Abstract
There are two very interesting aspects to the evolution of sex chromosomes: what happens after recombination between these chromosome pairs stops and why suppressed recombination evolves. The former question has been intensively studied in a diversity of organisms, but the latter has been studied largely theoretically. To obtain empirical data, we used codominant genic markers in genetic mapping of the dioecious plant Silene latifolia, together with comparative mapping of S. latifolia sex-linked genes in S. vulgaris (a related hermaphrodite species without sex chromosomes). We mapped 29 S. latifolia fully sex-linked genes (including 21 newly discovered from transcriptome sequencing), plus 6 genes in a recombining pseudo-autosomal region (PAR) whose genetic map length is ∼25 cM in both male and female meiosis, suggesting that the PAR may contain many genes. Our comparative mapping shows that most fully sex-linked genes in S. latifolia are located on a single S. vulgaris linkage group and were probably inherited from a single autosome of an ancestor. However, unexpectedly, our maps suggest that the S. latifolia PAR region expanded through translocation events. Some genes in these regions still recombine in S. latifolia, but some genes from both addition events are now fully sex-linked. Recombination suppression is therefore still ongoing in S. latifolia, and multiple recombination suppression events have occurred in a timescale of few million years, much shorter than the timescale of formation of the most recent evolutionary strata of mammal and bird sex chromosomes.
Keywords: Silene latifolia, sex chromosomes, pseudo-autosomal region, genetic map, recombination suppression
AN important question concerning the evolution of sex chromosomes is why recombination suppression evolves between the evolving Y and X chromosomes, leading to subsequent genetic degeneration of Y chromosomes in XY chromosome pairs (and of the W in ZW pairs) and to accumulation of repetitive sequences and, ultimately, to heterochromatinization (e.g., Charlesworth and Charlesworth 2000; Charlesworth et al. 2005; Bachtrog 2006; Bergero and Charlesworth 2009). It is now understood that the large nonrecombining regions of sex chromosomes in mammals have expanded over evolutionary time, forming “evolutionary strata” with differing levels of sequence divergence between X- and Y-gene pairs, from ancient highly diverged regions, to regions in which the genes started diverging much more recently, which are located closest to the pseudo-autosomal region (PAR) on the X chromosome genetic and physical maps (Lahn and Page 1999; Skaletsky et al. 2003; Marais et al. 2008). The more recent evolutionary strata were formed by recombination suppression in regions formed by an addition of autosomal genetic material to the sex chromosomes (Waters et al. 2001), demonstrating that suppressed recombination spread from the ancestral sex chromosomal region into initially recombining regions. The PAR is shared across Eutherian mammals (despite differences in its boundary and the recent addition of a second PAR, PAR2, in humans), which is a physically small remnant of the added region, which is still autosomal in marsupials (Waters et al. 2001).
The major current explanation for the evolution of expanded nonrecombining regions of sex chromosomes is sexual antagonism. According to this theory, sexually antagonistic (SA) alleles that arise in sex chromosomes’ recombining regions can be maintained polymorphically in populations because one allele benefits one sex, but harms the other (Jordan and Charlesworth 2012). Such a polymorphic situation selects for reduced recombination with the fully sex-linked (sex-determining) region, because this increases transmission of the male-benefit allele to males and reduces transmission of these alleles to females (Charlesworth and Charlesworth 1980; Rice 1987; Mank and Ellegren 2009; Jordan and Charlesworth 2012). However, alternatives exist [see Discussion in the accompanying article in this issue (Qiu et al. 2013)], and, no direct evidence currently connects SA alleles to the evolution of reduced recombination of Y or W sex chromosomes.
Testing the SA hypothesis for suppressed recombination requires studying genes in the PARs of sex chromosomes of species that are currently evolving suppressed recombination. Pseudo-autosomal genes are, however, notoriously difficult to find (Burgoyne 1982; Cooke et al. 1985; Schmitt et al. 1994) and only recently have multiple markers been available in the human PAR1 (Lien et al. 2000), despite the progress towards a complete genome sequence (International Human Genome Sequencing Consortium 2001). Eutherian PARs now include only a few genes (e.g., Van Laere et al. 2008), making it unlikely that SA polymorphisms will be found among them, so mammal PARs are no longer likely to be informative about what caused most genes on the sex chromosome pair to become included in the fully sex-linked region.
To study the evolution of sex chromosomes and test the SA polymorphism hypothesis for recombination suppression, a genetic map is essential, together with comparative maps from related species without sex chromosomes. Ideally such mapping work should be based on markers in functional genes to identify orthologous genes for comparative mapping, which can be helpful in testing for chromosome rearrangements in sex chromosome evolution (Filatov 2005).
Young sex chromosome systems, in which evolutionary strata are still evolving, are particularly relevant for testing the SA polymorphism hypothesis, because (unlike some ancient sex chromosome systems) they may often have physically large PAR regions, containing many genes that might harbor such balanced polymorphisms. It should therefore be ideal for mapping PAR genes and testing these ideas. PAR genes of such species, including several plants (Liu et al. 2004; McDaniel et al. 2007; Spigler et al. 2008), could thus be used to search for evidence of SA polymorphisms. However, detailed genetic maps are available in few dioecious plants. In dioecious crop, most work has aimed at finding reliable fully sex-linked markers, even anonymous ones such as AFLP markers, to allow individuals to be sexed before maturity, with potentially important cost savings for long-lived crop plants, including papaya (Liu et al. 2004), poplar (Gunter et al. 2003; Yin et al. 2008), and asparagus (Nakayama et al. 2006; Telgmann-Rauber et al. 2007).
Silene latifolia is a plant widely used for studying sex chromosome evolution, but the genetic map, even of its sex chromosomes, remains sparse (Bergero et al. 2007) because, until recently, very few genes on these chromosomes had been identified. Those that have been mapped and sequenced from the X and Y demonstrate the existence of at least two evolutionary strata (Bergero et al. 2007). Genic markers are preferable to anonymous ones such as AFLPs, the basis for the only previous dense genetic mapping in this species (Delph et al. 2010), because codominant markers, such as SNPs and intron-size variants, are more reliable (e.g., Vekemans et al. 2002) and allow integration of maps from both parents of a cross (e.g., Lorieux et al. 1995a) and construction of consensus maps from multiple families. It is also easier with codominant markers to map individual loci, even when duplicates exist. Finally, genic markers offer future prospects for identifying genes affecting the development of the sex phenotypes and affecting fitness. Studying fitness effects will be necessary if evidence is obtained that SA polymorphisms may be present; it will then be important to verify the fitness effects on the two sexes and to identify the genes involved. Our gene-based map of S. latifolia is much denser than has previously been available and will also be useful for other investigations using this species (Bernasconi et al. 2009).
Although whole-genome sequencing can identify many genes and locate them on chromosomes, it is impractical for S. latifolia, due to its large genome size of almost 3 Gb (Costich et al. 1991; Siroky et al. 2001; Meagher et al. 2005) and highly repetitive sequence content (Hobza et al. 2007; Cermak et al. 2008; Macas et al. 2008). Moreover, sequencing alone does not identify the nonrecombining regions of the sex chromosomes. To distinguish these regions from the recombining PAR, it is necessary also to show that variants are found only in males, and this requires a study of multiple individuals sampled from natural populations (i.e., population genetic data), particularly in a species with a young sex chromosome system where there may be a young stratum with slight Y-X divergence, making it difficult to distinguish from the PAR or even from autosomal regions.
Genes, including PAR genes, can, however, be identified by making a genetic map of the sex chromosomes. Ascertaining PAR genes, however, ideally needs a quite dense map without large gaps. To characterize the PAR region from the young S. latifolia sex chromosome system, we therefore carried out linkage analysis of hundreds of markers to minimize the possibility of large gaps. We identified linkage groups corresponding to the expected number of chromosomes, and we found several genes in a genetically large PAR region. Recently, several putative PAR genes were identified by sequencing a BAC clone after screening a BAC library for a known anonymous PAR marker (Blavet et al. 2012). These genes are necessarily restricted to a small region of the PAR, while our mapping (see below) supports a PAR location for one of these genes and identifies further PAR genes in more distal regions of the PAR.
We also describe comparative mapping in the related species S. vulgaris to extend the previous sparse map of the orthologues of S. latifolia sex-linked genes (Filatov 2005) by mapping several more genes. Comparing the X chromosome genes and their genetic map with the map locations of their S. vulgaris orthologues reveals that chromosome regions have been added to the S. latifolia sex chromosomes to form the PAR region and that some of these genes have become fully sex-linked, suggesting recent formation of a new evolutionary stratum. In the accompanying article in this issue, Qiu et al. (2013), we describe population genetic data that confirm the PAR locations of the PAR genes inferred by genetic mapping, and we use these genes in tests for sexual antagonism that employ sequence analyses.
Materials and Methods
S. latifolia and S. vulgaris families and genetic mapping
To define the major linkage groups and start to identify pseudo-autosomal genes, we used a single F2 family (which we refer to as the “mapping family”) derived from a cross between plants from two European natural populations (Supporting Information, Table S1). To study genes that lacked variants in the mapping family, further families were genotyped (see Table 3 and Table S1).
