, 5).
M. tuberculosis is rarely detected by blood tests despite the potential advantages of this approach. TB blood culture usually takes several weeks to become positive and requires appropriately equipped facilities (15, 16). NATs such as the PCR have been investigated, but sensitivity with blood has been poor (20 to 55%) (4, 17–21). This could be due in part to the presence of PCR inhibitors (22, 23). The poor sensitivity of blood-based NATs could also be due to the small volume of blood (500 μl to <5 ml) that is usually collected for these tests. These small volumes could make it difficult or impossible to detect M. tuberculosis circulating in blood if the bacteria are present at very low densities. However, 5 to 10 ml of blood is typically used for blood cultures (24, 25). NATs for TB might have sensitivities equal to or greater than that of blood culture if similar volumes of blood could be tested by improved sample-processing methods.
The Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA) has greatly simplified TB detection in sputum samples (26–28). This completely automated real-time PCR assay simultaneously detects the presence of M. tuberculosis and rifampin resistance (29). The assay is approximately 98.3% sensitive and 99% specific for detecting M. tuberculosis in sputum samples (26, 29). Sensitivity with extrapulmonary samples has varied from 25 to 100%, depending on the body site and study (17–20, 30). The ability of the Xpert MTB/RIF assay to detect M. tuberculosis in blood has not been investigated, and the current sample-processing protocol used in the Xpert MTB/RIF assay has not been previously optimized for blood. For example, the current Xpert MTB/RIF protocol uses 1 ml of sputum, while a protocol for blood should arguably start with larger sample volumes.
Here, we present a novel lysis and centrifugation approach that permits relatively large volumes of blood to be tested with the Xpert MTB/RIF assay. Analytic results suggest that our new approach could be usefully applied to the clinical testing of blood samples from patients suspected of having TB.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the University of Medicine and Dentistry of New Jersey (UMDNJ) Institutional Review Board in protocols 0120080060 (for use of discarded blood tests), 0120060015 (for use of discarded blood bank blood), and 012010406 (for drawing and testing blood from healthy human volunteers). Blood from human volunteers was obtained with informed consent.
Bacterial strain and culture conditions.
The Mycobacterium bovis BCG Pasteur strain used as a surrogate test strain for M. tuberculosis was a kind gift from William Jacobs, Jr., Albert Einstein College of Medicine, Bronx, NY. BCG was propagated as described previously (31). Experiments used prequantitated BCG that was frozen in aliquots. Aliquots were sonicated three times (Branson ultrasonic cleaner 1510; Branson Ultrasonics, Danbury, CT) for 30 s each before serial dilutions were made with Middlebrook 7H9 broth (BD, Franklin Lakes, NJ).
Blood sample collection and storage.
Expired blood (with anticoagulant citrate-phosphate-dextrose-adenine [CPDA]) from the UMDNJ—University Hospital (UH; Newark, NJ) blood bank that would normally have been discarded was collected and stored at 4°C until use. Hematocrit values were adjusted to 40% by diluting the banked blood (hematocrit, 60 to 80%) in phosphate-buffered saline (PBS; 0.01 M, pH 7.4) to simulate a normal adult blood hematocrit value (32, 33). Studies to examine the effect of anticoagulants, as well as limit-of-detection (LOD) experiments, were performed with fresh blood collected from healthy human volunteers. This blood was drawn directly into 8-ml BD Vacutainer tubes containing either potassium EDTA or acid citrate dextrose (ACD-A, ACD-B) as an anticoagulant and analyzed immediately or stored at 4°C for no longer than 24 h until use.
Blood sample processing.
Red blood cell (RBC) lysis solution 2.2.1 was prepared to contain 10% (wt/vol) sucrose, 0.5% (wt/vol) magnesium chloride, and 5% (vol/vol) Triton X-100 in 0.01 M Tris HCl solution adjusted to pH 7.2. When processing blood sample volumes of ≥5 ml, RBC lysing solution 2.2.1 was added at 15% (vol/vol) to blood, mixed well, and incubated at room temperature (RT; 20 to 25°C) for 10 min. The blood tubes were centrifuged for 30 min at 3,000 × g. The supernatant was carefully decanted, and the pellet was resuspended in 1 ml of PBS (pH 7.2). Xpert MTB/RIF assay sample reagent (SR; Cepheid) was added at a 1:1 ratio to the resuspended sample and mixed well, and the sample was loaded into Xpert MTB/RIF assay cartridges (version G3, research use only) after 15 min of incubation at RT (Fig. 1). When a 1-ml blood volume was being processed, the blood sample was mixed with 1 ml of Xpert SR and incubated for 15 min. The blood-SR mixture was then loaded into the sample loading chamber of an Xpert MTB/RIF assay cartridge. Subsequent sample processing and PCR were then performed in accordance with the manufacturer's recommendations for sputum samples (Cepheid).
