Abstract
Deep sequencing technologies enable the study of the effects of rare variants in disease risk. While methods have been developed to increase statistical power for detection of such effects, detecting subtle associations requires studies with hundreds or thousands of individuals, which is prohibitively costly. Recently, low-coverage sequencing has been shown to effectively reduce the cost of genome-wide association studies, using current sequencing technologies. However, current methods for disease association testing on rare variants cannot be applied directly to low-coverage sequencing data, as they require individual genotype data, which may not be called correctly due to low-coverage and inherent sequencing errors. In this article, we propose two novel methods for detecting association of rare variants with disease risk, using low coverage, error-prone sequencing. We show by simulation that our methods outperform previous methods under both low- and high-coverage sequencing and under different disease architectures. We use real data and simulation studies to demonstrate that to maximize the power to detect associations for a fixed budget, it is desirable to include more samples while lowering coverage and to perform an analysis using our suggested methods.
Keywords: rare variants, low-coverage sequencing, association test, sequencing, pooling
OVER the last decade, many genome-wide association studies (GWAS) have been conducted for a wide range of diseases and phenotypes (Easton et al. 2007; Wellcome Trust Case Control Consortium 2007; Schunkert et al. 2011) that have successfully identified associations with hundreds of single-nucleotide variants (SNVs). However, for many conditions, only a small fraction of the heritability is currently explained by these SNVs (Manolio et al. 2009).
There are several possible explanations for this missing heritability, including undiscovered gene–gene and gene–environment interactions, inaccurate phenotyping, and disease heterogeneity. One of the most appealing hypotheses is that a large portion of the missing heritability may be explained by rare SNVs, which have not been explored by GWAS due to technological limitations. Most GWAS have been performed on a set of a few hundred thousand common SNVs with a minor allele frequency (MAF) of at least 1% in populations of European ancestry, and often low-frequency SNVs were simply discarded from the analysis due to power considerations. However, if rare variants do in fact contribute to disease status, it is likely that each individual will carry different rare variants with such effects (Cohen et al. 2004; Kryukov et al. 2007; Gorlov et al. 2008).
Recent advances in sequencing technologies allow us to explore the entire genome for several thousand dollars, and thus whole-genome, sequence-based association studies are becoming feasible. Using these technologies, we can perform association studies on all SNVs in the genome, including rare SNVs. However, the analysis of such new studies is complicated by the fact that the power to detect association with a single SNV depends on its minor allele frequency—the higher the frequency is, the higher the power. SNVs with very low MAF require the sequencing of tens of thousands of individuals to achieve reasonable power in association studies.
To circumvent this problem, statistical tests have been suggested that aggregate the rare SNV information across a genomic region (Li and Leal 2008). The general principle behind all of these methods is that in a gene or region of interest that is associated with the disease, we expect to observe substantially more (or less) rare alleles across the region in cases compared with controls, particularly with rare SNVs. For instance, in the analysis of Ahituv et al. (2007), the gene SIM1 had six rare mutations in obese individuals and no rare mutations in lean individuals. This approach has two advantages: first, it reduces the burden of multiple hypotheses, as the number of regions is smaller than the number of SNVs; second, the aggregated frequencies of all SNVs in each region are much higher than the frequency of individual rare SNVs. Both of these advantages increase statistical power.
The cost of sequencing technologies, although considerably cheaper than a decade ago, still prohibits GWAS on tens of thousands of samples necessary for the discovery of subtle associations. To perform studies with large numbers of samples, researchers may compromise on the sequencing accuracy to reduce costs. One strategy is the use of low-coverage sequencing, where the amount of sequencing per sample is reduced. Particularly, this strategy has been adapted by the 1000 Genomes Project Consortium (2010), where the majority of individuals were sequenced at 5× coverage. This approach obviously reduces cost considerably. However, it increases the complexity of the downstream analysis due to missing and erroneous variant calls. Unfortunately, existing methods for aggregate rare SNV statistics assume that genotype calls contain no errors. Thus, these methods are not designed to work well with sequencing data with low coverage or sequencing errors, and they cannot be applied directly to most data collected as of today.
In this article, we propose a strategy for implicating rare variants in disease, utilizing low-coverage sequencing data. Our approach leverages on two novel methods for the analysis of rare SNVs in the context of low-coverage sequencing and sequencing errors. The first method we present is based on a likelihood-ratio test (LRT) in which the alternative hypothesis assumes that there exists a set of specified causal SNVs, together with their effect sizes. This approach extends the method by Sul et al. (2011a) to address low-coverage sequencing, and it explicitly models sequencing errors. The second method we present is based on an aggregate weighted sum of variance-stabilizing transformations (VSTs) of the difference of the allele frequencies between cases and controls. Several previous methods have suggested (Madsen and Browning 2009; Sul et al. 2011b) calculating statistics based on the weighted sum of the allele counts in the cases vs. the controls. Implicitly, many such methods assume that allele frequency counts are normally distributed. However, even though this assumption holds in the limit, it is a well-known fact in probability theory that a binomial distribution X ∼ B(n, p) with a small constant np is not well approximated by a normal distribution, but much better approximated by a Poisson distribution with parameter λ = np. Thus, the normality approximation is too crude for rare SNVs, where the minor allele counts more closely follow a Poisson distribution. Additionally, when modeling the distribution of minor allele counts in the population, the variance of the distribution often depends on the minor allele frequency, whose misestimation may impair the accuracy of modeling and subsequent analysis. Therefore, the proposed VST method considers a weighted sum of appropriate transformations of the counts, which have a variance-stabilizing effect, instead of a weighted sum of the counts themselves. We show that both approaches provide higher power than previous methods for both low and high sequencing coverage.
