Identification of peptides that form complexes with beryllium and class II HLA molecules and are recognized by CD4+ T cells from patients with chronic beryllium disease.
Abstract
Chronic beryllium disease (CBD) is a granulomatous disorder characterized by an influx of beryllium (Be)-specific CD4+ T cells into the lung. The vast majority of these T cells recognize Be in an HLA-DP–restricted manner, and peptide is required for T cell recognition. However, the peptides that stimulate Be-specific T cells are unknown. Using positional scanning libraries and fibroblasts expressing HLA-DP2, the most prevalent HLA-DP molecule linked to disease, we identified mimotopes and endogenous self-peptides that bind to MHCII and Be, forming a complex recognized by pathogenic CD4+ T cells in CBD. These peptides possess aspartic and glutamic acid residues at p4 and p7, respectively, that surround the putative Be-binding site and cooperate with HLA-DP2 in Be coordination. Endogenous plexin A peptides and proteins, which share the core motif and are expressed in lung, also stimulate these TCRs. Be-loaded HLA-DP2–mimotope and HLA-DP2–plexin A4 tetramers detected high frequencies of CD4+ T cells specific for these ligands in all HLA-DP2+ CBD patients tested. Thus, our findings identify the first ligand for a CD4+ T cell involved in metal-induced hypersensitivity and suggest a unique role of these peptides in metal ion coordination and the generation of a common antigen specificity in CBD.
CD4+ T cells play a critical role in the development of chronic beryllium disease (CBD), a fibrotic lung disease characterized by mononuclear cell interstitial infiltrates and granulomatous inflammation (Fontenot and Maier, 2005). Proliferation of blood CD4+ T cells in response to beryllium (Be) salts in vitro defines sensitization (Rossman et al., 1988; Mroz et al., 1991), and progression to CBD is heralded by the accumulation of Be-specific, Th1 cytokine–secreting CD4+ T cells in the lung (Tinkle et al., 1997; Fontenot et al., 2002). These Be-responsive cells are characterized by oligoclonally expanded T cell subsets that share a CDR3 motif among multiple patients with active disease (Fontenot et al., 1999), and the vast majority of these T cells recognize antigen in an HLA-DP–restricted manner (Fontenot et al., 2000; Lombardi et al., 2001). Importantly, genetic susceptibility to CBD is strongly linked to HLA-DP alleles that contain a glutamic acid at the 69th position of the β-chain (βGlu69; Richeldi et al., 1993). Depending on susceptibility and exposure, CBD develops in up to 18% of Be-exposed workers (Kreiss et al., 1993a,b, 1996; Richeldi et al., 1993). Thus, CBD is a classical example of a human disease resulting from gene–environment interactions.
The peptide-binding groove of HLA-DP2, the most prevalent βGlu69-containing HLA-DP molecule, is wider than that of other MHC class II (MHCII) proteins (Dai et al., 2010). The gap between the peptide backbone and the HLA-DP2 β-chain α-helix opens a solvent-exposed acidic pocket composed of three HLA-DP2 β-chain glutamic acid residues (including βGlu69) that could easily accommodate a Be-containing compound. Mutation of βGlu69 or either of the other two glutamic acid residues present in this pocket, βGlu26 and βGlu68, eliminates Be presentation (Dai et al., 2010), suggesting that this acidic gap represents the putative Be-binding site within the footprint of the TCR. However, the role of peptide in coordinating a Be moiety and which peptides are recognized by Be-specific CD4+ T cells remain unknown.
To investigate the spectrum of peptides that permit Be recognition by particular TCRs, we used positional scanning libraries (Pinilla et al., 1992, 1994; Hemmer et al., 1998) to screen hybridomas expressing Be-specific TCRs derived from the lung of an HLA-DP2–expressing CBD patient. We identified multiple mimotopes and endogenous self-peptides, including those derived from plexin A proteins, that are recognized by the T cell hybridomas only in the presence of Be. These peptides share a core motif of acidic amino acids adjacent to the putative Be-binding site in HLA-DP2 and participate in metal ion capture. Be-loaded HLA-DP2–mimotope and HLA-DP2–plexin A4 tetramers detected CD4+ T cells that recognize these complexes in the lungs of many CBD patients, strongly suggesting that these related ligands play a key role in disease. Thus, the current study is the first to identify a complete MHCII–peptide–metal ion complex recognized by pathogenic CD4+ T cells in CBD and provides insight into the role of MHC-bound peptide in metal-induced hypersensitivity.