Fig 1.
Flow chart describing our large-volume blood sample-processing methodology.
Effect of blood volume on detection of very low levels of M. tuberculosis in blood.
To determine the effect of blood sample volume on sensitivity for M. tuberculosis, BCG cells were spiked at concentrations of 0.25, 1, 5, and 10 CFU/ml into various volumes of reconstituted banked blood. Blood samples were processed as described above. All of the samples were run in replicates of seven.
LOD of mycobacteria in ACD-B-treated blood.
ACD-B proved to be the best-performing anticoagulant in our tests. Therefore, the LOD was established with the blood from healthy volunteers collected in ACD-B anticoagulant tubes (BD). M. bovis BCG was spiked at concentrations of 0.5, 1, 5, 10, and 50 CFU/ml into 10 ml of ACD-B-treated blood. All of the samples were processed in accordance with the lysis centrifugation protocol. Seven and 8 replicates were run for each cell concentration in two separate experiments or runs, for a total of 15 replicates.
Stability of solution 2.2.1.
RBC lysis solution 2.2.1 was prepared and stored at both RT and under refrigerated conditions (4 to 8°C) for up to 3 months. The aliquoted tubes were removed at days 1, 7, and 15, followed by 1, 2, 3 months, and tested as follows. For each time point, reconstituted banked blood was spiked with 10 CFU/ml of BCG per 10 ml of blood. RBC lysis solution 2.2.1 was added at 15% vol/vol (1.5 ml per 10 ml) and the blood was processed as described above. Stability was assessed by the average cycle threshold (CT) of the first rpoB probe measured in the Xpert MTB/RIF assay, with increasing CTs indicative of degraded performance.
Storage of BCG-infected blood at refrigerated temperature.
Tests were performed to understand how long ACD-B-anticoagulated blood could be stored in the refrigerator before processing. BCG was spiked into 10 ml of ACD-B-treated blood at 10 CFU/ml and stored in a refrigerator (2 to 8°C) for up to 8 days. The sample was removed and processed with solution 2.2.1 in accordance with the protocol described above. The effect of storage time was evaluated by observing any change in the CT or the endpoint fluorescence values in the first rpoB probe measured in the Xpert MTB/RIF assay.
Statistical analysis.
Analysis (average, standard deviation, and t test) was performed with Microsoft Excel 2000 for Windows. One-way analysis of variance (ANOVA) for independent or correlated samples was carried out to evaluate the P values with the online ANOVA calculator ay http://faculty.vassar.edu/lowry/anova1u.html. Curve fitting for the LOD curve was done by sigmoid linear regression analysis with SigmaPlot V8.0.
RESULTS
Large volumes of blood allow M. tuberculosis detection and outperform small volumes.