We simulate rare variant disease association studies under a variety of disease models under both high and low coverage. From these simulations, we demonstrate that VST and LRT methods are either superior to other methods or comparable under a variety of disease models. We also show that moving from high coverage to low coverage only moderately reduces the power of a study. Thus, for the same budget in terms of sequencing cost, low-coverage sequencing of a larger number of individuals has higher power than high-coverage sequencing of fewer individuals.
While low-coverage sequencing reduces the cost of sequencing considerably, barcoding of the samples is still required for each sample, which can be a costly procedure. We can eliminate the cost of barcoding through the use of DNA pools, where DNA from several individuals is mixed and sequenced together in each sequencing run without barcoding. Our methods above are also applicable to the scenario in which the study population is sequenced by partitioning to small groups of 5–10 individuals each, and the samples from each group are pooled and sequenced in a single run. Clearly, this results in lower-coverage sequencing, with the added complexity introduced by the loss of individual information. DNA pooling has been successfully applied to GWAS data that reduce costs by one or two orders of magnitude (Hanson et al. 2007; Brown et al. 2008; Skibola et al. 2009). However, pooling DNA from a large number of individuals can introduce a great deal of background noise in the data that may reduce the reliability of and increase the difficulty in the downstream analysis.
In contrast to pooling strategies in GWAS data where a small number of pools are genotyped, each consisting of a large number of samples, here we consider a strategy in which a small number of individuals are sequenced in each pool, making the noise amenable to explicit modeling. Moreover, DNA pooling has been successfully applied to next generation sequencing (Erlich et al. 2009) and we therefore argue that the proposed application is feasible. To assess the feasibility of applying the proposed methods in the context of DNA pools, we first measured the coverage, the sequencing error rate, and pooling accuracy in data from a pooled sequencing study of non-Hodgkin lymphoma and then used the measured parameters to simulate a study in which the budget allows for sequencing of 80 pools, where we vary the number of individuals per pool. We observed that the power of the proposed methods increases considerably when the number of individuals per pool increases. The immediate conclusion from this simulated study is that in a given study, it is generally preferable to perform the sequencing on the DNA of all available individuals, even if this requires samples to be sequenced with low coverage or pooled in small groups due to budget constraints.
Materials and Methods
Rare variants disease model
The methods described below are optimized for the disease model proposed by Madsen and Browning (2009). In this model, rarer causal variants have larger effect sizes than common ones. We use and to denote true MAF in cases and controls, respectively, and and are
| (1) |
| (2) |
where γi is the relative risk of variant i, and pi is the MAF in the population, which can be estimated by the frequency in the controls. The methods used to estimate p are described in the section Estimating allele frequencies in the LRT framework and in supporting information, File S1.
In the disease risk model, each group of variants has group population attributable risk (PAR), and each variant has marginal PAR denoted as ω, which is the group PAR divided by the number of causal variants in the group. Then, the relative risk of variant i, γi, is defined as
| . | (3) |
We compute using Equations 1 and 3.
We note that our methods can still be applied if this model does not reflect the reality, and although the power is reduced in this case, the power remains greater than for previous methods under some of the conditions that we tested in the Results section, e.g., in the case where some of the variants are protective.
LRT statistic
Sul et al. proposed a likelihood-ratio test to detect an association of a group of rare variants (Sul et al. 2011a). The method assumes that the true genotype of each individual is known; in other words, it requires high coverage for each individual. We propose a new likelihood-ratio test that can be applied with low-coverage sequencing and where the sequencing errors are modeled explicitly, and thus we account for uncertainty in sequencing reads.
Assume that for every individual k, we are given observations of the major and minor alleles at each of the M variant positions. Each is of the form , where and are the numbers of observed minor and major alleles of variant i in individual k. We denote . Let D+ and D− be the sets of observations in the cases and controls, respectively. In the likelihood-ratio test, we calculate L1/L0, the likelihood ratio of the alternative vs. null models, where
| (4) |
| (5) |
and scenario vj is a binary vector indicating which variants are causal among M variants: . v0 is the scenario in which all variants are noncausal. The priors P(vj) are given by
| (6) |
where ci is the probability that variant i is causal. Additionally, the probability P(D+; D−|vj) is given by
where r is the (hidden, unobserved) minor allele count of variant i in chromosomes in individual k (and so r ∈ {0, 1, 2}, where a heterozygous SNP is modeled by having r = 1).
Given the true value of r, , and the error rate, e, we have that follows a binomial distribution with trials and probability of success fk(r) as defined in File S1, Equation 3. Then, the probability of observing given the true r minor alleles is
| . |
Next, we show how to efficiently compute P(r|+; vj) and P(r|−; vj).
Decomposition of likelihood function and efficient permutation test
To avoid iterating over all 2M possible vj’s in Equation 5, we assume that there is no linkage disequilibrium between rare variants, which is reasonably justified in the case of rare variants (Pritchard 2001; Pritchard and Cox 2002) and has been generally assumed by previous aggregation methods. It is easy to see that P(r|−; vj) follows a binomial distribution with two trials and probability if vi = 1 or pi if vi = 0. Similarly, P(r|+; vj) follows a binomial distribution with two trials and either probability if vi = 1 or pi if vi = 0. Let B(k; n, p) be the probability mass function of the binomial distribution, . Then, L0 in Equation 4 can be expanded as
| (7) |
To compute L1, we instead compute L0 + L1 to simplify our formula. We first denote ζk as the case–control status of the kth individual. If ζk = 1, the individual is a case, and otherwise it is a control. We can randomly permute case–control status and set values of ζk in the permutation test. Let P be the total number of individuals. Then, L0 + L1 can be computed as
where
and
| (8) |
(see the Appendix of Sul et al. 2011a for the derivation). For cases we compute , and for controls we compute ; moreover, in Equations 7 and 8 do not change in the permutation test. Hence, we can precompute values of all these variables. The number of Ai values is M, and the number of and values is P*M, which is the number of individuals times the number of variants and should not be too large to store in memory.