RESULTS
Specific peptide requirement for T cell recognition of Be
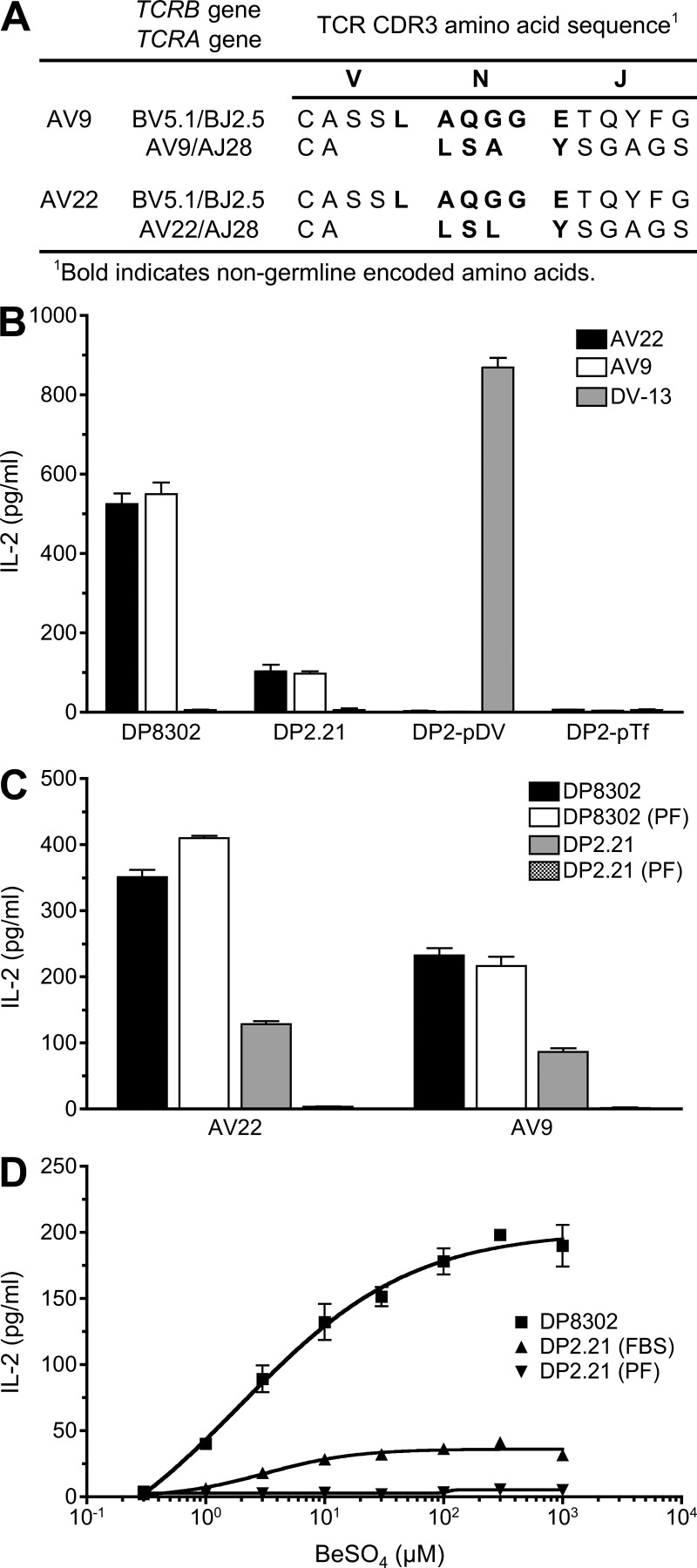
Using T cell hybridomas AV22 and AV9 that express related Be-specific TCRs derived from the lung of a CBD patient (Fig. 1 A; Bowerman et al., 2011), we sought to identify Be-dependent peptides capable of stimulating these TCRs. AV22 and AV9 respond to BeSO4 presented by mouse DAP3.L cells transfected with HLA-DP2 (designated DP8302) independently of the addition of exogenous protein or peptide (Fig. 1 B). To investigate the role of peptide in the recognition of Be, we created HLA-DP2–expressing antigen-presenting cell lines that are incapable of peptide exchange and present only a single peptide. An HLA-DP2 β-chain gene was constructed that utilizes a genetically encoded linker to covalently attach individual HLA-DP2–binding peptides (Okamoto et al., 1998; Díaz et al., 2005) to the N terminus of the HLA-DPB1*0201 gene. The DPA1*0103 and DPB1*0201 peptide gene constructs were transfected into a kidney fibroblast line derived from C57BL/6 mice that are genetically deficient for MHCII and invariant chain (B6 DK10; Huseby et al., 2005). Equivalent cell surface expression of the HLA-DP2/peptide molecules was confirmed by staining with an anti–HLA-DP mAb (not depicted). In contrast to B6 DK10 fibroblasts transfected with native HLA-DP2 (designated DP2.21), fibroblasts expressing covalently attached HLA-DP2–binding peptides derived from transferrin receptor (p6-17) or a dengue virus NS3 peptide (p254-265; Díaz et al., 2005) failed to stimulate Be-specific T cell hybridomas in the presence of BeSO4 (Fig. 1 B). Conversely, a dengue virus NS3–specific hybridoma responded to HLA-DP2 with the covalently linked dengue peptide (Fig. 1 B).