Table 3. Partial sex-linkage of the putative pseudo-autosomal genes.
| Family |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| The “mapping family” H2005-1F2 (92 events)a |
G2008-3 (22 events) |
G2008-9 (24 events) | D2009-2 (36 events) |
D2009-1 (36 events) | J2010-1 (58 events) |
|||||
| Gene | Xb | Y | X | Y | Y only | X | Y | Y only | X | Y |
| E219 | —c | 20.3 (13.1, 29.4) | — | — | — | 22.2 (10.1, 39.1) | 25 (12.1, 42.2) | 27.8 (14.2, 45.2) | — | — |
| E592 | 14.2 (7.7, 22.9) | 8.3 (3.9, 15.2) | — | — | — | — | — | — | — | — |
| E352 | 0 (0, 4.0) | 0 (0, 4.0) | — | — | — | — | — | 14.7 (4.9, 31.1) | 9.5 (3.9, 21.2) | 10.5 (5.0, 23.3) |
| E200 | 1.1 (0, 5.9) | 0 (0, 4.0) | 4.5 (0.1, 22.8) | — | 4.2 (0.1, 21.1) | — | — | — | — | — |
| E241 | 1.1 (0, 5.9) | 0 (0, 4.0) | — | 9.1 (1.1, 29.2) | — | —d | 2.9 (0.1, 14.5) | — | — | — |
| E284 | — | 0 (0, 4.0) | — | 0 (0, 15.4) | 4.2 (0.1, 21.1) | — | 2.8 (0.1, 14.5) | — | — | — |
The table gives the Kosambi-corrected map distances of each gene from the Y PAR boundary and/or from its inferred map position on the X in six S. latifolia families. Ninety-five percent binomial confidence intervals, computed following Clopper and Pearson (1934), are given in parentheses. The genes are ordered from the highest distance from the Y PAR boundary in the map in Figure 1, which is based on the mapping family (note that E284 was mapped in other families and does not appear in Figure 1).
Numbers of meiotic events.
Distances are from X or Y pseudo-autosomal boundary.
Map distances are not given when informative markers for either or both male and female meioses were not found in a family.
The distance of this gene from the PAR boundary on the X is not estimated in this family, because suitable markers were not found for gene E247.
Genes that mapped to the S. latifolia sex chromosome pair (see Results), and a few autosomal ones, were also mapped in S. vulgaris, a related species that does not have sex chromosomes, but whose populations are gynodioecious (with females and hermaphrodites; see Marsden-Jones and Turrill 1957; Baker 1966; Charlesworth and Laporte 1998). This species is believed to have separated from S. latifolia before the S. latifolia X and Y chromosomes evolved (Marais et al. 2011). Our mapping used 87 progeny plants from a single S. vulgaris family, obtained by crossing a female plant (E2000 5/9 from Dijon, France) with an unrelated hermaphrodite (99K-10-4 from Sussex, United Kingdom; see Table S1). The map was estimated only for genic (codominant) markers that were heterozygous in one parent or the other, and the results from the two parents were integrated into a single map. Some genes could not be mapped in this species due to lack of suitable variants.
Genes studied
To obtain genes for mapping (Table 1, Table S2), we used a large set of EST sequences from 454 transcriptome sequencing of S. latifolia male and female flower buds or from RNA-Seq (Bergero and Charlesworth 2011).
Table 1. Summary of the linkage groups inferred in S. latifolia and the numbers of markers used in the analyses.
| Linkage group | No. of markers | No. of genic markers | Mean intermarker distance (cM) | Length (cM) | LOD scores |
|---|---|---|---|---|---|
| 1 | 60 | 13 | 1.1 | 70.4 | 9 |
| 2 | 38 | 11 | 2.1 | 81.8 | 4 |
| 3 | 23 | 10 | 3.1 | 72.1 | 8 |
| 4 | 27 | 8 | 1.9 | 50.8 | 7 |
| 5 | 12 | 9 | 2.9 | 35.2 | 10 |
| 6 | 26 | 14 | 1.4 | 36.0 | 10 |
| 7 | 13 | 9 | 3.5 | 46.1 | 6 |
| 8 | 12 | 7 | 3.4 | 40.5 | 5 |
| 9 | 41 | 23 | 1.3 | 52.8 | 10 |
| 10 | 32 | 10 | 2.3 | 75.5 | 5 |
| 11 | 34 | 14 | 1.0 | 41.5 | 10 |
| 12 (X) | 32 | 25a | 2.6 | 75.0 | 5 |
| 12 (Y) | 57 | 15 | — | 29.4 | 6 |
Including partially sex-linked genes.
To obtain the EST sequences, flower buds were collected from unrelated male and female S. latifolia plants growing in greenhouse condition, snap-frozen in dry ice, and stored at –80°. The buds were ground in a mortar and pestle under liquid nitrogen, and messenger RNA (mRNA) was extracted using the Roche Diagnostics (Mannheim, Germany) mRNA isolation kit, which uses streptavidin-coated magnetic beads and a biotin-labeled oligo(dT)20, and cDNA was prepared. Double-stranded cDNA was obtained using the Roche cDNA synthesis kit and amplified with phi29 DNA polymerase (Qiagen) following the manufacturer’s recommendations. The amplified library was checked for representation of cDNA sequences by cloning in a T-tailed pBS vector (Stratagene) before 454 sequencing. A total of 223,112 reads were obtained and assembled using Roche’s Newbler, with the default settings, into 13,345 contigs of an average length of 318 bp.
To search for genes to provide codominant markers, sequences >300 bp were selected. Amino acid sequences from open reading frames (ORFs) of these EST sequences were aligned to the translated Arabidopsis thaliana transcriptome (Swarbreck et al. 2008) using FASTA (Pearson 1990). ORFs with e values <10-5 with single-copy A. thaliana orthologues or orthologues belonging to small gene families were retained, and Sequencher v.4.5 (GeneCodes, Ann Arbor, MI) was used with the A. thaliana genomic sequence of the putative orthologues to suggest intron position order to obtain codominant intron size variants (ISVS markers; see Bergero et al. 2007) for our genetic mapping described below. PCR-amplified introns from a total of 160 genes showed suitable variants in our mapping family, i.e., variants that are informative in either male and female meiosis, or both. Table S2 lists the genes mapped in S. latifolia with their GenBank accession numbers.
To increase the marker density, segregating Miniature Inverted-repeat Transposable Element (MITE) insertions identified by Bergero et al. (2008) were mapped. These dominant MITE markers are denoted in the maps by names beginning with either El_ or To_ followed by numbers identifying individual markers.
DNA extraction, PCR reactions, and cloning
Genomic DNA for sequencing and marker genotyping was extracted from leaves using the FastDNA kit (Q-BIOgene, Carlsbad, CA) following the manufacturer’s instructions. The seedlings’ sexes were determined by PCR amplification of intron 2 of the SlCyp gene, which contains a fixed 293-bp indel in the Y copy (Bergero et al. 2007).
PCR amplifications were done in a Finnzymes’ Piko Thermal Cycler with Phire hot-start DNA polymerase (Finnzymes, Thermo Scientific Genomics, Epsom, UK) under the following conditions: 1 cycle of initial denaturation at 98° for 30 sec, 10 cycles of DNA denaturation at 98° for 5 sec, primer annealing varying from 60° to 70° for 5 sec, and DNA amplification 72° for 30 sec, 25 cycles at 98° for 5 sec, 57° for 5 sec, 72° for 60 sec, and finally 1 cycle at 72° for 5 min. For genotyping polymorphic intron size variants, PCR products from parents and offspring were run on capillary electrophoresis according to Bergero et al. (2007), except that the 1200 Genescan LIZ DNA ladder (Applied Biosystems, Carlsbad, CA) was used to size the PCR products.
To obtain sequences of the genes studied, and search for SNP variants when length variants (see above) were not available, PCR products were cleaned with ExoSAP-IT (Amersham Biosciences, Tokyo) and sequenced on an ABI 3730 capillary sequencer (Applied Biosystems). SNPs were obtained by direct sequencing from both strands. DNAs producing PCR amplicons with heterozygous indels in intron regions were re-amplified with the proofreading Phusion DNA polymerase (Finnzymes, Thermo Scientific Genomics) using the PCR conditions given above and cloned into a T-tailed pBSKS+ vector (Stratagene, Santa Clara, CA) before sequencing. The final sequences were aligned in Sequencher 4.8 (Gene Codes; http://www.genecodes.com), including sequences of the putative orthologous genes from S. vulgaris, and manually adjusted by using Se-al v. 2.0 (http://tree.bio.ed.ac.uk/software/seal/).
Linkage analysis
Because genetic maps can be affected by null alleles, making individuals appear to be homozygotes when they are not, we checked the segregation patterns of markers from putative PAR genes carefully within multiple families to ensure that they were always consistent with segregation of alleles at a single locus and that all parents classified as heterozygotes segregated (which excludes the presence of paralogous copies). The accompanying article in this issue, Qiu et al. (2013), describes further tests that can distinguish between partial and complete sex linkage based on population genetic evidence.