Estimating allele frequencies in the LRT framework
We use the following approach to estimate allele frequencies used in the LRT method. First, we use the maximum-likelihood approach discussed in File S1 to detect SNVs whose minor allele frequency is 0. These are SNVs whose minor alleles are all errors, and hence they are nonpolymorphic sites. We remove these SNVs from subsequent analysis. We then use the LRT statistic itself to estimate the allele frequencies. To estimate pi in the Ai term in Equation 7, we perform a grid search on pi to find the pi value that maximizes the Ai term for each SNV. To estimate and , we use the following approach. First we note that we can calculate from and the PAR, using Equation 1. Next, we note that if the PAR value is fixed, we can perform an independent grid search for each SNV, so that we find the value of (and therefore also ) that maximizes the expression of Equation 8. Thus, we perform a double-grid search; we search over the space of PAR values, and for each PAR we compute the LRT statistic by searching over the space of for each SNV.
We note that the LRT algorithm can be easily extended to deal with pools by replacing each individual in the above description by a pool, and thus r ∈ {0, 1, 2, … , hk}, where hk is the number of haplotypes in the kth pool. The assumption is that the genotype of a pool is chosen from a binomial distribution B(hk, p+) for a case pool or from B(hk, p−) for a control pool.
VST-based method
Based on the Neyman–Pearson lemma (Neyman and Pearson 1933), the likelihood-ratio test proposed above should be the most powerful possible test when the assumed model accurately represents reality. However, the utility of the LRT method may be impaired because it relies on prior knowledge of the proportion of causal SNVs in the region, which may be difficult to estimate. LRT also requires the grid search to estimate allele frequencies, which may require a search over a large space, resulting in an increased runtime. Moreover, our proposed LRT method differs from previous methods for rare variants such as that of Madsen and Browning (2009), where a weighted sum of minor allele counts is compared between the cases and the controls, and weights are adjusted according to the disease model. We therefore present a simpler method based on VST, with power similar to that of the LRT, but that directly uses the allele counts of the SNVs.
The VST method is based on an aggregate weighted sum of variance-stabilizing transformations of the difference of the allele frequencies between cases and controls. It has been previously suggested (Madsen and Browning 2009; Sul et al. 2011b) to calculate statistics based on the weighted sum of the allele counts in the cases vs. the controls. Implicitly or explicitly, many such methods assume that allele frequency counts are normally distributed. However, this assumption does not hold for rare SNVs, where the counts more closely follow a Poisson distribution. Furthermore, many methods implicitly rely on the accuracy of estimating minor allele frequency and are thus vulnerable to its misestimation. To correct for this, in general terms, the proposed VST method considers a weighted sum of appropriate transformations of the counts instead of a weighted sum of the minor allele frequency counts.
Improved approximation of minor allele count normal distribution
Let X be a random variable corresponding to the number of minor alleles at a particular genomic position in a population of n/2 diploid individuals. X follows a binomial distribution: X ∼ B(n, p), where p is the minor allele frequency. For sufficiently large n and p, the distribution is approximately normal. However, when p is small, this is no longer a good approximation, and an approximation to a Poisson distribution with parameter np is more accurate. Statistics such as Rare variant Weighted Aggregate Statistic (RWAS) (Sul et al. 2011b) that directly compare the counts of the minor alleles in the cases and the controls typically ignore the different distribution of rarer variants (particularly RWAS uses a z-score for each SNV). We correct for this by dividing the variants into approximately normally distributed and approximately Poisson distributed, according to a threshold determined by n and p (we use the threshold suggested by Decker and Fitzgibbon 1991: when n0.31P > 0.47, the normal approximation is used; otherwise the Poisson approximation is used). We denote the two sets of SNVs as rare SNVs and common SNVs. Furthermore, for both types of variants, it is desirable to approximate their minor allele count distributions so that the effect of misestimating minor allele frequency on the accuracy of approximation is minimal. We achieve this using variance-stabilizing transformations, as shown below.
For rare SNVs, we use the f0 transformation of X,
| (9) |
where and . Bar-Lev and Enis (1988) showed that this function is a variance-stabilizing transformation of the Poisson-distributed X to a normal distribution, so that . For common SNVs, we use the f1 transformation:
| . | (10) |
Bromiley and Thacker (2001) showed that f1 is a variance-stabilizing transformation of the binomially distributed X to a normal distribution, so that . We show below how to adapt the standard z-score test statistic to make use of the f0 and f1 approximations for rare and common variants.
Definition of VST statistic
In GWAS, an association statistic of a variant or z-score is computed from an allele frequency difference between case and control individuals to determine whether the variant is associated with a disease (Eskin 2008). Specifically, the z-score for a variant is calculated as
which utilizes the (approximately) normal distribution under the null hypothesis of the difference , scaled so that the variance under both hypotheses is 1 ( and denote the estimated MAFs in cases and controls, respectively, and denotes their average). We apply the same motivation and use f0 and f1 to define a new statistic ρi for each variant I,
| (11) |
where and are the estimated minor allele counts of variant i in cases and controls, respectively, and and are the estimated MAFs of variant i in cases and controls, respectively. The method used to estimate minor allele counts is described in File S1. From the properties of f0 and f1 the ρi-statistic is normally distributed, around a mean of 0 and with variance 1 under the null. Under the alternative, the variance is also 1, and the mean is equal to the value of ρi when and , where p+ and p− are the population MAFs in cases and controls, respectively. For example, this expectation is equal to when both and are small.