Figure 1.
Be-specific CD4+ T cells require Be and a specific peptide for antigen recognition. (A) TCR gene segment usage and junctional region amino acid sequence of the Be-specific T cell hybridomas AV22 and AV9. (B) Be-specific (AV22 and AV9) and dengue virus–specific (DV-13) T cell hybridomas were stimulated with the following antigen-presenting cell lines: HLA-DP2–transfected mouse DAP3.L cells (DP8302), fibroblasts (B6 DK10) derived from MHCII- and invariant chain-deficient mice and transfected with native HLA-DP2 (DP2.21), and B6 DK10 cells transfected with HLA-DP2 with a transferrin receptor peptide (DP2-pTf, EPLSYTRFSLAR) or a dengue virus NS3–derived peptide (DP2-pDV, REIVDLMCHATF) covalently attached to the N terminus of the DP2 β-chain. 200 µM BeSO4 was added to all antigen-presenting cells except DP2-pDV–expressing fibroblasts, and the IL-2 response (mean ± SEM pg/ml) by the hybridomas was measured by ELISA. (C) AV22 and AV9 T cell hybridomas were stimulated with 200 µM BeSO4 presented by HLA-DP2–transfected fibroblasts grown in either FBS or protein-free (PF) medium. IL-2 secretion (mean ± SEM pg/ml) was measured by ELISA. (B and C) Data shown are representative of three experiments performed in triplicate. (D) AV22 hybridoma cells were stimulated over a range of BeSO4 concentrations (0.3–1,000 µM) using DP8302 cells grown in FBS-containing medium compared with DP2.21 cells grown in FBS and protein-free conditions. Data shown (mean IL-2 ± SD pg/ml) are representative of three experiments performed in triplicate.
In assessing the role of peptide in mediating Be recognition, the AV22 and AV9 hybridomas consistently secreted 75% less IL-2 when using DP2.21 fibroblasts as antigen-presenting cells compared with DP8302 as a result of restriction of the peptide repertoire (Fig. 1, B and C). Adapting DP2.21 to protein-free medium completely abrogated the Be-specific hybridoma response, whereas this medium had no effect on cell viability or growth (not depicted) or the ability of DP8302 cells to present BeSO4 to the Be-specific T cell hybridomas (Fig. 1, C and D). Because this cell line does not naturally present Be to the Be-specific hybridomas, all antigen presentation assays using peptides were conducted using DP2.21 grown in protein-free medium.
Next, we tested a larger set of 25 known HLA-DP2–binding peptides (Díaz et al., 2005) in the presence of BeSO4 using DP2.21 as antigen-presenting cells. None of these peptides induced IL-2 secretion by the AV22 or AV9 hybridomas (not depicted), demonstrating that HLA-DP2–restricted T cell recognition of Be depends on a limited number of specific peptides.
Identification of Be-dependent mimotopes using a decapeptide positional scanning library
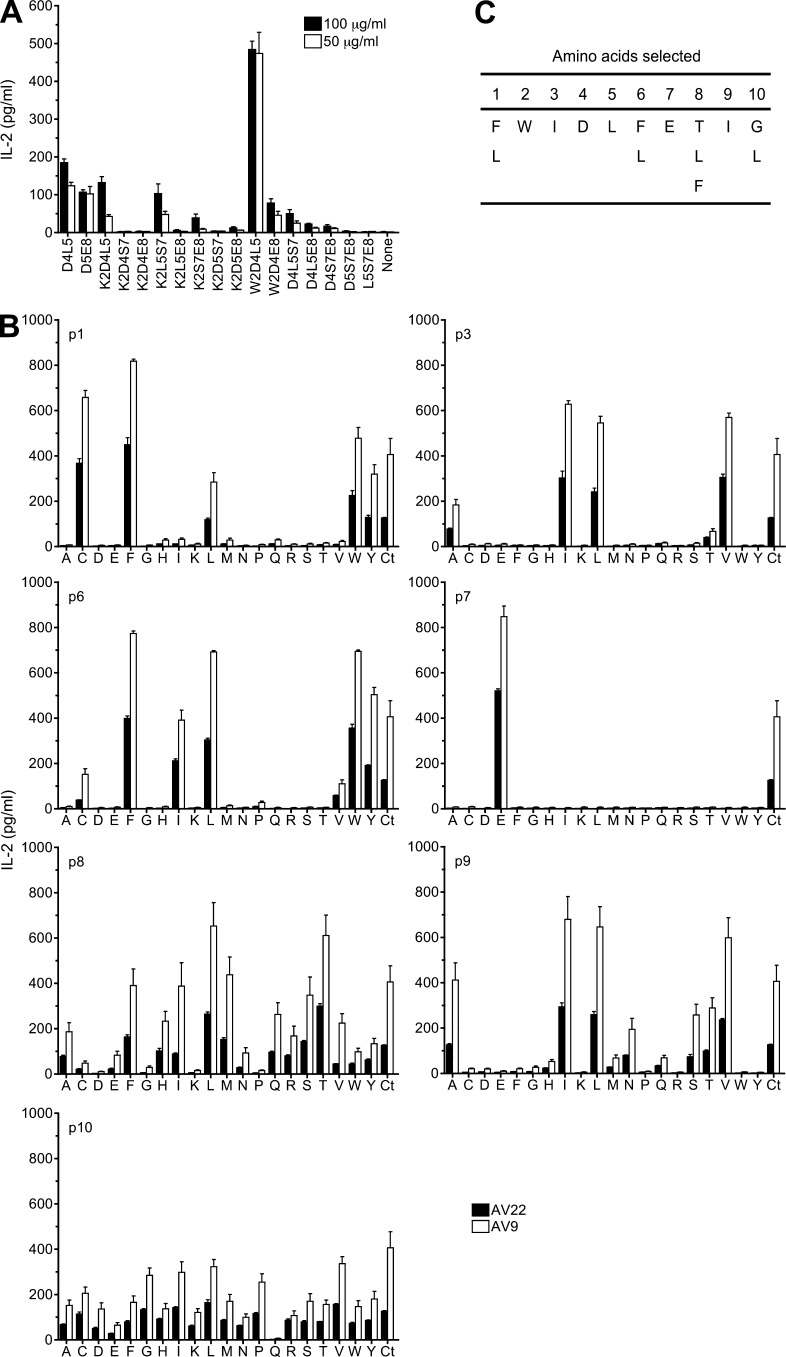