Linkage groups were inferred using the JoinMap software version 4.0 (van Ooijen 2006) with a minimum LOD (logarithm of odds) score of 3. JoinMap searches for the best-fitting order of markers by a trial-and-error procedure. The map for each linkage group was then estimated in JoinMap with the regression mapping procedure, Kosambi mapping function, a recombination threshold of 0.4, and a goodness-of-fit jump threshold of 5.0. The genotypes of markers exhibiting significantly distorted segregation ratios were checked and excluded when they were not clear. A consensus map of the X chromosome (Figure 2) was estimated with JoinMap to include another family, J2010-1 (whose female parent is from the mapping family; see Table S1), in which seven more fully sex-linked genes were mapped, as well five genes in common with the mapping family, including one putative PAR gene (see Results). Unfortunately, the only other family with segregation data for E219 from the female parent has too few mapped markers to produce a reliable consensus map.
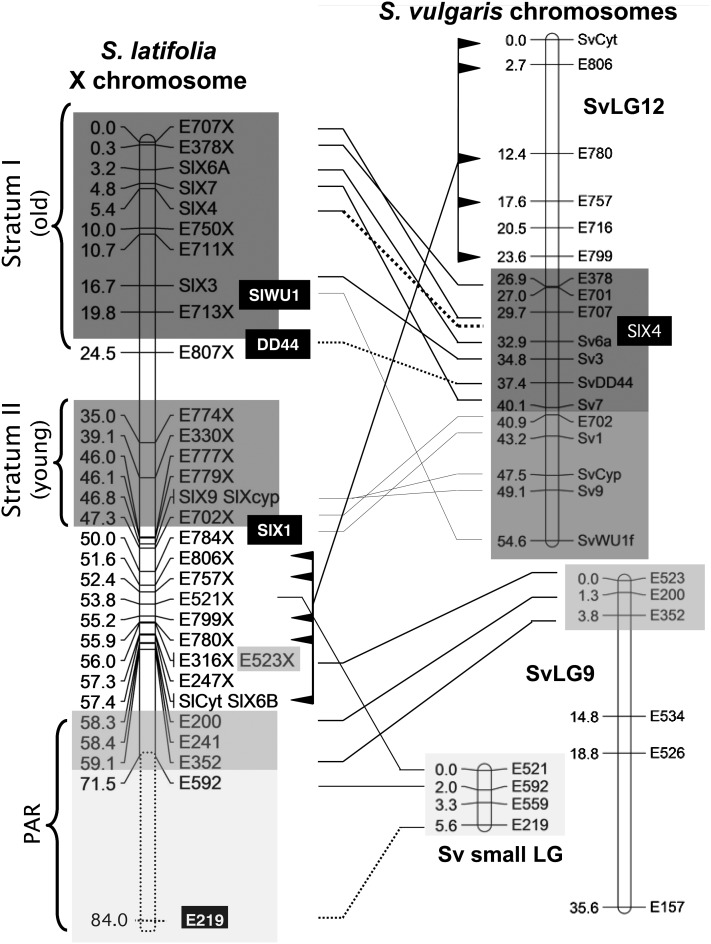
Figure 2.
Consensus genetic map of the S. latifolia X chromosome, including the PAR, and map of the homologous genes in S. vulgaris. All fully X-linked genes have markers showing that they are sex linked, either in the mapping family or, when variants were not found in the female parent, in another family. The PAR is indicated, and also the two strata previously described (Bergero et al. 2007); genes mapping close to genes in one of these strata are assumed to belong to the same stratum, even when X-Y divergence values have not been estimated. Genes mapped in both species are given the same names in S. vulgaris as in S. latifolia and are connected with lines (or lines and flags), and genes mapped in only one species, or mapped in S. latifolia families different from ours, are indicated by solid boxes and dashed lines to the locations in the species for which information is missing in our families. Boxes of different shades indicate genes in distinct S. vulgaris linkage groups that correspond to syntenic blocks in the S. latifolia X. We named one S. vulgaris linkage group Sv9 because it carries homologs of three genes mapped to S. latifolia LG 9 (see text), but did not assign a linkage group number to the very small group (“SV small LG”) because all four genes are in the S. latifolia PAR or closely linked to it. Two such genes (SlX1 and DD44-X) were previously mapped to the S. latifolia X (Filatov 2005; Nicolas et al. 2005), while one S. latifolia gene (SlX4) could not be mapped in our S. vulgaris family.
To test for linkage with known genes on the sex chromosomes, we also mapped genes that had previously been shown to be sex-linked, including OPA, a nongenic RAPD pseudo-autosomal marker (DiStilio et al. 1998). OPA does not appear in the Y map in Figure 1 due to lack of suitable variants in the mapping family. For the fully sex-linked region, our maps for the Y and the X (Figure 1) are based on segregation of markers that are informative for male and female parent meioses, respectively, as the X and Y maps cannot be integrated across the PAR and fully sex-linked regions.
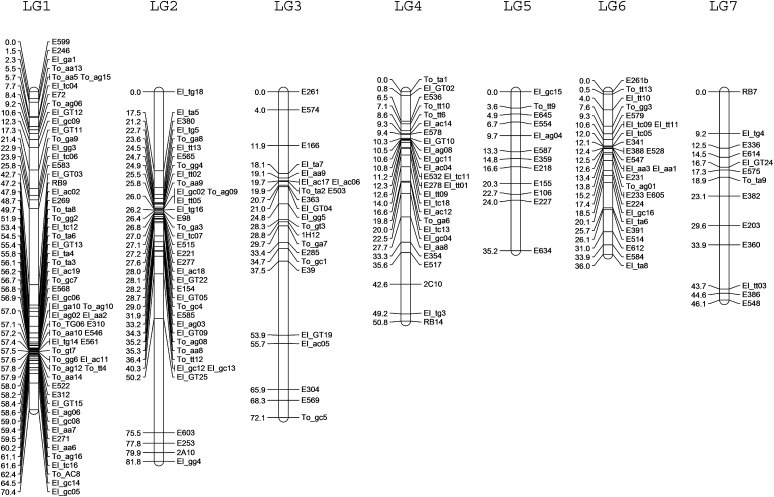
Figure 1.
S. latifolia genetic map. The map (estimated for the S. latifolia mapping family using JoinMap; see Materials and Methods) includes a total of 156 genic markers (153 excluding 3 genic markers mapped only on the Y, 2 of them in the nonrecombining part), and 239 segregating MITE insertions (197 excluding 42 on the Y). The analysis yielded 12 major linkage groups, corresponding to the expected 11 autosomes, plus the sex chromosome pair, all with LOD scores >4 (Table 1). For linkage group 4, integration of female-informative and male-informative markers was achieved using an additional family (J2010-1; see Table 3), and the map shown for this linkage group is therefore a consensus map. Five MITE markers were not detectably linked to any others, and there is also a small linkage group (12 cM) containing four MITE markers. Numbers to the left of the linkage groups indicate genetic distances in centimorgans, estimated using the Kosambi mapping function.
The genetic map distances of pseudo-autosomal genes from the Y PAR boundary shown in Table 3 were estimated from segregation patterns of heterozygous variants in the putative pseudo-autosomal genes in the male parent and a Y-specific variant in the fully Y-linked SlCypY gene (as a marker for the plant’s sex even when the individual had not flowered; variants in this gene were available in all the families in Table 3). We also scored the segregation of markers that were heterozygous in the maternal parents of our families and estimated distances between the putative genes and variants in the fully X-linked gene E247, which is closely linked to PAR genes (see Figure 2). In family D2009-2, this gene could not be used, and E219 was mapped using the pseudo-autosomal gene E241; given the possible map locations for E241 (see Results), the distance of E219 from the PAR boundary on the X may be underestimated.
To estimate the physical size (in megabases) of a centimorgan in S. latifolia, we used the estimated genome size of 2879 Mb (Vagera et al. 1994) and assumed that 14% of the female genome is represented by the X chromosome of ∼350 Mb and that 86% is autosomal, representing ∼2076 Mb (Matsunaga et al. 1994).
Results
Genetic map of S. latifolia
Segregation analyses of 407 markers from our S. latifolia mapping family yielded a genetic map with 12 linkage groups, corresponding to the expected 11 autosomes, plus the pair of sex chromosomes (Table 1, Figure 1). The total genetic length, excluding the Y, estimated by adding twice the mean marker spacing to the length of each linkage group (Fishman et al. 2001) is 731.5 cM. Method 4 of Chakravarti et al. ( 1991) yields 690.8 cM; i.e., our map includes ∼98% of these estimated map lengths. Using this genetic length value, we estimated, following Fishman et al. ( 2001), that 99% of the genome should be within 5 cM of a marker and 64% within 1 cM; with nongenic markers also available (Table 1), the map of this species is quite well covered. Overall, using an estimated total autosomal genome size of 2076 Mb (see Materials and Methods), the estimated physical size per centimorgan is 3.4 Mb for the autosomes, among the higher values for plants (Rees and Durrant 1986; Laurie et al. 1993; King et al. 2002).