We now consider a set of SNVs, s1, … , sM, and compute a weighted sum of ρi across all SNVs. Thus, the VST statistic is defined as
| , | (12) |
where wi is the weight assigned to variant i, as described below. As each ρi is normally distributed with variance 1, then assuming they are independent, ρ is also normally distributed with variance 1 and also has an expectation of 0 under the null hypothesis. We use this property in the selection of weights.
Optimal weights for VST under a disease model
The ρ-statistic can be used without making any further assumptions by setting wi = 1 for every i. However, it is desirable to set the weights according to a disease model (for example, placing more weight on rarer variants or on variants that are more common in cases than in controls). To maximize power, the weights have to be chosen so that they maximize the expectation of the statistic under the alternative; that is, maximize . According to the Cauchy–Schwarz inequality, this is maximized when wi = E[ρi].
Using the disease model described in Materials and Methods, we can use the estimated and to calculate the expected value E[ρi] and set wi accordingly. For rare SNVs (with small ), the optimal weight would be
| , | (13) |
and for common SNVs, the optimal weight would be
| (14) |
Recall that ω is the marginal PAR. Additionally, by assuming that ω is small and using a first-order Taylor series expansion of f0 and f1, we obtain the following weights for rare SNVs,
| (15) |
and for common SNVs,
| (16) |
As the ω-factor is constant in all weights, we remove it and obtain weights independent of the PAR value. We artificially set wi = 0 for variants whose minor allele is not observed. Finally, we use a permutation test to derive a P-value for the VST statistic.
Adjustment for covariates
Since the statistical frameworks of LRT and VST do not directly allow for covariate adjustment, we perform a parametric bootstrap (Davison and Hinkley 1997) discussed in Lin and Tang (2011) to correct for covariates. In this approach, the logistic regression model is fitted to estimate regression coefficients for covariates. Let be coefficients for covariates and Qj and Yj be covariates and disease status (0 for controls and 1 for cases) for the jth individual, respectively. Then, we compute , the probability that the jth individual is a case while taking into account covariates as follows:
| . |
The parametric bootstrap is similar to the permutation test except that an individual becomes a case or a control depending on the probability rather than by a random assignment in the permutation test. We implement this bootstrap method in our software.
Sequencing data from a study of non-Hodgkin lymphoma
We sequenced individuals from a case–control study of non-Hodgkin lymphoma (L. Conde, I. Eskin, F. Hormozdiari, P. M. Bracci, E. Halperin, and C. Skibola, unpublished data). The samples were genotyped in a GWAS in which a total of 312,563 markers were genotyped in 1431 individuals that included 213 cases of follicular lymphoma (FL) and 750 controls after a set of quality-control criteria was applied (see Conde et al. 2010 for details). Among the 213 FL cases for which GWAS data were available, a subset of 5 FL males and 5 FL females, all HIV-negative white non-Hispanics, was selected for pooling in this pilot study.
DNA pool construction for whole-genome sequencing
Genomic DNA for FL cases was extracted from whole blood (DNeasy Blood and Tissue Kit; QIAGEN, Valencia, CA). DNA integrity was checked for absence of fragmentation by gel electrophoresis on 2% agarose (MetaPhor; BioWhitaker, Rockland, ME) in TAE buffer, stained with ethidium bromide. Concentration was measured by PicoGreen (Quant-iT PicoGreen dsDNA reagent; Invitrogen, Carlsbad, CA) in triplicate and adjusted to 60 ng/μl in TE. To ascertain purity, a UV absorption spectrum was obtained using an ND-1000 NanoDrop spectrophotometer; all samples had 260/280 ratios between 1.84 and 1.92. To construct each pool, equal amounts of DNA (1320 ng) were combined from five individuals in a total volume of 110 μl.
Sequencing and primary analysis
Sequencing was outsourced to Illumina FastTrack Services (San Diego). gDNA samples were used to generate short-insert (target 300 bp) paired-end libraries and a HiSeq2000 instrument was used to generate paired 100-base reads according to the manufacturer’s instructions. The software ELAND was used for sequence alignment, and the coverage was 44 per base.
Results
VST and LRT outperform existing methods
We compared the power of the proposed VST and LRT methods with that of existing methods to detect a disease-associated set of SNVs via simulation of low- and high-coverage sequencing-based disease association studies, with and without considering sequencing error. To evaluate the power of the methods, we generate for each combination of parameters 1000 data panels fitting the alternative hypothesis, according to the disease model discussed in Materials and Methods. In this disease model, rarer variants have higher effect sizes. Each panel simulates sequencing data from N = 1000 cases and 1000 controls (unless otherwise specified), with expected coverage g, given as a parameter. To clarify, g equals the expected number of times each variant position is read during sequencing. For example, if g = 10, then each of a diploid individual’s two alleles at a particular genomic position will be read 10/2 = 5 times on average. The generated simulated data consist of a pair of integers for each individual and each SNV, indicating how many times we observe the major and minor alleles of that SNV in that individual. For each individual, we first generate haplotypes and then generate observations for each position on those haplotypes (i.e., how many times we see the major and minor alleles of each SNV). First, haplotypes are generated as in Madsen and Browning (2009). There are 100 SNVs; MAFs are sampled from Wright’s formula with the same parameters; each SNV has a 0.1 chance of being causal; i.e., ci = 0.1; and PAR = 0.02. Then, the number of observations for each position is sampled from a binomial distribution with and g ⋅ 100,000 trials, since each individual has two haplotypes. We add errors by randomly changing the observation at each read with probability e (where e differs across experiments).