Having observed that common HLA-DP2–binding peptides do not stimulate T cell hybridomas expressing Be-specific TCRs, we used a decapeptide positional scanning library, which makes no assumptions regarding peptide composition and allows a systematic assessment of all peptides of a given length in a standard T cell activation assay. The initial library screen and a subsequent screen using less diverse mixtures containing fixed amino acids at two positions showed that the most potent mixtures contained an aspartic acid (D) at p4 and a leucine (L) at p5 or a D at p5 and a glutamic acid (E) at p8 (not depicted). We next synthesized select decapeptide mixtures with three positions fixed. As shown in Fig. 2 A, the only mixture that enhanced IL-2 secretion in the presence of BeSO4 above the control D4L5 and D5E8 mixtures contained a tryptophan (W) at the p2 position, D at p4, and L at p5. Importantly, none of these mixtures stimulated an HLA-DP2–restricted, dengue virus–specific hybridoma (DV-13) or Be-specific hybridomas in the absence of BeSO4 (not depicted).
Figure 2.
Deconvolution strategy to define Be-dependent peptides that stimulate T cell hybridomas AV22 and AV9. (A) AV22 cells were stimulated with selected decapeptide mixtures with three positions fixed in the presence of 75 µM BeSO4. Two concentrations of mixtures were used, and IL-2 response (mean ± SEM pg/ml) was measured by ELISA. Mixtures with two fixed positions (D4L5 and D5E8) were included as positive controls. Data are representative of three separate experiments performed in triplicate. (B) IL-2 response of three separate experiments (mean ± SEM) of AV22 and AV9 to a biased decapeptide positional scanning library (W2D4L5) is shown. Peptide mixtures were fixed at W2, D4, and L5, and one additional peptide position (p1, p3, p6, p7, p8, p9, and p10) was scanned with each of 20 amino acids (140 mixtures). Each panel shows the response of hybridomas to 20 µg/ml mixtures and 75 µM BeSO4 compared with the W2D4L5 mixture with no additional fixed positions (Ct). For A and B, the x axis denotes the amino acid (single letter code) fixed at each defined position, and mixtures did not stimulate hybridomas in the absence of BeSO4. (C) Selection of amino acids for each peptide position based on the most active mixtures in the presence of BeSO4.
With preferred amino acids defined at three positions of the decapeptide, we performed a positional scan of the remaining seven positions. All mixtures included three fixed positions (W2D4L5) and a fourth position fixed with 1 of 20 amino acids at each remaining position of the peptide. As shown in Fig. 2 B, most positions showed a distinct profile of allowed amino acids with identical preferences for AV22 and AV9. For example, known anchor residues for HLA-DP2–binding peptides (e.g., phenylalanine [F] and L) were preferred at p1 and p6. At positions p3 and p9, related, nonpolar amino acids (isoleucine [I], valine [V], and L) were selected. The only amino acid at p7 that enhanced IL-2 secretion by AV22 and AV9 above the control W2D4L5 mixture was E. Nonrelated amino acids, including threonine (T), L, and F, were selected at p8. In contrast, no definitive selection of amino acids was seen at p10.