The genetic lengths of the S. latifolia chromosomes (excluding the Y) increase with the numbers of genes assigned to them (y = 47.5 + 0.69x), but the relationship is nonsignificant by a Spearman rank correlation (Figure S1). The intercept of a linear regression on the y-axis suggests that there is always one crossover per chromosome. This is surprising because the chromosomes are metacentric (e.g., Siroky et al. 2001), and a crossover on each arm would be expected.
Genetic map of the sex chromosome pair and the PAR:
Our new results considerably increase the number of fully sex-linked genes mapped to locations on the S. latifolia X chromosome (Bergero et al. 2007; Kaiser et al. 2009, 2011). All these genes have markers showing that they are sex linked, but not all have variants in our mapping family; Figure 2 shows a consensus map based on two families with several markers mapped, plus inferred locations for three genes not mapped in these families; this map does not differ in any important respect from that of the mapping family alone in Figure 1. Table 2 lists the PAR genes, plus three genes that map close to PAR genes and were initial candidate PAR genes, but are now assigned to the fully sex-linked region of the X chromosome (see below). The cytological length of the X, relative to the set of autosomes in females, is known to be ∼14% of the genome (Vagera et al. 1994). On this basis, our mapping results yield large estimated physical sizes per centimorgan for both the X and autosomes (4.65 and 3.4 Mb, respectively). This result suggests a large repetitive content of the genome, and perhaps especially for the X, much of which recombines only in females and therefore has a lower recombination rate than the autosomes.
Table 2. Pseudo-autosomal genes and three newly identified fully sex-linked genes mapped close to the S. latifolia PAR boundary.
| S. latifolia gene name | Locationa | Accession no. of A. thaliana orthologue | Putative function or protein encoded |
|---|---|---|---|
| E200 | PAR | at5g06160 | Protein with similarity to pre-mRNA splicing factor SF3a60b |
| E219 | PAR | at2g13360 | Peroxisomal photorespiratory enzyme catalyzing transaminationc |
| E241 | PAR | at4g27700 | Rhodanese-like domain-containing protein |
| E284 | PAR | at5g52440 | Proton motive force dependent protein transmembrane transporter |
| E592 | PAR | at3g22960 | Chloroplast pyruvate kinase α-subunit |
| E352 | PAR | at2g43640 | Chloroplast RNA binding |
| E559 | XY | at3g21865 | Interacts with PEX4d |
| E521 | XY | at4g27450 | Unknown function |
| E523 | XY | at5g55220 | Peptidyl-prolyl cis-trans isomerase |
The putative orthologues in A. thaliana were used to suggest the genes’ likely functions.
For the fully sex-linked genes, we indicate those for which both X and Y alleles have been sequenced, or just X-linked ones.
Involved in gametic cell fate determination and expressed more strongly in gametophytes than sporophytes.
Reactions with multiple substrates.
Suggested to be involved in remodeling peroxisome matrix contents during glyoxysome transition to leaf peroxisomes.
As well as fully sex-linked genes, we found four new partially sex-linked genes (E200, E352, E219, and E592; see Figure 2 and Table 3) in addition to the two PAR loci previously described without genetic details, E241 and E284 (Qiu et al. 2010). Segregation patterns indicate that the variants used to infer partial sex linkage are allelic (despite the existence of a paralogous copy of E284; see the accompanying article in this issue, Qiu et al. 2013), and they indicate linkage to the sex-determining region of the sex chromosomes and/or to other X-linked or PAR genes (Table 3 shows recombination distances in various families). For gene E200, partial sex linkage is well supported by recombinants in three different families (with small estimated recombination fractions with the X or Y PAR boundary). E352 appeared fully sex linked in our results from the mapping family based on an intron size variant that consistently segregated from the male parent to male progeny, but not to females, but recombinants were present at much higher frequencies in two other families in both female and male meiosis (Table 3). Further testing of all six candidate PAR genes using samples from natural populations confirmed partial sex linkage (see the accompanying article in this issue, Qiu et al. 2013). One of the PAR genes, E241, was also found in a recently described PAR BAC (Blavet et al. 2012). As expected, this gene is closely linked to the OPA marker that was used to screen the PAR BAC (Figure 1). However, in our mapping family our estimated recombination distance from the PAR boundary is considerably smaller than estimated by Blavet et al. (2012) from the male parent of their family or from the male parent of another of our families (Table 3). The difference seems unlikely to reflect a higher recombination rate in male than female meiosis, as it was similar in meiosis of both parents of our mapping family, and there is no consistent difference in rates for the few other intervals that we were able to compare between the sexes (Table 3). Another possibility is heterozygosity of the maternal parent for an X chromosome rearrangement in the region. This is consistent with the fact that another gene, E352, also recombines rarely with fully sex-linked markers in the mapping family, but much more often in two other families; in family J2010-1, the rate estimates are similar in meiosis of both sexes (Table 3), again arguing against a higher male than female recombination rate.
S. latifolia PAR:
Unlike the previously published map, which used dominant AFLP markers and inferred two PARs (Scotti and Delph 2006; Delph et al. 2010), our map, with a new set of mostly codominant markers, finds only a single PAR. This suggests that more study is needed before the existence of a second PAR is accepted, particularly as cytological studies consistently report terminal pairing of the X and Y chromosomes (e.g., Zluvova et al. 2007; Filatov et al. 2008), except in chromosome mutants (e.g., Westergaard 1946, 1948a; Zluvova et al. 2007). Both the previous mapping and ours, including families other than the mapping family, find a PAR of ∼25–30 cM (Figure 2, Table 3). The small number of PAR genes currently mapped could have caused under-estimation of its total map length, but, given the high level of polymorphism in this species (see above and Qiu et al. 2013), it seems unlikely that we failed to detect and map a single gene in a second PAR of comparable or larger size (e.g., due to all genes located in the region completely lacking any variants).
Our genetic map length for the S. latifolia PAR is double the estimated map distance of <11 cM for the anonymous OPA marker from fully sex-linked markers, supported by recent mapping of four genes identified from a BAC clone that contains the OPA sequence (Blavet et al. 2012); from the standard errors in table S3 of Blavet et al. (2012), those data are compatible with a rate of up to 18 cM. There is no reason to believe that this BAC contains the whole of the PAR, or the distal part of it, and a larger PAR map is supported by our location of one gene (E219) at least 20 cM from the fully sex-linked region in three independent families, one with data from both the parental plants (the lowest recombination rate compatible with the 95% confidence intervals in Table 3 is 10.1). In the mapping family, the LOD for the linkage of the E219 marker to the Y-linked allele of E592 is 7.3. In S. vulgaris, the ortholog of E219 is also linked to the orthologs of two S. latifolia fully sex-linked genes, E559 and E592 (LOD value for linkage with E559 = 28.12).
A physically very small PAR, like those of mammalian sex chromosomes, would be expected to have a larger genetic map length in male than female meiosis. The higher male crossing-over rate in such regions in other organisms, such as humans and mice, is thought to be caused by the need for a crossover to ensure correct chromosome segregation (Burgoyne et al. 1992; Matsuda et al. 1992; Mohandas et al. 1992); this leads to an elevated number of crossovers within this small physical region in males, whereas, in females, other parts of the sex chromosome pair can crossover (Lien et al. 2000). In S. latifolia, however, the genetic map distances between PAR loci appear to be similar in female and male meiosis (Table 3). This is consistent with a physically large PAR or with crossovers restricted to a physically small region of this chromosome arm in both sexes.
To further test whether crossing over is unusually high in the PAR, we also analyzed the GC content of the PAR genes. A very high recombination rate in PAR genes in male meiosis is likely to increase their CG content through the effect of biased gene conversion (Marais and Galtier 2003). However, GC3 in the coding sequences of the S. latifolia PAR genes is 0.328 ± 0.083, lower than for 11 X-linked genes (0.391 ± 0.006, excluding the 3 genes E559, E521, and E523—which, as explained below, probably became fully sex linked only recently—or 0.403 ± 0.024 including these genes), and slightly higher than the value for the available autosomal genes (0.371 ± 0.027). When all sites are analyzed, the GC contents are almost identical (0.387 for fully X-linked versus 0.384 for PAR genes), and the value for autosomal genes is higher (0.424 ± 0.008). Thus, consistent with the conclusion of Blavet et al. (2012), the GC values do not suggest an elevated recombination rate in the PAR genes, although more genes should be studied in the future.