The power of existing methods was assessed using PLINK/SEQ software (v0.07, http://atgu.mgh.harvard.edu/plinkseq/). While the proposed VST and LRT methods take allele counts directly from the read data as input, other methods require genotype data for each individual. Hence, we used a simple maximum-likelihood approach to determine the genotype of a variant from the allele counts and provided the most likely genotype call as an input for the other methods. To ensure that the estimation of genotypes did not adversely affect the performance of competing methods, a comparison was also performed given the true genotypes and produced similar power to that obtained with 20× coverage, for all methods. Tested methods from the PLINK/SEQ package included the C-alpha test (Neale et al. 2011), a frequency-weighted test similar to the Madsen–Browning test, and the variable threshold (VT) test of Price et al. (2010). In addition, we compared our methods with RWAS (Sul et al. 2011b), which is based on a weighted sum of Z-scores, and with the previous LRT method (Sul et al. 2011a), denoted as “LRT_G” in the tables, which requires genotypes of individuals. Table 1 summarizes the power of each method and shows that VST and LRT have greater sensitivity than existing methods both with and without sequencing error and with different budgets, particularly in scenarios with low coverage and a realistic rate of sequencing error (1%). Furthermore, LRT outperforms VST in all scenarios. However, we note that the difference between the two is negligible compared to the improvement over previous methods. This makes the VST method particularly appealing due to its simplicity and yet high power.
Table 1. Power of different methods, on 1000 data sets and a region with 100 rare SNVs.
| Error rate | Coverage per person | C-alpha | Freq-weight | VT | RWAS | LRT_G | VST | LRT |
|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 0.062 | 0.659 | 0.872 | 0.668 | 0.817 | 0.973 | 0.975 |
| 10 | 0.062 | 0.713 | 0.903 | 0.688 | 0.854 | 0.98 | 0.99 | |
| 20 | 0.067 | 0.722 | 0.903 | 0.692 | 0.858 | 0.987 | 0.988 | |
| 0.01 | 4 | 0.051 | 0.174 | 0.208 | 0.269 | 0.286 | 0.634 | 0.745 |
| 10 | 0.066 | 0.479 | 0.674 | 0.610 | 0.71 | 0.965 | 0.972 | |
| 20 | 0.067 | 0.686 | 0.887 | 0.690 | 0.848 | 0.988 | 0.993 |
Two error rates (0% and 1%) were considered with three different coverages (4, 10, and 20). Population attributable risk of the region was 0.02, with ci = 0.1 and a significance threshold of 0.05. Tested methods were C-alpha (Neale et al. 2011), Freq-weight similar to the Madsen–Browning method (Madsen and Browning 2009), variable threshold (VT) (Price et al. 2010), RWAS (Sul et al. 2011b), LRT_G (Sul et al. 2011a), and our proposed methods (VST and LRT).
Low-coverage sequencing for rare variant association incurs only moderate power loss
Interestingly, the results of Table 1 show that the power loss comparing high-coverage (20) sequencing to low-coverage (4) sequencing is moderate for VST and LRT. Thus, we can reduce the cost of the study by a factor of 5, using pooling, with only a minor power reduction. Because of sample preparation costs, an equivalent-cost, low-coverage study can increase the number of individuals sequenced by a factor of <5. Nonetheless, even by increasing the number by a factor of 2 or 3, the power gain of the low-coverage sequencing study is greater than the power loss due to the low coverage.
VST and LRT are robust under different disease models
We evaluated the power of VST, LRT, and existing methods under two additional disease models. In the first disease model, causal variants must have MAF ≤ 1%, and have the same relative risk of 5 regardless of their MAF (ci = 0.2). This framework simulates a disease model where only rare variants are causal with the fixed effect size. The second disease model is similar to the PAR disease model discussed in Materials and Methods (PAR = 2%, ci = 0.1), but it includes additional protective variants. After choosing d deleterious variants, we selected an additional 25% of d variants as protective to produce a ratio of 8:2 deleterious to protective variants. Table 2 shows the power of methods in the first disease model, and the VT method generally has the greatest power in this model. It is because VT assumes there exists an unknown MAF threshold such that variants whose MAF is less than the threshold are more likely to be involved in a disease, which is consistent with the first disease model. Note that the power of LRT is very close to that of VT in this disease model. In the disease model with protective variants, Table 3 shows that LRT is the most powerful method.
Table 2. Power of different methods in a disease model in which only rare variants (MAF ≤ 1%) are causal with relative risk of 5 (ci = 0.2).
| Error rate | Coverage per person | C-alpha | Freq-weight | VT | RWAS | LRT_G | VST | LRT |
|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 0.731 | 0.786 | 0.879 | 0.425 | 0.6 | 0.473 | 0.867 |
| 10 | 0.778 | 0.801 | 0.898 | 0.43 | 0.623 | 0.481 | 0.872 | |
| 20 | 0.782 | 0.806 | 0.896 | 0.433 | 0.631 | 0.489 | 0.866 | |
| 0.01 | 4 | 0.5 | 0.68 | 0.802 | 0.376 | 0.524 | 0.257 | 0.838 |
| 10 | 0.759 | 0.809 | 0.898 | 0.446 | 0.628 | 0.479 | 0.896 | |
| 20 | 0.784 | 0.815 | 0.908 | 0.438 | 0.621 | 0.475 | 0.887 |
Tested methods were C-alpha (Neale et al. 2011), Freq-weight similar to the Madsen–Browning method (Madsen and Browning 2009), variable threshold (VT) (Price et al. 2010), RWAS (Sul et al. 2011b), LRT_G (Sul et al. 2011a), and our proposed methods (VST and LRT).