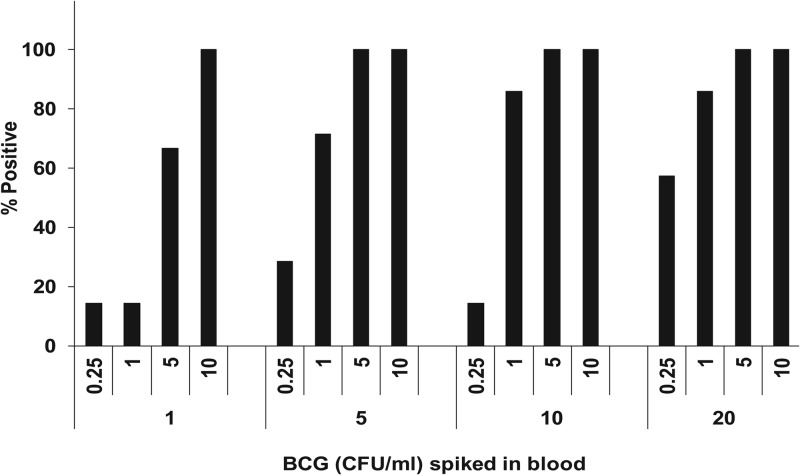
We developed a protocol that enabled us to lyse up to 20 ml of blood, concentrate any M. tuberculosis present in the sample by centrifugation, and resuspend the resulting pellet in Xpert SR. The resulting sample was then suitable for testing by the standard sputum protocol. We tested this approach by spiking BCG at 0.25 to 10 CFU/ml into different volumes of (blood-banked) blood ranging from 1 to 20 ml. The 1-ml blood samples were processed without lysis and centrifugation by being mixed with equal volumes of SR, and then the 2-ml mixtures were loaded into Xpert MTB/RIF assay cartridges. The larger blood volume samples were processed by our novel lysis-centrifugation protocol. As shown in Fig. 2, increasing the volume of blood that was tested resulted in a substantially increased rate of positive results, especially for tests performed with 0.25-, 1-, and 5-CFU/ml samples. Blood volumes of 20 ml that were spiked with as few as 0.25 and 1 CFU/ml could be detected consistently 57 and >80% of the time, respectively. However, in a smaller blood volume of 1 ml, the same cell concentrations were detected <20% of the time. Thus, 1-ml blood samples needed to be spiked with 5 to 10 times more M. tuberculosis CFU than 20-ml blood samples to achieve similar rates of test positivity.
Fig 2.
Effect of blood volume on the probability of mycobacterium detection. Various numbers of BCG CFU per milliliter were spiked into different volumes of blood. The percentages of samples among seven samples per concentration that were positive at each concentration are shown.
We considered the possibility that our lysis and centrifugation method might concentrate PCR inhibitors at the same time that it concentrated M. tuberculosis bacilli. The Xpert MTB/RIF assay includes a sample-processing control assay for Bacillus globigii that can also be used to indicate the presence of PCR inhibitors (34). Inhibition is detected by an increase in the B. globigii CT. We did not observe any increase in the B. globigii CT with increasing blood volumes, suggesting that our procedure did not concentrate significant amounts of inhibitors of the Xpert MTB/RIF assay.
Role of anticoagulants in M. tuberculosis detection in blood.
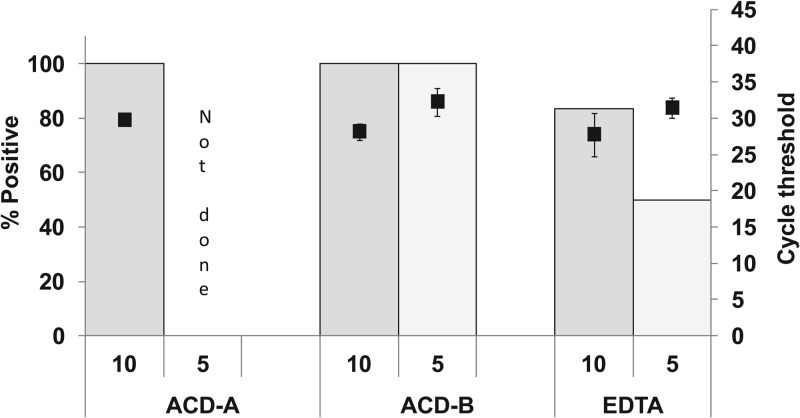
Our processing protocol requires anticoagulated blood samples. However, certain anticoagulants such as heparin or EDTA can inhibit PCR assays. We tested the impact of various anticoagulants on assay sensitivity after large-volume lysis and centrifugation. We selected anticoagulants for testing on the basis of their use in commonly available commercial blood collection tubes. Blood was drawn from human volunteers into collection tubes containing ACD-A, ACD-B, or EDTA. BCG was spiked into 5-ml aliquots of each sample at 5- and 10-CFU/ml concentrations. The samples were then processed by our lysis and centrifugation protocol and tested with the Xpert MTB/RIF assay. As shown in Fig. 3, the ACD-anticoagulated blood showed the best overall performance. M. tuberculosis was detected in 100% of the test aliquots containing 10 CFU/ml that were anticoagulated with either ACD-A or ACD-B and in 100% of the test aliquots containing 5 CFU/ml that were anticoagulated with ACD-B (ACD-A was not tested at 5 CFU/ml). This is in contrast to the EDTA-anticoagulated samples, which had substantially fewer positive test results. Comparing the rpoB values of the ACD-A- and ACD-B-anticoagulated samples containing 10 CFU/ml, we noted that ACD-A anticoagulation resulted in a delayed mean CT (29.7 ± 0.5 versus 28.1 ± 1, P = 0.015). This suggested that ACD-B anticoagulation would provide the best assay sensitivity.