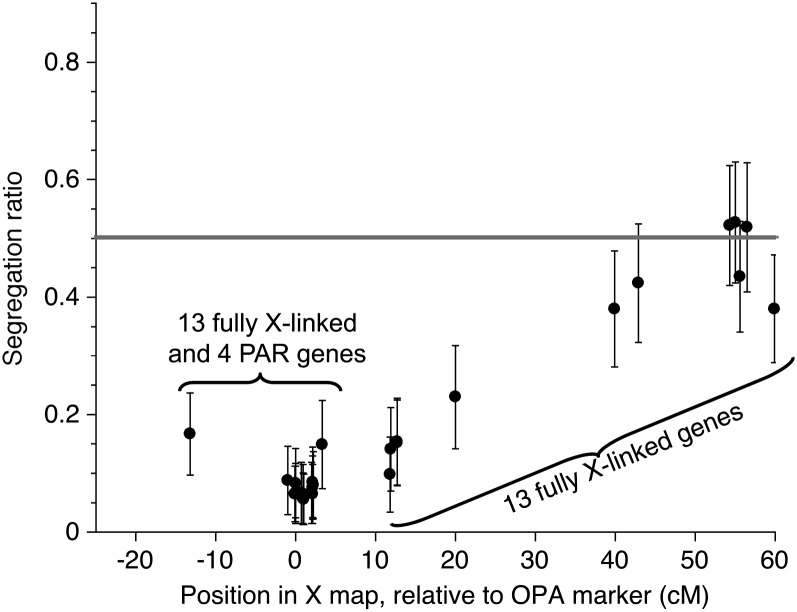
Aberrant segregation ratios:
The expected segregation ratios for X-linked genes heterozygous in the maternal parent are 1:1, but significantly deviating ratios (based on chi square tests of the 1:1 null hypothesis) are observed for several X-linked genes in the mapping family (Figure 3). The ratios are strongly aberrant for several genes that we inferred to be closely linked to the PAR boundary, with a deficiency of one maternal haplotype; a lesser effect is detected for some genetic distance on both sides of the boundary. A highly aberrant segregation ratio in the mapping family was previously found for the SlCyt X-linked gene and other closely linked genes (Kaiser et al. 2009). Other families showed normal segregation ratios for these genes, but the distorted segregation phenotype is heritable, since female progeny in the mapping family that are heterozygous for the two PAR boundary haplotypes also showed non-Mendelian segregation.
Figure 3.
Segregation ratios in the mapping family. Binomial confidence intervals were calculated based on the numbers of progeny plants genotyped. The horizontal shaded line indicates the expected 1:1 ratio.
The cause of the aberrant ratios is unknown. Given the evidence for chromosome rearrangements affecting the PAR region of the X chromosome (see below), a translocation that is not yet fixed in the species might be involved. Translocation heterozygotes commonly produce gametes with gene duplications and deficiencies, and inviable seeds are produced at high frequencies (Swanson 1957) that could potentially explain the ratios that we observe. More work will be needed in the future to determine whether our plants showing distorted X-chromosome segregation are heterozygous for rearrangements of the X in the PAR boundary region. The fully sex-linked region shows no sign of X-chromosome polymorphisms. Similar gene orders (with only minor differences involving closely linked loci) have been found in genetic maps using four independent crosses involving unrelated plants (Filatov 2005; Nicolas et al. 2005; Bergero et al. 2007; Fujita et al. 2011).
Clustering of loci near the PAR boundary:
The X chromosome map shows a cluster of markers near the PAR boundary in which the ordering is not reliable. For the genic markers, population genetics approaches (see Qiu et al. 2013) confirm partial, as opposed to complete, sex linkage of the genes listed as PAR genes in Table 2; full sex linkage was previously demonstrated for SlCyt (Kaiser et al. 2009) and Sl6B (Bergero et al. 2007). There is, however, no way to further test the MITE markers in this region.
Apparent clustering of markers can potentially be due to the presence of a gene with distorted segregation, but, although this is indeed the case in our family (see above), this should not be the cause of the clustering because our markers in these genes are codominant (Lorieux et al. 1995b). Strongly non-Mendelian segregation ratios in a parent, including ratios caused by differential mortality, however, can produce inaccurate genetic map intervals and create spurious linkages of markers when using standard mapping approaches employed in widely used software (Cloutier et al. 1995). Unbiased pairwise rate estimates using Bailey’s (1949) formula can be applied to our genotype data, using the approach designed for a backcross (Lorieux et al. 1995a), since both maternal alleles can be scored in each progeny individual. Using the appropriate tests of the null hypothesis of independent segregation, all the pairs of adjacent markers show highly significant evidence for linkage. The true orders of the loci, however, could differ somewhat from those inferred by JoinMap. Bailey’s (1949) formula yields zero recombination rates for all the PAR intervals in the mapping family. However, these loci clearly recombine in families that segregate normally (Table 3), supporting their PAR locations. Of the genes mapped in family J2010-1 (see above), at least six map within the region near the PAR boundary where the mapping family shows significantly non-1:1 ratios, but their segregation ratios are compatible with 1:1 in family J2010-1. This suggests that the clustering is unlikely to be an artifact due to non-Mendelian ratios.
Comparative map of S. latifolia sex chromosomal markers in S. vulgaris
It is of interest to test whether the orthologues of our S. latifolia fully sex-linked genes map to a single chromosome in the outgroup species S. vulgaris (with no history of having sex chromosomes) because the initial evolution of sex-determining genes is hypothesized to have occurred on a single ancestral chromosome (Charlesworth and Charlesworth 1978). Linkage groups shared with an outgroup species such as S. vulgaris are likely to be ancestral, unless chromosome rearrangements are very frequent.
Orthologues of four fully sex-linked genes have previously been mapped in S. vulgaris (Filatov 2005), and all indeed proved to be located on a single linkage group. The results of our mapping of 14 additional S. vulgaris orthologues of genes from the two X-chromosome evolutionary strata, using 85 progeny of a full-sib family, are completely consistent with this conclusion, although we now detect some rearrangements between the homologous chromosomes of the two species (Figure 2). Genetic mapping in S. vulgaris initially suggested that the SlCyt gene belongs to a linkage group different from that carrying orthologues of the other S. latifolia X-linked genes, suggesting that this gene had moved onto the X (Kaiser et al. 2009). However, our new mapping, with more loci, now locates SlCyt to the S. vulgaris linkage group carrying the other S. latifolia fully X-linked genes, but at the end of the linkage group farthest from the S. latifolia PAR gene homologs; in S. latifolia, however, this gene is in the cluster of genes closely linked to the PAR boundary (Figure 2). Four other genes in a 24-cM region that includes the S. vulgaris SlCyt orthologue also have different map positions in the two species (Figure 2), and we infer that a large region, probably containing many genes, has been moved in a single rearrangement event in one lineage or the other. In the S. latifolia genetic map, these genes map to only 2.4 cM; they may have been added to a physically large pericentromeric region with limited recombination, as is commonly found in plant chromosomes (reviewed in King et al. 2007). However, we cannot currently exclude a rearrangement of the mapping family X in the PAR boundary region (see above).
A surprising finding is that homologs of three S. latifolia PAR genes and four fully X-linked genes closely linked to the PAR boundary map of two distinct S. vulgaris linkage groups, both apparently unlinked to the major linkage group homologous to the S. latifolia X chromosome (Figure 2). The larger of these S. vulgaris linkage groups (whose top part carries the orthologues of the S. latifolia pseudo-autosomal genes E200 and E352) is probably homologous to S. latifolia linkage group 9 (Figure 1 and Figure 2), as it includes three genes, E157, E526, and E534, from that autosomal linkage group.
A smaller S. vulgaris syntenic block (labeled “SV small LG” in Figure 2) also combines S. latifolia PAR genes (E219 and E592) and fully sex-linked ones (E559 and E521). Both of the latter have copies on the S. latifolia X and the Y (see Figure S2), but unfortunately heterozygosity of both parents for variants prevented our mapping E559 on the X in our families. Segregation of E559 variants indicates complete Y-linkage in our D2009-2 family, but only 36 meioses were studied, which cannot exclude partial sex linkage (see Table 3). However, our diversity survey using samples from natural populations (see Qiu et al. 2013) detected numerous Y-specific SNP variants (Figure S2A). Complete Y-linkage of copies of E521 and E523 was inferred from segregation of intron size variants in the mapping family, and their complete Y-linkage is confirmed by male-specific SNPs in E523 (Figure S2B) and intron size variants in both genes.