Table 3. Power of different methods in a disease model in which 20% of causal variants are protective.
| Error rate | Coverage per person | C-alpha | Freq-weight | VT | RWAS | LRT_G | VST | LRT |
|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 0.054 | 0.417 | 0.653 | 0.471 | 0.646 | 0.456 | 0.956 |
| 10 | 0.069 | 0.46 | 0.712 | 0.472 | 0.689 | 0.506 | 0.987 | |
| 20 | 0.07 | 0.473 | 0.713 | 0.478 | 0.699 | 0.515 | 0.99 | |
| 0.01 | 4 | 0.053 | 0.14 | 0.164 | 0.17 | 0.191 | 0.222 | 0.685 |
| 10 | 0.066 | 0.304 | 0.468 | 0.376 | 0.495 | 0.425 | 0.955 | |
| 20 | 0.06 | 0.458 | 0.673 | 0.465 | 0.67 | 0.491 | 0.976 |
PAR was 0.02 with ci = 0.1 Tested methods were C-alpha (Neale et al. 2011), Freq-weight similar to the Madsen–Browning method (Madsen and Browning 2009), variable threshold (VT) (Price et al. 2010), RWAS (Sul et al. 2011b), LRT_G (Sul et al. 2011a), and our proposed methods (VST and LRT).
Applying VST and LRT to pooling of samples
We consider study designs in which a large number of pools are sequenced, where each pool includes the DNA of h/2 individuals and thus h haplotypes. Typically, h is relatively small (e.g., h < 20). This study design avoids the need for barcoding, and thus it further reduces the cost of the study. We compared the sensitivity of the proposed methods in scenarios with and without pooling of individuals’ DNA samples and also with and without considering sequencing error.
We generated simulation data using the scheme described above but in addition, simulated sequencing in pools. We first generated haplotypes as described above, but then pooled case and control individuals separately into pools, with each pool containing the haplotypes of h/2 individuals. Thus, each pool contains h haplotypes. Coverage is spread among these haplotypes. For example, in a pool containing DNA from five diploid individuals (i.e., 10 haplotypes), coverage of g = 50 will yield ∼50/10 = 5 reads of each base on each haplotype. In other words, in these data, the number of observations of a particular base on a particular haplotype is sampled from a binomial distribution with and g ⋅ 100,000 trials.
Table 4 shows the effects of pooling and errors on sensitivity on both methods. Both LRT and VST show high sensitivity, even when pooling and sequencing errors are present. Again, LRT outperformed VST in all tested scenarios, but the difference is not substantial.
Table 4. Power with pooling and errors, on 1000 data sets and a region with 100 rare SNVs.
| Error rate | Pool size | Coverage per person | VST | LRT |
|---|---|---|---|---|
| 0 | 1 | 4 | 0.973 | 0.975 |
| 10 | 0.98 | 0.99 | ||
| 20 | 0.987 | 0.988 | ||
| 5 | 4 | 0.977 | 0.981 | |
| 10 | 0.98 | 0.989 | ||
| 20 | 0.982 | 0.988 | ||
| 0.01 | 1 | 4 | 0.634 | 0.745 |
| 10 | 0.965 | 0.972 | ||
| 20 | 0.988 | 0.993 | ||
| 5 | 4 | 0.376 | 0.557 | |
| 10 | 0.85 | 0.924 | ||
| 20 | 0.981 | 0.987 |
PAR of the region was 0.02, with ci = 0.1 and a significance threshold of 0.05.
To obtain a realistic characterization of the parameters used in our simulations (i.e., error rate and pooling accuracy), and to assess the implications on study design under budget constraints, we evaluated the characteristics of a real data set taken from a case–control study of non-Hodgkin lymphoma (L. Conde, I. Eskin, F. Hormozdiari, P. M. Bracci, E. Halperin, and C. Skibola, unpublished data). We sequenced two pools, each containing a mixture of DNA from fvie case individuals for whom GWAS data were available (see Materials and Methods for details). We then measured the coverage, pooling accuracy and error rate (see File S1). Particularly, we found that the pooling was highly accurate in terms of the number of reads coming from each sample, that the average coverage was 44 per base, and that the sequencing error rate was ∼0.235%. This is consistent with error rate reported in other sequencing studies (Minoche et al. 2011; Hufford et al. 2012). We used these parameters to simulate a full study, as described below.
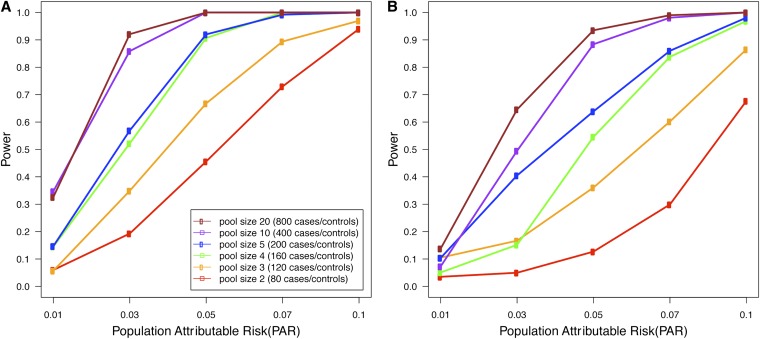
Pooling-based studies offer a trade-off between the introduction of uncertainty due to pooling and increased sample size. It is unclear what would be an optimal study design under a budget constraint. To explore this issue, we simulated a rare variants disease association study with realistic parameters, estimated from the study data. We assumed the budget allowed for 80 runs of the sequencing platform that produces reads at an average coverage of 44 per base per pool, with an error rate of 0.235%. We considered study designs involving an equal number of case and control pools. Figure 1 shows the expected power of LRT with various pool sizes for different values of PAR on two different data sets, where one data set has 20 SNVs in a region with ci = 50% while the other data set has 100 SNVs with ci = 10%. The results show that the power increases dramatically as we increase the pool size from pool size 2 (80 cases and 80 controls) to pool size 20 (800 cases and 800 controls). This suggests that for a given budget, it is generally better to increase the number of individuals per pool as a means of increasing the sample size and that this outweighs the detrimental effects of pooling.