Fig 3.
Comparison of various anticoagulants. Blood samples anticoagulated with ACD-A, ACD-B, or EDTA were spiked with 5 and 10 CFU/ml of BCG in 5 ml whole blood. The proportion of samples that were positive (bars) and the CT value of the rpoB assay (■) are shown for each condition. The number of replicates run with each anticoagulant type was 4 to 7.
LOD with ACD-B-anticoagulated blood.
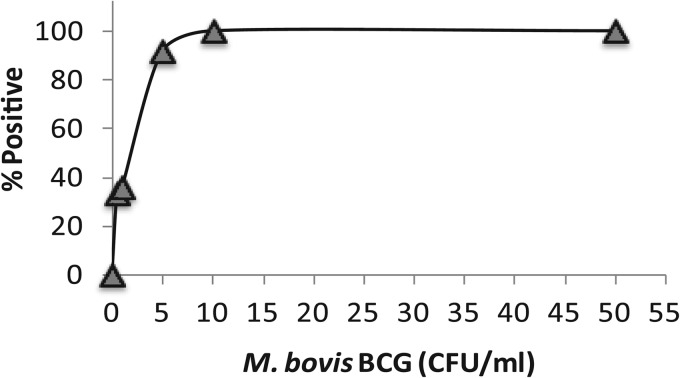
We determined the LOD of our lysis and centrifugation protocol with blood anticoagulated with ACD-B, since this appeared to be the best candidate for eventual clinical studies. We found that our approach identified M. tuberculosis 100% of the time in aliquots containing 50 and 10 CFU/ml, defining the LOD as 10 CFU/ml. However, the assay continued to perform well even with lower inoculum concentrations, detecting M. tuberculosis in 91.6% of the blood samples spiked with 5 CFU/ml, 36% of those spiked with 1 CFU/ml, and 33% of those spiked with 0.5 CFU/ml (Fig. 4). Overall, the performance of the ACD-B-anticoagulated blood samples was similar to what we observed with CPDA-anticoagulated banked blood (Fig. 2), suggesting that banked blood can be used for future analytic studies.
Fig 4.
LOD of mycobacteria in ACD-B-anticoagulated blood. BCG was spiked into 5-ml blood aliquots. The proportion of positive samples for each CFU concentration out of 15 samples tested per concentration is shown. Each replicate is indicative of a blood sample from one healthy individual.
Stability studies.
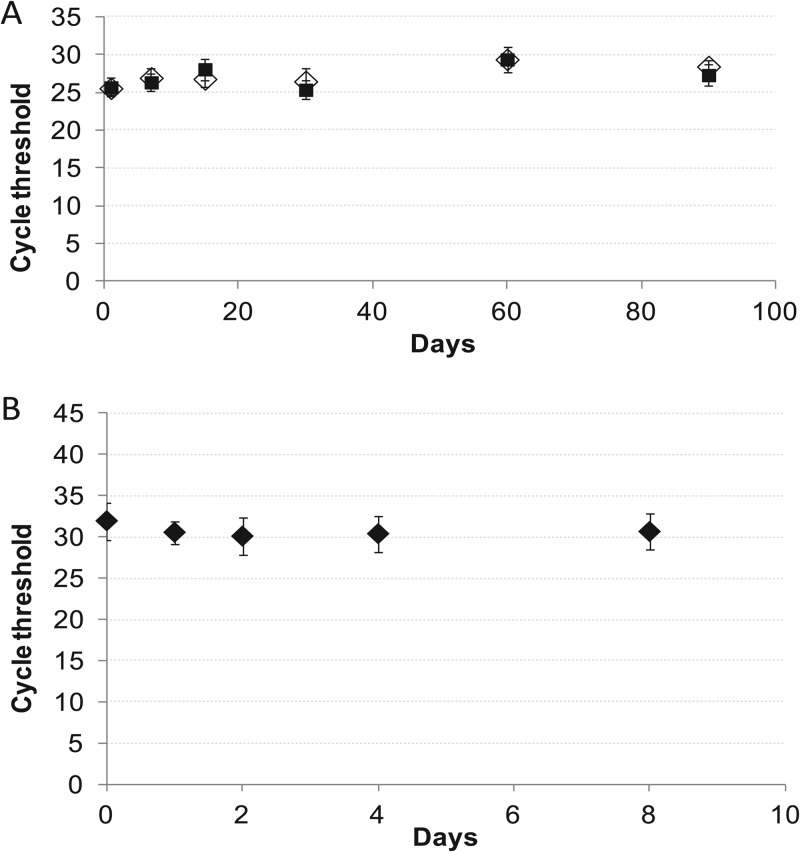
Our assay might be useful in clinics or laboratories that do not have the capacity to easily make our lysis solution. Therefore, we performed stability studies to determine how long the lysis solution could be stored without affecting assay performance. The solution was stored at both RT (20 to 25°C) and 4°C for up to 3 months. Aliquots were removed periodically to process blood samples spiked with known numbers of BCG CFU/ml. The CT of the first rpoB probe was used to asses assay performance (Fig. 5A). The storage temperature of the lysis buffer did not appear to affect the rpoB CT (95% confidence interval, P = 0.8).