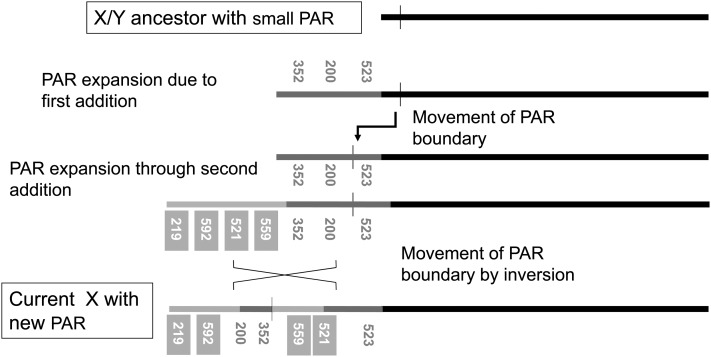
Figure 4 shows an interpretation of the additions to the S. latifolia sex chromosomes. These events presumably happened after the evolution of the younger of the two previously known strata (Bergero et al. 2007), and cessation of recombination must have been even more recent. Most likely, a first event added three genes, E200, E352 and E523, and the latter recently became fully sex linked. Consistent with this, silent site divergence between the E523 X and Y sequences is low (1.79%, based on 527 sites); as diversity is very low among the X- and the Y-linked sets of alleles, net divergence for all site types is slightly lower (based on all 662 sites, it is 1.35%, with standard deviation 0.3%). The distal location on the X genetic map of the two genes, E219 and E592, suggests that they were added in the most recent event involving the S. vulgaris linkage group labeled “Sv small LG” in Figure 2, along with E521 and E559, which have also become fully sex linked. For the latter genes, net divergence between the Y- and X-linked sequences from our natural population sample (see Qiu et al. 2013) is 1.2% for E559 (based on 777 bp and a similar value for silent sites), but much higher for E521 (3.5% for all site types, based on 640 bp, with standard deviation 1.4%, and 6.6% for 336 silent sites). We discuss these results further in Qiu et al. (2013).
Figure 4.
Stages in the evolution of the PAR region of the S. latifolia sex chromosomes. Horizontal lines symbolize X chromosomes at several intermediate stages during its evolution, from the initial state, before additions and rearrangements, to the current state (indicated by the text in boxes), and the additions and rearrangements needed to account for the conclusions described in the text. The ancestral sex chromosome is solid, and the two added regions and genes are shaded. The regions and genes that have been added to the X must also have been added to the Y (see the text), but the Y is not shown in the figure.
Discussion
Pseudo-autosomal regions are centrally important in the theory of sex chromosome evolution because it is the evolution of suppressed recombination in former PARs that leads to the genetic isolation of the Y from the X and, over time, to the special properties of Y chromosomes. After selection for closer linkage between sterility factors that initiated the evolutionary change from hermaphroditism or cosexuality to the separate sex state (Charlesworth and Charlesworth 1978), sex chromosomes are predicted to acquire SA mutations, some of which may be in PAR genes, and may establish polymorphisms. Selection for closer linkage between the gene with such a polymorphism and the Male-Specific Y region (MSY) can lead to an increased MSY, while the formerly pseudo-autosomal region(s) shrink. This second stage may be repeated several times, creating evolutionary strata.
Sex chromosome strata can also evolve by additions of genome regions to an existing sex chromosome pair, and such additions have occurred in many sex chromosome systems and can be followed by loss of recombination in the added region. In mammals, strata 1 and 2 are X-linked in marsupials, but strata 3–5 and one PAR derive from the “X added region,” which was added by Eutherian-specific translocation of previously autosomal regions (Waters et al. 2001). A second human PAR, PAR2, also evolved through later additions of several genes to the Xq tip during mammalian evolutionary history; the Y lacked these genes until the region was duplicated onto the Y in a primate ancestor of humans, forming a new recombining PAR (Charchar et al. 2003).
In S. latifolia, two strata of fully sex-linked genes have previously been inferred (Atanassov et al. 2001; Bergero et al. 2007) and two genes from each stratum of X-Y divergence were mapped to a single S. vulgaris chromosome (Filatov 2005), suggesting that the younger X chromosome stratum was not derived by an addition to the sex chromosome pair, unlike the situation in the mammalian XY pair.
Our comparative mapping now shows that none of the S. latifolia PAR genes maps to this S. vulgaris linkage group and suggests that two previously autosomal regions have been added to the S. latifolia sex chromosome pair by translocations or fusions. Part of one of the two S. vulgaris LGs involved corresponds to S. latifolia autosomal LG 9, yet three genes of this LG map to the S. latifolia PAR, or close by (see Figure 2). Moreover, this addition probably occurred first, before that involving the small S. vulgaris linkage group, because genes with homologs on the Sv small LG are at the end of the PAR map (see above and Figure 4). Therefore, if the small S. vulgaris linkage group is indeed part of the same chromosome that carries orthologues of S. latifolia fully sex-linked genes, the addition of the LG9-orthologous genes must involve a further rearrangement. Ideally, a dense map of all S. vulgaris linkage groups should be estimated, as our present data cannot definitively exclude the possibility that all S. latifolia sex-linked genes belong to a single S. vulgaris chromosome that includes the small linkage group in Figure 2. However, large gaps in our map seem unlikely. It is implausible that genes in a region all lack variants, as the species has high sequence diversity, and gaps due to errors in genetic distance estimates are unlikely with our mapping population of 85 plants. Because our S. vulgaris map includes largely only genes known to be on the S. latifolia sex chromosomes, we cannot exclude gaps due to failure to ascertain sex-linked genes in some regions. Such gaps could be caused by absence of Y-linked homologs of S. latifolia X-linked genes or by absence of fixed Y-linked variants that allowed us to detect and map S. latifolia sex-linked genes. The absence of a gap in the S. latifolia map might argue against this, but it could simply be due to the fact that this region is one of close linkage, with a potentially large physical distance but only small genetic map distances. Further mapping in S. vulgaris is needed to identify other genes belonging to the small linkage group and to find which S. latifolia chromosome carries their orthologues.
Size of the S. latifolia PAR
The large genetic map length of the PAR suggests that it probably carries many genes. It is consistent with this conclusion that the PAR genes that we have identified, which clearly recombine with one another, include only one of the genes in the sequenced BAC (Blavet et al. 2012).The size of the S. latifolia pairing region between the X and Y chromosomes is not well estimated from the cytologically visible pairing and is described as “arbitrary” (Westergaard 1948b; Lengerova et al. 2003), although the region appears to be small in FISH preparations (Filatov et al. 2008). If the PAR of our map corresponds with the PAR-bearing Xp arm of the X, as established by Lengerova et al. (2003), the map lengths would correspond roughly with the physical size of the two X arms (Lengerova et al. 2003).
Finally, additions to the PAR are consistent with the large cytological size of the S. latifolia sex chromosome pair, relative to the autosomes, while the chromosomes of S. vulgaris and other outgroup species are all roughly similar in size (Siroky et al. 2001), which suggests that this difference is due to changes in the S. latifolia lineage, rather than in the S. vulgaris one (which would imply the reverse order of events in Figure 4). Additions to the XY chromosome pair are consistent with the location, by FISH mapping, of the SlX1 gene to a position that is not close to the end of the chromosome (Howell et al. 2011). This appeared inconsistent with the low X-Y divergence estimated for this fully sex-linked gene, which suggested a location near the PAR boundary. If the PAR is a physically large region, the inconsistency disappears. In S. vulgaris, the SlX1 homolog is closely linked to the orthologues of two other genes in the youngest S. latifolia stratum previously identified, SlCyp (Bergero et al. 2007) and SlX9 (Kaiser et al. 2011). In S. vulgaris, the mapping predicts that the physical locations of these genes are near the tip of the chromosome corresponding to SvLG12.
Origins of the new S. latifolia sex chromosome strata
Additions to the sex chromosomes (rather than breakup of the homologous chromosome in the S. vulgaris lineage) are also suggested by the finding that some genes from both the syntenic blocks that we infer to be added are currently fully sex linked, while others are not. In other words, a new stratum has formed since the formation of the younger of the two strata previously identified (Bergero et al. 2007). Two events presumably added regions onto the S. latifolia Xp arm that carries the single PAR detected in our mapping (see Figure 2). Both additions probably expanded an initially small PAR, and then recombined, so that both the X and the Y now carry copies of these genes. Alternatively, these genes must have been in a PAR larger than the current one, and chromosome rearrangements moved some to the fully sex-linked region. From either view, genes from both regions have become fully sex linked. As described in Qiu et al. (2013), the E559 sequences, located in the second added region (Figure 4), contain X- and Y-specific SNPs that clearly indicate complete sex linkage, but X and the Y alleles also share variants, supporting a very recent event suppressing recombination (we found no polymorphisms shared by the X and Y in either the E523 or the E521 regions sequenced).
It thus appears that multiple losses of recombination have happened within an evolutionary time much shorter than that since the most recent strata were established in the sex chromosome systems of mammals (see Introduction) and birds (Lawson-Handley et al. 2004; Nam and Ellegren 2008; Moghadam et al. 2012). The most recent mammal stratum (5) is estimated to have formed ∼30 MYA, ∼5 MY before the divergence of the human and Old World monkey lineages (Hughes et al. 2012), and stratum 4 ∼10 MY earlier (Ross et al. 2005), after PAR boundary shifts earlier in Eutherian mammal evolution (Van Laere et al. 2008; Das et al. 2009). Several recent shifts have been discovered within the genus Mus (reviewed in White et al. 2012). The evidence that multiple events causing loss of recombination have occurred in S. latifolia within a short evolutionary time strongly suggests that selection for recombination suppression has acted, although it is not yet possible to definitively exclude some nonselective process. Interestingly, a closely related species, S. diclinis, has also undergone a rearrangement involving the sex chromosomes. A reciprocal Y-autosome translocation has added material to the Y, but not to the X, chromosome; in male meiosis, the S. diclinis X therefore pairs with the ancestral Y region (the majority of the translocated Y1), while the rest of Y1 pairs with the unrearranged autosome (which would segregate as a neo-X), which, in turn, pairs with the Y2 (Howell et al. 2009). This rearrangement must differ from the ones that we infer here because the S. diclinis X is not rearranged, and therefore it would not explain our observation that previously autosomal genes have become linked to X-linked genes in female meiosis.