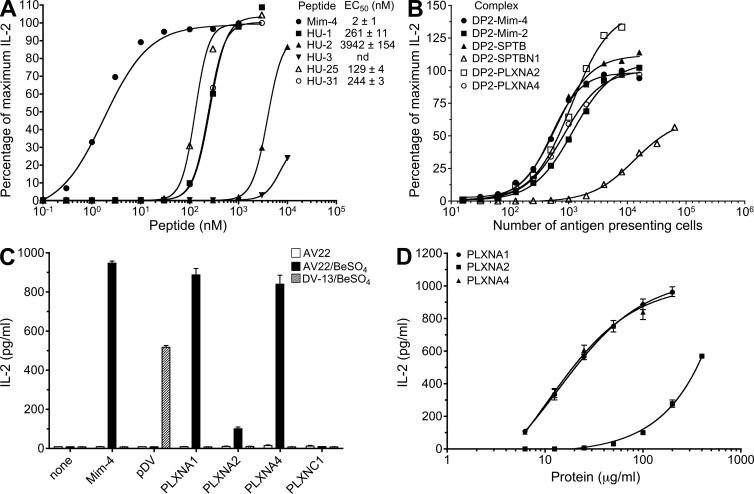
Based on the defined amino acids at each position of the peptide (Fig. 2 C), we synthesized a set of 24 mimotopes and tested their ability to stimulate Be-specific T cell hybridomas. All of the mimotopes stimulated IL-2 secretion by AV22 in the presence of BeSO4. In Table 1, mimotopes were ranked by EC50 values (peptide concentration inducing half-maximal IL-2 response), and the ranking of the mimotopes was similar for AV9 (not depicted). Of note, the three highest ranking mimotopes only differed at the p8 position and had similar EC50 values; thus, we focused on mimotopes 2 and 4.
Table 1.
Mimotopes that stimulate the Be-specific T cell hybridoma AV22
| Mimotope | Peptide sequence | AV22 | ||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | EC50 | SEM | |
| ng/ml | ||||||||||||
| 4 | F | W | I | D | L | F | E | L | I | G | 143 | 7 |
| 2 | F | W | I | D | L | F | E | T | I | G | 152 | 22 |
| 6 | F | W | I | D | L | F | E | F | I | G | 201 | 21 |
| 3 | F | W | I | D | L | F | E | L | I | L | 211 | 43 |
| 8 | F | W | I | D | L | L | E | T | I | G | 219 | 34 |
| 18 | L | W | I | D | L | F | E | F | I | G | 229 | 19 |
| 10 | F | W | I | D | L | L | E | L | I | G | 251 | 33 |
| 12 | F | W | I | D | L | L | E | F | I | G | 251 | 20 |
| 16 | L | W | I | D | L | F | E | L | I | G | 260 | 7 |
| 14 | L | W | I | D | L | F | E | T | I | G | 293 | 27 |
| 15 | L | W | I | D | L | F | E | L | I | L | 418 | 109 |
| 20 | L | W | I | D | L | L | E | T | I | G | 428 | 24 |
| 22 | L | W | I | D | L | L | E | L | I | G | 737 | 26 |
| 9 | F | W | I | D | L | L | E | L | I | L | 961 | 127 |
| 11 | F | W | I | D | L | L | E | F | I | L | 1,266 | 364 |
| 7 | F | W | I | D | L | L | E | T | I | L | 1,448 | 137 |
| 5 | F | W | I | D | L | F | E | F | I | L | 1,480 | 53 |
| 24 | L | W | I | D | L | L | E | F | I | G | 1,882 | 137 |
| 19 | L | W | I | D | L | L | E | T | I | L | 1,913 | 17 |
| 1 | F | W | I | D | L | F | E | T | I | L | 2,071 | 286 |
| 17 | L | W | I | D | L | F | E | F | I | L | 2,350 | 297 |
| 13 | L | W | I | D | L | F | E | T | I | L | 2,400 | 176 |
| 23 | L | W | I | D | L | L | E | F | I | L | 3,138 | 103 |
| 21 | L | W | I | D | L | L | E | L | I | L | 4,082 | 491 |
Mimotopes are ordered by most to least active (i.e., lowest to highest EC50 values). Bolded amino acids denote peptide positions that vary in the mimotopes.