Fig 5.
Solution and sample stability. Stability studies were performed by running Xpert MTB/RIF assays with lysis solution stored for 0 to 90 days at RT (♢) or under refrigerated conditions (4 to 8°C) (■) (A) and BCG-spiked blood samples stored for 0 to 8 days (B). Each time point shows the mean of five test results. An increase in the CT (not shown) would have indicated that a stability storage limit had been reached.
Laboratories might find it convenient to batch test blood samples, or blood might need to be transported to off-site locations. To study how long blood could be stored at 4°C prior to processing, we spiked 10 CFU/ml of BCG into 10-ml aliquots of ACD-B-anticoagulated blood and then stored the samples at 4°C for up to 8 days. Samples were processed and analyzed at 0, 1, 2, 4, and 8 days by comparing the number of positive samples and the assay CT values (Fig. 5B). All assays were positive at all time points, and no difference in assay CTs was noted even after the 8-day time point. This study suggests that M. tuberculosis is stable in human blood stored at 4°C for at least 8 days and that large-volume sample processing can be delayed for at least 1 week without affecting assay performance.
DISCUSSION
The major findings of this study were that a simple preprocessing step can be used to sensitively detect M. tuberculosis in large volumes of blood with the Xpert MTB/RIF assay. Important assay parameters affecting sensitivity of detection include the blood volumes and the anticoagulants used.
Our proposed lysis centrifugation protocol has several notable advantages over prior assays to detect M. tuberculosis in blood. First, our lysis buffer was more concentrated (used at a 0.15:1 ratio with blood) than most of the blood lysis buffers reported, which use much higher ratios, usually 1:1 to 10:1. The limited volume requirement of the lysis buffer made it possible to lyse large blood volumes in blood collection tubes or by transferring the collected blood sample into a simple test tube that was prefilled with lysis buffer. These procedures simplified the technical components of the assay. Second, we showed that our lysis buffer completely removes PCR inhibitors, in the context of the Xpert MTB/RIF assay. This property made it possible for us to test very large blood volumes in this study, up to 20 ml, without adding measurable inhibitors to the PCR mixture. The ability to test large blood volumes enabled us to achieve substantial gains in sensitivity. Finally, we show that our assay has acceptable reagent and sample stability.
Our goal was to develop the most sensitive blood PCR assay possible for M. tuberculosis. This was based on the assumption that M. tuberculosis bacteria are present in the blood of TB patients at very low concentrations, when they are present at all. We are not aware of any studies that have formally examined this question in the case of M. tuberculosis. However, for other microbial pathogens, bacterial loads of <1 CFU/ml have been reported among septic adult patients (35). Furthermore, the volume of blood tested by blood culture is considered to be the single most important factor in the detection of bacteria in the blood of adult patients (36–38). Each additional 1 ml of blood cultured increases microbial recovery by up to 3% (36, 39–41). Thus, we expect that our large-volume approach will substantially improve M. tuberculosis detection.
It is unlikely that our method of using the Xpert MTB/RIF assay with blood will diagnose all forms of TB with equal sensitivity. Rebollo et al. (20) noted that the sensitivity of PCR and culture of blood for M. tuberculosis detection was greatest in patients with disseminated TB and those with HIV confection. These are patient groups that frequently do not have a productive cough. Thus, it is likely that our approach will be especially useful in a subset of TB cases that are particularly poorly served by current diagnostics.