Rearrangements also appear to have occurred involving the added regions, possibly contributing to recombination suppression. Given the small number of recombinants among the genes clustered in this region of the X genetic map in our mapping family, double crossover events are rare, so we cannot reliably order the genes. However, the finding of full sex linkage for some, but not all, of the genes that were added to the X in each of the two separate events implies that at least one rearrangement must have occurred (see Figure 4 and above).
Because S. latifolia is closely related to other dioecious species, it will be interesting in the future to study orthologues of S. latifolia PAR genes in those species, to test whether the additional events pre- or post-date their recent split, and, if the former, whether the PAR boundary has shifted since these species split. As discussed above, the cluster of genes closely linked to the S. latifolia PAR boundary may recombine rarely simply because this is a pericentromeric region. It will therefore be interesting to map more genes in the two S. vulgaris linkage groups that appear to have contributed to recent PAR expansion in S. latifolia.
Supplementary Material
Acknowledgments
We thank the Natural Environment Research Council (NERC) Biomolecular Analysis Facility (Liverpool, United Kingdom) for 454 sequencing; GenePool for the Sanger sequencing and capillary electrophoreses; and Yusuke Kazama (RIKEN Nishina Center, Wako, Saitama, Japan) for information about the location of the SlWUS1 gene on the S. latifolia X chromosome map.
Note added in proof: See Qui et al. 2013 (pp. 663–672) in this issue, for a related work.
Footnotes
Communicating editor: S. I. Wright
Literature Cited
- Atanassov I., Delichère C., Filatov D. A., Charlesworth D., Negrutiu I., et al. , 2001. A putative monofunctional fructose-2,6-bisphosphatase gene has functional copies located on the X and Y sex chromosomes in white campion (Silene latifolia). Mol. Biol. Evol. 18: 2162–2168 [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2006. A dynamic view of sex chromosome evolution. Curr. Opin. Genet. Dev. 16: 578–585 [DOI] [PubMed] [Google Scholar]
- Bailey N., 1949. The estimation of linkage with differential viability II. The use of the product formula for the estimation of linkage in intercrosses when differential viability is present. Heredity 3: 215–225 [DOI] [PubMed] [Google Scholar]
- Baker H. G., 1966. The evolution of floral heteromorphism and gynodioecism in Silene maritima. Heredity 21: 689–692 [Google Scholar]
- Bergero R., Charlesworth D., 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24: 94–102 [DOI] [PubMed] [Google Scholar]
- Bergero R., Charlesworth D., 2011. Preservation of the Y transcriptome in a 10MY old plant sex chromosome system. Curr. Biol. 21: 1470–1474 [DOI] [PubMed] [Google Scholar]
- Bergero R., Forrest A., Kamau E., Charlesworth D., 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Forrest A., Charlesworth D., 2008. Active miniature transposons from a plant genome and its nonrecombining Y chromosome. Genetics 178: 1085–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi G., Antonovics J., Biere A., Charlesworth D., Delph L., et al. , 2009. Silene as a model system in ecology and evolution. Heredity 103: 5–14 [DOI] [PubMed] [Google Scholar]
- Blavet N., Blavet H., Cegan R., Zemp N., Zdanska J., et al. , 2012. Comparative analysis of a plant pseudoautosomal region (PAR) in Silene latifolia with the corresponding S. vulgaris autosome. BMC Genomics 13: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne P. S., 1982. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 61: 85–90 [DOI] [PubMed] [Google Scholar]
- Burgoyne P. S., Mahadevaiah S., Sutcliffe M., Palmer S., 1992. Fertility in mice requires X-Y pairing and a Y-chromosomal spermiogenesis gene-mapping to the long arm. Cell 71: 391–398 [DOI] [PubMed] [Google Scholar]
- Cermak T., Kubat Z., Hobza R., Koblizkova A., Widmer A., et al. , 2008. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Res. 16: 961–976 [DOI] [PubMed] [Google Scholar]
- Chakravarti A., Lasher L. K., Reefer J. E., 1991. A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charchar F. J., Svartman M., El-Mogharbel N., Ventura M., Kirby P., et al. , 2003. Complex events in the evolution of the human pseudoautosomal region 2 (PAR2). Genome Res. 13: 281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997 [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Phil. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 35: 205–214 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Laporte V., 1998. The male-sterility polymorphism of Silene vulgaris. Analysis of genetic data from two populations, and comparison with Thymus vulgaris. Genetics 150: 1267–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128 [DOI] [PubMed] [Google Scholar]
- Clopper C., Pearson E., 1934. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26: 404–413 [Google Scholar]
- Cloutier S., Cappadocia M., Landry B. S., 1995. Analysis of RFLP mapping inaccuracy in Brassica napus L. Theor. Appl. Genet. 96: 83–91 [DOI] [PubMed] [Google Scholar]
- Cooke H., Brown W., Rappold G., 1985. Hypervariable telomeric sequences from the human sex chromosomes are pseudoautosomal. Nature 317: 687–692 [DOI] [PubMed] [Google Scholar]
- Costich D. E., Meagher T. R., Yurkow E. J., 1991. A rapid means of sex identification in Silene latifolia by use of flow cytometry. Plant Mol. Biol. Rep. 9: 359–370 [Google Scholar]
- Das P., Chowdhary B., Raudsepp T., 2009. Characterization of the bovine pseudoautosomal region and comparison with sheep, goat, and other mammalian pseudoautosomal regions. Cytogenet. Genome Res. 126: 139–147 [DOI] [PubMed] [Google Scholar]
- Delph L., Arntz A., Scotti-Saintagne C., Scotti I., 2010. The genomic architecture of sexual dimorphism in the dioecious plant Silene latifolia. Evolution 64: 2873–2886 [DOI] [PubMed] [Google Scholar]
- DiStilio V. S., Kesseli R., Mulcahy D. L., 1998. A pseudoautosomal random amplified polymorphic DNA marker for the sex chromosomes of Silene dioica. Genetics 149: 2057–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov D. A., 2005. Evolutionary history of Silene latifolia sex chromosomes revealed by genetic mapping of four genes. Genetics 170: 975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov D., Howell E. C., Groutides C., Armstrong S., 2008. Recent spread of a retrotransposon in the Silene latifolia genome, apart from the Y chromosome. Genetics 181: 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Morgan E., Willis J. H., 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Torii C., Ishii K., Aonuma W., Shimizu Y., et al. , 2011. Narrowing down the mapping of plant sex-determination regions using new Y chromosome-specific markers and heavy-ion beam irradiation-induced Y deletion mutants in Silene latifolia G3 (Bethesda) 2: 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter L. E., Roberts G. T., Lee K., Larimer F. W., Tuskan G. A., 2003. The development of two flanking SCAR markers linked to a sex determination locus in Salix viminalis L. J. Hered. 94: 185–189 [DOI] [PubMed] [Google Scholar]
- Hobza R., Kejnovsky E., Vyskot B., Widmer A., 2007. The role of chromosomal rearrangements in the evolution of Silene latifolia sex chromosomes. Mol. Genet. Genomics 278: 633–638 [DOI] [PubMed] [Google Scholar]
- Howell E. C., Armstrong S., Filatov D., 2009. Evolution of neo-sex chromosomes in Silene diclinis. Genetics 182: 1109–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell E. C., Armstrong S., Filatov D., 2011. Dynamic gene order on the Silene latifolia Y chromosome. Chromosoma 120: 287–296 [DOI] [PubMed] [Google Scholar]
- Hughes J. F., Skaletsky H., Brown L., Pyntikova T., Graves T., et al. , 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Jordan C., Charlesworth D., 2012. The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66: 505–516 [DOI] [PubMed] [Google Scholar]
- Kaiser V. B., Bergero R., Charlesworth D., 2009. SlCyt, a newly identified sex-linked gene, has recently moved onto the X chromosome in Silene latifolia (Caryophyllaceae). Mol. Biol. Evol. 26: 2343–2351 [DOI] [PubMed] [Google Scholar]
- Kaiser V. B., Bergero R., Charlesworth D., 2011. A new plant sex-linked gene with high sequence diversity and possible introgression of the X copy. Heredity 106: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Armstead I. P., Donnison I. S., Thomas H. M., Jones R. N., et al. , 2002. Physical and genetic mapping in the grasses Lolium perenne and Festuca pratensis. Genetics 161: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Armstead I. P., Donnison S. I., Roberts L. A., Harper J. A., et al. , 2007. Comparative analyses between Lolium/Festuca introgression lines and rice reveal the major fraction of functionally annotated gene models is located in recombination-poor/very recombination-poor regions of the genome. Genetics 177: 597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn B. T., Page D. C., 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967 [DOI] [PubMed] [Google Scholar]