Figure 1.
(A and B) Expected power with 80 sequencing runs as a function of the population attributable risk, for regions with 20 (A) or 100 (B) rare SNVs. The 20-SNVs region has ci = 50% while the 100-SNVs region has ci = 10%. Sequencing error rate is 0.235%, and significance threshold is 0.05.
We note that the power of low-coverage sequencing studies is always at least as high as the power of a pooling study with the same number of samples and the same coverage per sample. The results demonstrated in Figure 1 also suggest that for a given budget, it is generally better to increase the number of individuals in the study and that this outweighs the detrimental effects of low-coverage sequencing.
LRT is robust to misspecified prior information
One of the drawbacks of the LRT statistic is that it uses prior probabilities ci for a variant to be causal. Although one may use bioinformatics tools such as PolyPhen (Adzhubei et al. 2010) or SIFT (Ng and Henikoff 2003) to estimate these priors, prior information may not always be accurate; therefore it is important to assess the sensitivity of LRT to incorrect prior information. To do so, we tested the LRT method with different prior information on the data that was generated using ci = 0.1, as described in Materials and Methods. Specifically, we provided the LRT method with priors ci = 0.02, ci = 0.5, ci = 0.5, and ci sampled from the uniform distribution U(0, 1). We used error rate e = 0.01, pool size of 5, and coverage of 20. The power of LRT with the correct prior information (ci = 0.1) is 0.987 (Table 4), while for the three tested scenarios, the power was 0.965, 0.968, and 0.943, respectively. The results demonstrate that even if prior information is incorrect, LRT still achieves high power, albeit lower than the power achieved by VST (0.981 in this case).
VST and LRT control type I error rates
To measure type I error rates, we generated 10,000 random data sets fitting the null hypothesis with a fixed read error rate of 1% and coverage of 4 per person. We measured the rate of spurious association detection at a confidence threshold of 0.05 with VST and LRT. The detection rate was in the range 0.0480–0.0513 for the two methods, showing that type I error is well controlled. Similar results were observed in the cases where pools of five samples each were analyzed. We also measured the error rates at a more stringent threshold of 0.001, using 100,000 random data sets with an error rate of 1% and coverage of 4. The type I error rates for LRT and VST were 0.00097 and 0.00088, respectively, and this indicates that VST and LRT control the type I error rates correctly at the stringent threshold.
Discussion
We proposed two new methods (VST and LRT) that identify associations of groups of rare variants. We show through simulations that our methods outperform previous methods under different disease models. Importantly, unlike previous methods that allow no errors in sequencing and require high-coverage sequencing of individual samples, our methods can be applied to low-coverage sequencing with errors. We demonstrate through simulations that in the presence of high error rates and low coverage, the power improvement of both LRT and VST over previous methods is substantial. These simulations are based on a disease risk model where rarer variants have higher effect sizes, and we also explored additional disease risk models, often used in previous methods. When only rare variants are causal with the same effect size, the VT method has the greatest power because the assumptions of the VT statistic are consistent with the assumption of the disease model. However, LRT has comparable power to that of VT in this model. When the disease risk model contains protective variants, we show that LRT is the most powerful method. We note that VST can be easily modified to incorporate protective variants by taking the square of its statistic and that the modified VST achieves similar power to LRT when protective variants are present (data not shown).
In addition to the simulated data, we used a real data set from a study of non-Hodgkin lymphoma to obtain parameters related to the sequencing technology and pooling strategy. We estimated coverage and error rates of sequencing, and we verified that pooling can be performed accurately on a small number of samples (five in this case). Our proposed methods can also be applied to the scenario in which a large set of small pools is sequenced, and we used the estimates of the sequencing error rates to simulate the power of a pooled association study, when the number of samples per pool varies (from 1 to 20), but the number of pools is fixed. Based on our experiments, we believe that sequencing-based association studies should include as many individuals as possible by applying low-coverage sequencing and pooling when necessary.
Whereas LRT is generally more powerful than VST in most simulations, VST has several advantages over LRT. Importantly, VST makes fewer assumptions on the model; particularly it does not require prior information on the probability of a variant to be causal, and when incorrect prior information is specified, VST achieves higher power than LRT. VST is also a natural extension of several previous methods such as RWAS (Sul et al. 2011b), VT (Price et al. 2010), and the Madsen–Browning test (Madsen and Browning 2009), which consider a weighted sum of differences in mutation counts; therefore, understanding the relation between VST and LRT provides an insight on the relation between LRT and previous methods. Additionally, VST is a simpler method than LRT, and it can be easily implemented and modified, allowing a more flexible framework.
Our methods directly utilize allele counts from sequencing data to perform rare variants association testing, and an alternative approach is to call genotypes from allele counts and perform the testing. One may use a linkage disequilibrium aware method for genotype calling (Duitama et al. 2011) or methods based on sophisticated models to improve the accuracy of calling. However, this approach has several drawbacks. First, it is considerably computationally intensive because aforementioned genotype calling methods generally require extensive computation. Second, in low-pass sequencing, it is very difficult to call rare variants correctly because of an insufficient number of reads covering rare variants. Third, rare variants are not usually in linkage disequilibrium (LD) with other variants, and hence the LD aware method for genotype calling may not be very accurate for calling rare variants. Hence, power loss may be inevitable if methods attempt to call genotypes from low-pass sequencing and perform rare variants association testing. In addition, we showed in our simulation that even when genotypes are called correctly in the high-coverage scenario, our methods are more powerful than other methods. This means that even if other methods are able to correctly infer genotypes from low-pass sequencing, our methods would be still more powerful.