Several anticoagulants that are used to collect blood from patients can be PCR inhibitory and will affect assay sensitivity if not properly removed from the sample (42, 43). Heparin is a known PCR inhibitor (42, 44). EDTA can chelate the Mg2+ present in PCR buffer, thus affecting PCR assay performance (45). Isolator tubes recommended for lysis and centrifugation of blood cultures contain high concentrations of saponin and sodium polyanetholesulfonate, which have been shown to be inhibitory to PCR-based blood assays (46). In the present study, we examined blood collected in EDTA, CPDA (banked blood), and two different commercial ACD-anticoagulated tubes (ACD-A and ACD-B). In ACD, citric acid prevents coagulation by binding to calcium and dextrose acts as an RBC nutrient and preservative by maintaining RBC viability. It contains no known PCR inhibitors and thus was a good candidate for our studies. Of the two different ACD formulations available in commercial blood-drawing tubes, ACD-A is more concentrated (BD Vacutainer venous blood collection tube guide). It did not perform as well as ACD-B in our study, possibly because high concentrations of citrate in ACD-A might have mildly inhibited the PCR.
In summary, we have shown that it is possible to test large volumes of blood for the presence of M. tuberculosis with the existing Xpert MTB/RIF platform. Our approach can test samples as large as 20 ml of blood and has a detection limit of 5 to 10 CFU/ml. Bacterial titers as low as 0.25 to 1 CFU/ml can also be routinely detected, although there will be some false-negative assay results at the low end of this titer range. The sample-processing protocol we developed takes approximately 1 h. Combined with the Xpert MTB/RIF on-machine time, the time to result of this assay can be as low as 3 h from the initial blood drawing. This time frame compares very favorably to the several-week delay that is typical for M. tuberculosis blood cultures and may enable rapid diagnosis for a patient group that will particularly benefit from rapid diagnosis and initiation of treatment. The high sensitivity that we observed in our analytic study suggests that our approach may have clinical utility. However, it is necessary to perform clinical trials with relevant patient populations before clinical sensitivity and specificity can be established.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI098713 and AI080653. We thank Fermina M. Mazzella, Department of Pathology and Laboratory Medicine, New Jersey Medical School—University of Medicine and Dentistry of New Jersey, for help in collecting discarded blood tubes. We acknowledge the blood bank staff at UMDNJ—University Hospital, Newark, NJ, for providing expired blood.
D.A. is one of a group of investigators who invented molecular beacon technology and who receive income from licensees, including Cepheid, which licenses the molecular beacon technology in the Xpert MTB/RIF assay. To manage potential conflicts of interest, D.A. has irrevocably limited the fees that can accrue to him from the Xpert MTB/RIF assay to $5,000/year.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575–1580 [DOI] [PubMed] [Google Scholar]
- 2. Golden MP, Vikram HR. 2005. Extrapulmonary tuberculosis: an overview. Am. Fam. Physician 72:1761–1768 [PubMed] [Google Scholar]
- 3. Peto HM, Pratt RH, Harrington TA, LoBue PA, Armstrong LR. 2009. Epidemiology of extrapulmonary tuberculosis in the United States, 1993-2006. Clin. Infect. Dis. 49:1350–1357 [DOI] [PubMed] [Google Scholar]
- 4. Folgueira L, Delgado R, Palenque E, Aguado JM, Noriega AR. 1996. Rapid diagnosis of Mycobacterium tuberculosis bacteremia by PCR. J. Clin. Microbiol. 34:512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. von Reyn CF. 1999. The significance of bacteremic tuberculosis among persons with HIV infection in developing countries. AIDS 13:2193–2195 [DOI] [PubMed] [Google Scholar]