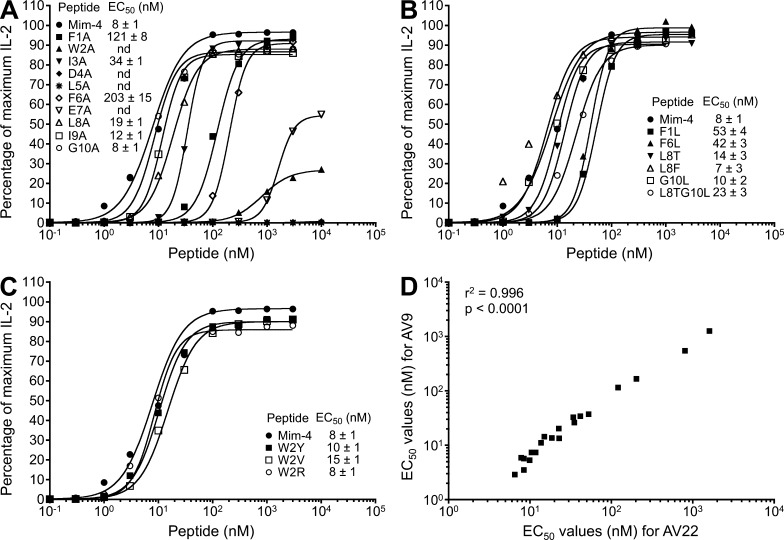
Alanine (A) and analogue scans of mimotope-4 were performed to determine the critical amino acids for anchoring to HLA-DP2, coordinating Be, or interacting with TCR. An F to A substitution at either p1 or p6 of the peptide shifted the dose–response curve to the right, resulting in a 16- and 26-fold reduction in the EC50, respectively (Fig. 3 A). The preferred anchor residue at p1 and p6 was an F compared with L (Fig. 3 B). Confirming the positional scanning library data, an A at p2, p4, p5, or p7 abrogated T cell recognition of the HLA-DP2–peptide/Be complex, whereas alanines at p8, p9, or p10 had minimal effects on activation of the Be-specific AV22 hybridoma (Fig. 3 A). In this regard, deletion of the p9 and p10 amino acids had minimal effects on EC50 values (not depicted), demonstrating that these positions are much less important for recognition of this ligand. In addition to W at p2, other amino acids, such as Y, V, and R, were also allowed at this position with no effect on IL-2 secretion (Fig. 3 C). EC50 values for AV22 and AV9 were strongly correlated (Fig. 3 D), confirming that these TCRs recognize the same ligand despite using different TCR Vα chains (Bowerman et al., 2011).
Figure 3.
Be-specific T cell hybridoma response to alanine substitutions and analogues of mimotope-4. Peptide dose–response curves were completed for T cell hybridoma AV22. (A–C) Equal numbers of AV22 cells and DP2.21 antigen-presenting cells were mixed with 75 µM BeSO4 and the following highly purified peptides: peptides with single alanine substitutions at each position of mimotope-4 (A), analogues of mimotope-4, selected based on the demonstrated potency of other mimotopes (B), and mimotope-4 variants at the p2 position of the peptide (C). IL-2 secretion was measured by ELISA after 22 h of culture, and data are plotted as the percentage of maximum IL-2 secretion against peptide concentration in the presence of BeSO4. EC50 values (mean ± SEM nM) for mimotope-4 and each variant peptide from four independent experiments are shown. Note that error bars have been left out for viewing clarity. (D) Correlation of EC50 values of all variants of mimotope-4 between hybridomas AV22 and AV9.