- Laurie D. A., Pratchett N., Devos K. M., Leitch I. J., Gale M. D., 1993. The distribution of RFLP markers on chromosome 2(2H) of barley in relation to the physical and genetic location of 5S rDNA. Theor. Appl. Genet. 87: 177–183 [DOI] [PubMed] [Google Scholar]
- Lawson-Handley L. J., Ceplitis H., Ellegren H., 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerova M., Moore R. C., Grant S. R., Vyskot B., 2003. The sex chromosomes of Silene latifolia revisited and revised. Genetics 165: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Szyda J., Schechinger B., Rappold G., Arnheim N., 2000. Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am. J. Hum. Genet. 66: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Moore P. H., Ma H., Ackerman C. M., Ragiba M., et al. , 2004. A primitive Y chromosome in Papaya marks the beginning of sex chromosome evolution. Nature 427: 348–352 [DOI] [PubMed] [Google Scholar]
- Lorieux M., Goffinet B., Perrier X., de Léon D. González, Lanaud C., 1995a Maximum-likelihood models for mapping genetic markers showing segregation distortion. 1. Backcross populations. Theor. Appl. Genet. 90: 73–80 [DOI] [PubMed] [Google Scholar]
- Lorieux M., Perrier X., Goffinet B., Lanaud C., Léon D. González de, 1995b Maximum-likelihood models for mapping genetic markers showing segregation distortion. 2. F2 populations. Theor. Appl. Genet. 90: 81–89 [DOI] [PubMed] [Google Scholar]
- Macas J., Kejnovsky E., Neumann P., Nova P., Koblizkova A., et al. , 2008. Next generation sequencing-based analysis of repetitive DNA in the model dioecious plant Silene latifolia. PLoS ONE 6: e27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J. E., Ellegren H., 2009. Sex-linkage of sexually antagonistic genes is predicted by female, but not male, effects in birds. Evolution 63: 1464–1472 [DOI] [PubMed] [Google Scholar]
- Marais G., Galtier N., 2003. Sex chromosomes: how X-Y recombination stops. Curr. Biol. 13: R641–R643 [DOI] [PubMed] [Google Scholar]
- Marais G., Nicolas M., Bergero R., Chambrier P., Kejnovsky E., et al. , 2008. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 18: 545–549 [DOI] [PubMed] [Google Scholar]
- Marais G. A. B., Forrest A., Kamau E., Käfer J., Daubin V., et al. , 2011. Multiple nuclear gene phylogenetic analysis of the evolution of dioecy and sex chromosomes in the genus Silene. PLoS ONE 6: e21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden-Jones E. M., Turrill W. B., 1957. The Bladder Campions. Ray Society, London [Google Scholar]
- Matsuda Y., Moens P., Chapman V., 1992. Deficiency of X-chromosomal and Y-chromosomal pairing at meiotic prophase in spermatocytes of sterile interspecific hybrids between laboratory mice (Mus domesticus) and Mus spretus. Chromosoma 101: 483–492 [DOI] [PubMed] [Google Scholar]
- Matsunaga S., Hizume M., Kawano S., Kuroiwa T., 1994. Cytological analyses in Melandrium album: genome size, chromosome size and fluorescence in situ hybridization. Cytologia (Tokyo) 59: 135–141 [Google Scholar]
- McDaniel S. F., Willis J. H., Shaw A. J., 2007. A linkage map reveals a complex basis for segregation distortion in an interpopulation cross in the moss Ceratodon purpureus. Genetics 176: 2489–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher T. R., Gillies A. C., Costich D. E., 2005. Genome size, quantitative genetics and the genomic basis for flower size evolution in Silene latifolia. Ann. Bot. (Lond.) 95: 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam H. K., Pointer M. A., Wright A. E., Berlin S., Mank J. E., 2012. W chromosome expression responds to female-specific selection. Proc. Natl. Acad. Sci. USA 109: 8207–8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas T., Speed R., Passage M., Yen P., Chandley A., et al. , 1992. Role of the pseudoautosomal region in sex-chromosome pairing during male meiosis: meiotic studies in a man with a deletion of distal Xp. Am. J. Hum. Genet. 51: 526–533 [PMC free article] [PubMed] [Google Scholar]
- Nakayama H., Ito T., Hayashi Y., Sonoda T., Fukuda T., et al. , 2006. Development of sex-linked primers in garden asparagus (Asparagus officinalis L.). Breed. Sci. 56: 327–330 [Google Scholar]
- Nam K., Ellegren H., 2008. Scrambled eggs: the chicken (Gallus gallus) Z chromsome contains at least three non-linear evolutionary strata. Genetics 180: 1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Marais G., Hykelova V., Janousek B., Laporte V., et al. , 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. A., 1990. Rapid and sensitive sequence comparison with FASTP and FASTA, pp. 63–98 in Methods in Enzymology, edited by Doolittle R., Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Qiu S., Bergero R., Forrest A., Kaiser V. B., Charlesworth D., 2010. Nucleotide diversity in Silene latifolia autosomal and sex-linked genes. Proc. Biol. Sci. 277: 3283–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S., Bergero R., Charlesworth D., 2013. Testing for the footprint of sexually antagonistic polymorphisms in the pseudo-autosomal region of a plant sex chromosome pair. Genetics 194: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees H., Durrant A., 1986. Recombination and genome size. Theor. Appl. Genet. 73: 72–76 [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution 41: 911–914 [DOI] [PubMed] [Google Scholar]
- Ross M. T., D. V. Grafham, A. J. Coffey, S. Scherer, K. McLay et al 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt K., Lazzeroni L., Foote S., Vollrath D., Fisher E., et al. , 1994. Multipoint linkage map of the human pseudoautosomal region based on single-sperm typing: Do double crossovers occur during male meiosis? Am. J. Hum. Genet. 55: 423–443 [PMC free article] [PubMed] [Google Scholar]
- Scotti I., Delph L. F., 2006. Selective trade-offs and sex-chromosome evolution in Silene latifolia. Evolution 60: 1793–1800 [PubMed] [Google Scholar]
- Siroky J., Lysak M. A., Dolezel J., Kejnovsky E., Vyskot B., 2001. Heterogeneity of rDNA distribution and genome size in Silene spp. Chromosome Res. 9: 387–393 [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P. J., Cordum H. S., Hillier L., et al. , 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837 [DOI] [PubMed] [Google Scholar]
- Spigler R., Lewers K., Main D., Ashman T.-L., 2008. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity 101: 507–517 [DOI] [PubMed] [Google Scholar]
- Swanson C. P., 1957. Cytology and Cytogenetics. Prentice Hall, New York [Google Scholar]
- Swarbreck D., Wilks C., Lamesch P., Berardini T., Garcia-Hernandez M., et al. , 2008. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acid Res. 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgmann-Rauber A., Jamsari A., Kinney M. S., Pires J. C., Jung C., 2007. Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol. Genet. Genomics 278: 221–234 [DOI] [PubMed] [Google Scholar]
- Vagera J., Paulikova D., Dolezel J., 1994. The development of male and female regenerants by in-vitro androgenesis in dioecious plant Melandrium album. Ann. Bot. (Lond.) 73: 455–459 [Google Scholar]
- Van Laere A.-S., Coppieters W., Georges M., 2008. Characterization of the bovine pseudoautosomal boundary: documenting the evolutionary history of mammalian sex chromosomes. Genome Res. 18: 1884–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen J. W., 2006. JoinMap 4.0 Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Plant Research International, Wageningen, The Netherlands [Google Scholar]
- Vekemans X., Beauwens T., Lemaire M., Roldán-Ruiz I., 2002. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 11: 139–151 [DOI] [PubMed] [Google Scholar]
- Waters P. D., Duffy B., Frost C. J., Delbridge M. L., Graves J. A. M., 2001. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80-130 million years ago. Cytogenet. Cell Genet. 92: 74–79 [DOI] [PubMed] [Google Scholar]
- Westergaard M., 1946. Structural changes of the Y-chromosomes in the offspring of polyploid Melandrium. Hereditas 32: 60–64 [DOI] [PubMed] [Google Scholar]
- Westergaard M., 1948a Aberrant Y-chromosomes and sex expression in Melandrium album. Hereditas 32: 419–443 [DOI] [PubMed] [Google Scholar]
- Westergaard M., 1948b The relation between chromosome constitution and sex in the offspring of triploid Melandrium. Hereditas 34: 257–279 [Google Scholar]
- White M. A., Ikeda A., Payseur B. A., 2012. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm. Genome 23: 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., DiFazio S., Gunter L., Zhang X., Sewell M., et al. , 2008. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 18: 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J., Georgiev S., Janousek B., Charlesworth D., Vyskot B., et al. , 2007. Early events in the evolution of the Silene latifolia Y chromosome: male specialization and recombination arrest. Genetics 177: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.