- 6. Chakravorty S, Dudeja M, Hanif M, Tyagi JS. 2005. Utility of universal sample processing methodology, combining smear microscopy, culture, and PCR, for diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 43:2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh KK, Muralidhar M, Kumar A, Chattopadhyaya TK, Kapila K, Singh MK, Sharma SK, Jain NK, Tyagi JS. 2000. Comparison of in house polymerase chain reaction with conventional techniques for the detection of Mycobacterium tuberculosis DNA in granulomatous lymphadenopathy. J. Clin. Pathol. 53:355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ait-Khaled N, Enarson DA. 2003. Tuberculosis: a manual for medical students, vol 99.272 World Health Organization, Geneva, Switzerland [Google Scholar]
- 9. Tortoli E, Russo C, Piersimoni C, Mazzola E, Dal MP, Pascarella M, Borroni E, Mondo A, Piana F, Scarparo C, Coltella L, Lombardi G, Cirillo DM. 2012. Clinical validation of Xpert MTB/RIF for the diagnosis of extrapulmonary tuberculosis. Eur. Respir. J. 40:442–447 [DOI] [PubMed] [Google Scholar]
- 10. Gous N, Scott LE, Wong E, Omar T, Venter WDF, Stevens W. 2012. Performance of the Roche LightCycler real-time PCR assay for diagnosing extrapulmonary tuberculosis. J. Clin. Microbiol. 50:2100–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mehta PK, Raj A, Singh N, Khuller GK. 2012. Diagnosis of extrapulmonary tuberculosis by PCR. FEMS Immunol. Med. Microbiol. 66:20–36 [DOI] [PubMed] [Google Scholar]
- 12. Tortoli E, Urbano P, Marcelli F, Simonetti MT, Cirillo MD. 2012. Is real-time PCR better than conventional PCR for Mycobacterium tuberculosis complex detection in clinical samples? J. Clin. Microbiol. 50:2810–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heysell S, Thomas T, Gandhi N, Moll A, Eksteen F, Coovadia Y, Roux L, Babaria P, Lalloo U, Friedland G, Shah S. 2010. Blood cultures for the diagnosis of multidrug-resistant and extensively drug-resistant tuberculosis among HIV-infected patients from rural South Africa: a cross-sectional study. BMC Infect. Dis. 10:344. 10.1186/1471-2334-10-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varma JK, McCarthy KD, Tasaneeyapan T, Monkongdee P, Kimerling ME, Buntheoun E, Sculier D, Keo C, Phanuphak P, Teeratakulpisarn N, Udomsantisuk N, Dung NH, Lan NT, Yen NT, Cain KP. 2010. Bloodstream infections among HIV-infected outpatients, Southeast Asia. Emerg. Infect. Dis. 16:1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mendelson M. 2007. Diagnosing tuberculosis in HIV-infected patients: challenges and future prospects. Br. Med. Bull. 81–82:149–165 [DOI] [PubMed] [Google Scholar]
- 16. Perkins MD, Cunningham J. 2007. Facing the crisis: improving the diagnosis of tuberculosis in the HIV era. J. Infect. Dis. 196:S15–S27 [DOI] [PubMed] [Google Scholar]
- 17. Ahmed N, Mohanty AK, Mukhopadhyay U, Batish VK, Grover S. 1998. PCR-based rapid detection of Mycobacterium tuberculosis in blood from immunocompetent patients with pulmonary tuberculosis. J. Clin. Microbiol. 36:3094–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Condos R, McClune A, Rom WN, Schluger NW. 1996. Peripheral-blood-based PCR assay to identify patients with active pulmonary tuberculosis. Lancet 347:1082–1085 [DOI] [PubMed] [Google Scholar]
- 19. Lima JF, Montenegro LM, de Albuquerque Montenegro R, Cabral MM, Lima AS, Abath FG, Schindler HC. 2009. Performance of nested PCR in the specific detection of Mycobacterium tuberculosis complex in blood samples of pediatric patients. J. Bras. Pneumol. 35:690–697 [DOI] [PubMed] [Google Scholar]
- 20. Rebollo MJ, San Juan Garrido R, Folgueira D, Palenque E, Díaz-Pedroche C, Lumbreras C, Aguado JM. 2006. Blood and urine samples as useful sources for the direct detection of tuberculosis by polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 56:141–146 [DOI] [PubMed] [Google Scholar]
- 21. Rolfs A, Beige J, Finckh U, Kohler B, Schaberg T, Lokies J, Lode H. 1995. Amplification of Mycobacterium tuberculosis from peripheral blood. J. Clin. Microbiol. 33:3312–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Al-Soud WA, Jönsson LJ, Rådström P. 2000. Identification and Characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 38:345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Soud WA, Rådström P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanscheid T, Monteiro C, Cristino JM, Lito LM, Salgado MJ. 2005. Growth of Mycobacterium tuberculosis in conventional BacT/ALERT FA blood culture bottles allows reliable diagnosis of mycobacteremia. J. Clin. Microbiol. 