HLA-DP2–mimotope-2/Be tetramer staining of CD4+ T cells from CBD patients
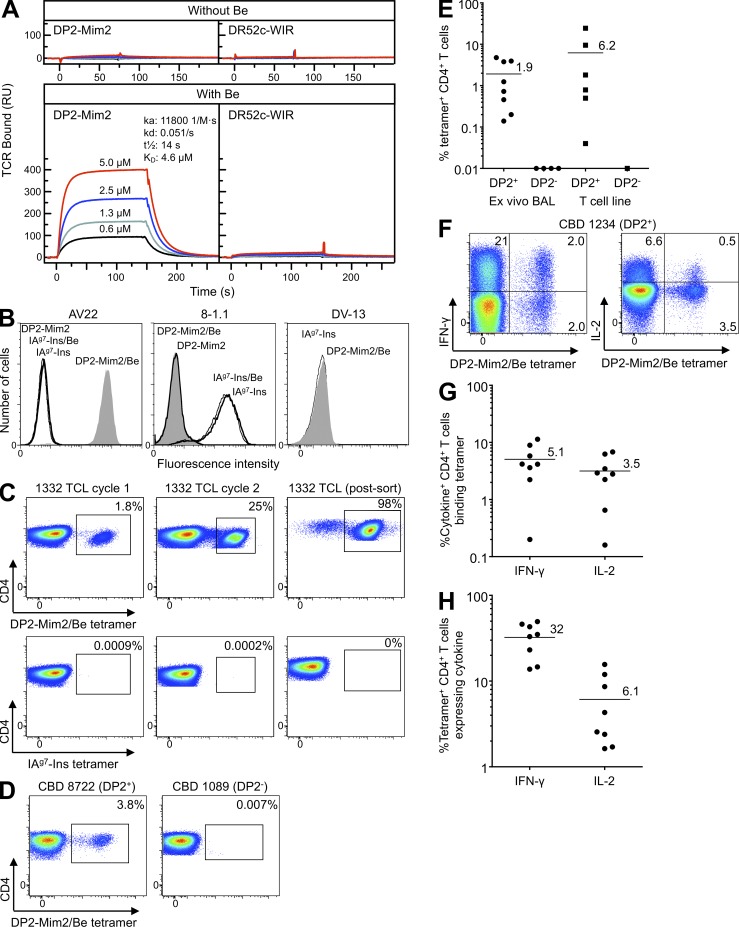
Next, we purified soluble recombinant HLA-DP2–mimotope-2 and AV22 TCR from baculovirus-infected insect cells to determine the affinity of the TCR for the HLA-DP2–mimotope-2/Be complex, using surface plasmon resonance. The AV22 TCR did not bind to either HLA-DP2–mimotope-2 or HLA-DR52c–WIRVNIPKRI (denoted HLA-DR52c–WIR; Yin et al., 2012) in the absence of BeSO4 (Fig. 4 A). However, after exposing the flow cells to 200 µM BeSO4, the AV22 TCR bound to HLA-DP2–mimotope-2, but not HLA-DR52c–WIR, with high affinity (Kd = 4.6 µM; Fig. 4 A), confirming that Be is a necessary component for T cell recognition.
Figure 4.
HLA-DP2–mimotope-2/Be tetramer staining of Be-specific T cell lines and ex vivo BAL cells from CBD patients. (A) Approximately 2,000 resonance units (RU) of biotinylated HLA-DP2–mimotope-2 and control HLA-DR52c–WIR complex were immobilized in flow cells of a BIAcore streptavidin biosensor chip. Various concentrations of soluble AV22 TCR were injected through the flow cells before and after loading with 200 µM BeSO4, and the surface plasmon resonance signal was obtained. Data shown are representative of three independent experiments. (B) Staining of T cell hybridomas (Be-specific AV22, insulin B:9-23–specific 8-1.1, and dengue virus–specific DV-13) with HLA-DP2–mimotope-2 (DP2-Mim2) and IAg7-insulin (IAg7-Ins) tetramers prepared into complexes either in the presence or absence of BeSO4. Cells were stained with 20 µg/ml of tetramers, and fluorescence intensity was evaluated by flow cytometry. Representative results from three independent experiments are shown. All hybridomas expressed high levels of TCR (not depicted). (C) Detection of HLA-DP2–mimotope-2/Be tetramer–binding T cells from parental T cell line derived from CBD patient 1332. BAL T cells after one and two cycles of stimulation with BeSO4 and after Vβ5.1 sorting were stained with the HLA-DP2–mimotope-2/Be (top) and IAg7-insulin (bottom) tetramers, and the frequency of CD4+ T cells stained with each tetramer is shown in each density plot. Representative staining results from two independent experiments are shown. (D) HLA-DP2–mimotope-2/Be tetramer staining of ex vivo BAL cells from CBD patients who are HLA-DP2+ (8722) and HLA-DP2− (1089). Density plots show tetramer staining of CBD patients, excluding cells staining with CD8, CD14, and CD19, and gating for CD3 and CD4 expression. Results are representative of ex vivo BAL cells from eight HLA-DP2+ and four HLA-DP2− CBD subjects. (E) Frequency (mean) of CD4+ T cells staining with the HLA-DP2–mimotope-2/Be tetramer is shown for ex vivo BAL cells (n = 12) and BAL T cell lines (n = 7) derived from DP2+ and DP2− CBD patients. Frequency was determined by subtracting nonspecific staining observed with the control tetramer from staining with the HLA-DP2 tetramer. (F) Flow cytometric analysis of dual intracellular cytokine and tetramer staining of ex vivo BAL cells from a DP2+ CBD patient is shown. Density plots show IFN-γ (left) and IL-2 (right) expression relative to HLA-DP2–mimotope-2/Be tetramer staining. Data are representative of the eight HLA-DP2+ CBD subjects studied. (G and H) For ex vivo BAL cells, the mean frequency of IFN-γ– and IL-2–expressing CD4+ T cells that bind the HLA-DP2–mimotope-2/Be tetramer (G) and the frequency of tetramer-positive cells that secrete these cytokines (H) are shown. (E, G, and H) Mean values for each group are indicated with horizontal bars.