Supplementary Material
Acknowledgments
E.H. is a faculty fellow of the Edmond J. Safra Bioinformatics Center at Tel-Aviv University. O.N. was supported by the Edmond J. Safra fellowship. E.H. and O.N. were also supported by the Israel Science Foundation, grant 04514831. J.H.S. and E.E. are supported by National Science Foundation (NSF) grants 0513612, 0731455, 0729049, 0916676, and 1065276 and National Institutes of Health (NIH) grants K25-HL080079, U01-DA024417, P01-HL30568, and P01-HL28481. B.H. is supported by NIH–National Institute of Arthritis and Musculoskeletal and Skin Diseases grant 1R01AR062886-01. C.F.S., L.C., and J.R. are supported by NIH grants CA154643 and CA104682. P.B. is supported by NIH grants CA87014, CA45614, and CA89745. This research was also supported in part by the German-Israeli Foundation, grant 109433.2/2010 and in part by NSF grant III-1217615.
Footnotes
Communicating editor: E. Stone
Literature Cited
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahituv N., Kavaslar N., Schackwitz W., Ustaszewska A., Martin J., et al. , 2007. Medical sequencing at the extremes of human body mass. Am. J. Hum. Genet. 80: 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Lev S. K., Enis P., 1988. On the classical choice of variance stabilizing transformations and an application for a Poisson variate. Biometrika 75: 803 [Google Scholar]
- Bromiley, P. A., and N. A. Thacker, 2001 The effects of a square root transform on a Poisson distributed quantity. TINA Memo No. 2001–010. Available at http:www.tina-vision.net/docs/memos.php
- Brown K. M., Macgregor S., Montgomery G. W., Craig D. W., Zhao Z. Z., et al. , 2008. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 40: 838–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., et al. , 2004. Multiple rare alleles contribute to low plasma levels of hdl cholesterol. Science 305: 869–872 [DOI] [PubMed] [Google Scholar]
- Conde L., Halperin E., Akers N. K., Brown K. M., Smedby K. E., et al. , 2010. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat. Genet. 42: 661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison, A. C., and D. V. Hinkley, 1997 Bootstrap Methods and Their Application, Vol. 1. Cambridge University Press, Cambridge/London/New York. [Google Scholar]
- Decker, R., and D. Fitzgibbon, 1991 The normal and Poisson approximations to the binomial: a closer look. Technical Report 82.3. Department of Mathematics, University of Hartford, Hartford, CT.
- Duitama J., Kennedy J., Dinakar S., Hernández Y., Wu Y., et al. , 2011. Linkage disequilibrium based genotype calling from low-coverage shotgun sequencing reads. BMC Bioinformatics 12(Suppl 1): S53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton D. F., Pooley K. A., Dunning A. M., Pharoah P. D. P., Thompson D., et al. , 2007. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich Y., Chang K., Gordon A., Ronen R., Navon O., et al. , 2009. Dna sudoku–harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 19: 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin E., 2008. Increasing power in association studies by using linkage disequilibrium structure and molecular function as prior information. Genome Res. 18: 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov I. P., Gorlova O. Y., Sunyaev S. R., Spitz M. R., Amos C. I., 2008. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J. Hum. Genet. 82: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson R. L., Craig D. W., Millis M. P., Yeatts K. A., Kobes S., et al. , 2007. Identification of pvt1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 56: 975–983 [DOI] [PubMed] [Google Scholar]
- Hufford M. B., Xu X., van Heerwaarden J., Pyhäjärvi T., Chia J.-M. M., et al. , 2012. Comparative population genomics of maize domestication and improvement. Nat. Genet. 44: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov G. V., Pennacchio L. A., Sunyaev S. R., 2007. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 80: 727–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.-Y. Y., Tang Z.-Z. Z., 2011. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 89: 354–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B. E., Browning S. R., 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoche A. E., Dohm J. C., Himmelbauer H., 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina hiseq and genome analyzer systems. Genome Biol. 12: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M., Rivas M. A., Voight B. F., Altshuler D., Devlin B., et al. , 2011. Testing for an unusual distribution of rare variants. PLoS Genet. 7: e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyman J., Pearson E. S., 1933. On the problem of the most efficient tests of statistical hypotheses. Philos. Trans. R. Soc. Lond. A Contain. Pap. Math. Phys. Character 231: 289–337 [Google Scholar]
- Ng P. C., Henikoff S., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Kryukov G. V., de Bakker P. I. W., Purcell S. M., Staples J., et al. , 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., 2001. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69: 124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Cox N. J., 2002. The allelic architecture of human disease genes: common disease-common variant…or not? Hum. Mol. Genet. 11: 2417–2423 [DOI] [PubMed] [Google Scholar]
- Schunkert H., König I. R., Kathiresan S., Reilly M. P., Assimes T. L., et al. , 2011. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola C. F., Bracci P. M., Halperin E., Conde L., Craig D. W., et al. , 2009. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat. Genet. 41: 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul, J. H., B. Han, and E. Eskin, 2011a Increasing power of groupwise association test with likelihood ratio test, pp. 452–467 in Proceedings of the 15th Annual International Conference on Research in Computational Molecular Biology, RECOMB’11 edited by V. Bafna and C. Sahinalp. Springer-Verlag, Berlin/Heidelberg, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul J. H., Han B., He D., Eskin E., 2011b An optimal weighted aggregated association test for identification of rare variants involved in common diseases. Genetics 188: 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium , 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.