43:890–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tenney JH, Reller LB, Mirrett S, Wang WL, Weinstein MP. 1982. Controlled evaluation of the volume of blood cultured in detection of bacteremia and fungemia. J. Clin. Microbiol. 15:558–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT, Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L, Whitelaw A, Sagadevan K, Alexander H, Albert H, Cobelens F, Cox H, Alland D, Perkins MD. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 377:1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawn SD, Kerkhoff AD, Vogt M, Ghebrekristos Y, Whitelaw A, Wood R. 2012. Characteristics and early outcomes of patients with Xpert MTB/RIF-negative pulmonary tuberculosis diagnosed during screening before antiretroviral therapy. Clin. Infect. Dis. 54:1071–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-Lopez EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taci N, Yurdakul AS, Ceyhan I, Berktas MB, Ogretensoy M. 2003. Detection of Mycobacterium tuberculosis DNA from peripheral blood in patients with HIV-seronegative and new cases of smear-positive pulmonary tuberculosis by polymerase chain reaction. Respir. Med. 97:676–681 [DOI] [PubMed] [Google Scholar]
- 31. Banada PP, Sivasubramani SK, Blakemore R, Boehme C, Perkins MD, Fennelly K, Alland D. 2010. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J. Clin. Microbiol. 48:3551–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bensinger TA, Zuck TF. 1976. Additional studies concerning the metabolism of packed erythrocytes in CPD adenine. Transfusion 16:353–356 [DOI] [PubMed] [Google Scholar]
- 33. Billett HH. 1990. Hemoglobin and hematocrit, p 718–719 In Walker HK, Hall WD, Hurst JW. (ed), Clinical methods: the history, physical, and laboratory examinations, 3rd ed Butterworth Publishers, Boston, MA: [PubMed] [Google Scholar]
- 34. Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kellogg JA, Manzella JP, McConville JH. 1984. Clinical laboratory comparison of the 10-ml isolator blood culture system with BACTEC radiometric blood culture media. J. Clin. Microbiol. 20:618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bouza E, Sousa D, Rodríguez-Créixems M, Lechuz JG, Muñoz P. 2007. Is the volume of blood cultured still a significant factor in the diagnosis of bloodstream infections? J. Clin. Microbiol. 45:2765–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reimer LG, Wilson ML, Weinstein MP. 1997. Update on detection of bacteremia and fungemia. Clin. Microbiol. Rev. 10:444–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weinstein MP, Mirrett S, Wilson ML, Reimer LG, Reller LB. 1994. Controlled evaluation of 5 versus 10 milliliters of blood cultured in aerobic BacT/Alert blood culture bottles. J. Clin. Microbiol. 32:2103–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunne J, Nolte F, Wilson M. 1997. Cumitech 1B, Blood cultures III. Coordinating ed, Hindler JA., Jr American Society for Microbiology, Washington, DC [Google Scholar]
- 40. Ilstrup DM, Washington JA, II 1983. The importance of volume of blood cultured in the detection of bacteremia and fungemia. Diagn. Microbiol. Infect. Dis. 1:107–110 [DOI] [PubMed] [Google Scholar]
- 41. Mermel LA, Maki DG. 1993. Detection of bacteremia in adults: consequences of culturing an inadequate volume of blood. Ann. Intern. Med. 119:270–272 [DOI] [PubMed] [Google Scholar]
- 42. Satsangi J, Jewell DP, Welsh K, Bunce M, Bell JI. 1994. Effect of heparin on polymerase chain reaction. Lancet 343:1509–1510 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Z, Kermekchiev MB, Barnes WM. 2010. Direct DNA amplification from crude clinical samples with a PCR enhancer cocktail and novel mutants of Taq. J. Mol. Diagn. 12:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yokota M, Tatsumi N, Nathalang O, Yamada T, Tsuda I. 1999. Effects of heparin on polymerase chain reaction for blood white cells. J. Clin. Lab. Anal. 13:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, Zumla A. 2008. Differential susceptibility of PCR reactions [sic] to inhibitors: an important and unrecognised phenomenon. BMC Res. Notes 1:70. 10.1186/1756-0500-1-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Levett PN, Morey RE, Galloway RL, Turner DE, Steigerwalt AG, Mayer LW. 2005. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol. 54:45–49 [DOI] [PubMed] [Google Scholar]