Having identified an MHCII/peptide/Be ligand for TCRs from a single CBD patient, we tested whether this ligand could engage TCRs on T cells from other HLA-DP2–expressing CBD patients. We constructed a tetramer consisting of HLA-DP2 with mimotope-2 covalently attached to the N terminus of the HLA-DP2 β-chain with and without saturating BeSO4. This reagent specifically stained the AV22 hybridoma only when Be was included in the complex and did not bind to either an HLA-DP2–restricted, dengue virus–specific hybridoma DV-13 or a T cell hybridoma specific for insulin B:9-23 in the context of IAg7 (8-1.1; Fig. 4 B). In contrast to 8-1.1, which bound to the IAg7-insulin B:9-23 tetramer (Crawford et al., 2011), AV22 did not stain with this reagent (Fig. 4 B). Identical findings were seen with AV9 (not depicted). The bronchoalveolar lavage (BAL)–derived CD4+ T cell line from which AV22 and AV9 were cloned also brightly stained with this tetramer. After one and two cycles of Be stimulation, 1.8 and 25% of the CD4+ T cells specifically recognized this αβTCR ligand and not the IAg7-insulin tetramer (Fig. 4 C). After cell sorting for TCR Vβ5.1 expression, the vast majority of T cells bound tetramer.
Next, additional Be-specific T cell lines and ex vivo BAL cells from CBD patients were stained. Remarkably, all HLA-DP2–expressing patients (n = 12) who had evidence of a Be-specific adaptive immune response in the lung had CD4+ T cells in BAL that bound the HLA-DP2–mimotope-2/Be tetramer (Fig. 4, D and E). For the ex vivo BAL samples (n = 8), the range was 0.1–4.8% of CD4+ T cells, with an overall frequency of 1.9 ± 0.7% (mean ± SEM; Fig. 4 E). Similar findings were seen with the T cell lines (Fig. 4 E). None of the HLA-DP2− CBD patients had detectable T cell staining with tetramer (Fig. 4 E), nor did four HLA-DP2+ Be-sensitized subjects who had no evidence of a Be-specific immune response in BAL (not depicted), further validating the specificity of the reagent.
To examine the frequency of tetramer-staining CD4+ T cells relative to the overall frequency of Be-responsive cells in the BAL of CBD patients, we coupled tetramer staining with intracellular cytokine staining for Be-induced IFN-γ and IL-2 expression (representative example shown in Fig. 4 F). We focused on these Th1-type cytokines because they are the most abundantly expressed T cell–derived cytokines in CBD lung (Tinkle et al., 1997; Fontenot et al., 2002). For ex vivo BAL cells, 5.1 ± 1.3% of IFN-γ–expressing and 3.2 ± 0.8% of IL-2–expressing CD4+ T cells bound tetramer (Fig. 4 G). The majority of tetramer-binding CD4+ T cells did not express either IFN-γ or IL-2 (Fig. 4, F and H). Although other Be-dependent specificities were present, our data showing the presence of CD4+ T cells specific for this ligand in all HLA-DP2–expressing CBD patients tested suggest that the HLA-DP2–mimotope-2/Be complex likely represents a major ligand in the lung for Be-specific CD4+ T cells.
Delineation of endogenous peptides that stimulate Be-specific αβTCRs
To discover endogenous peptides capable of stimulating the Be-specific AV22 and AV9 hybridomas, we performed a biometric analysis (Hemmer et al., 1999; Zhao et al., 2001; Pinilla et al., 2003) using a matrix derived from the stimulatory potency of each mixture defined with an amino acid at every peptide position of the W2D4L5 positional scanning library. As shown in Table 2, we screened 35 human peptides using IL-2 secretion from AV22 and AV9 as a readout for T cell response. Of the 11 peptides that gave positive results at low concentrations, 2 were derived from spectrin and 4 were derived from plexins. The most potent peptides (i.e., lowest EC50 values) were derived from plexin A2 (HU25), plexin A4 (HU31), and erythrocytic spectrin (HU1), although none were as good as mimotope-4 (Fig. 5 A).
Table 2.
Human peptides screened for the ability to stimulate the AV22 and AV9 Be-specific T cell hybridomas
| ID/rank | Ratio to max | Peptide sequence | GenBank accession no. | AV22 | AV9 | Protein | |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | IL-2 | IL-2 | ||||
| pg/ml | pg/ml | ||||||||||||||
| HU1 | 0.89 | M | W | A | D | L | L | E | L | I | D | BAD92652.1 | 997 | 1,117 | SPTB |
| HU2 | 0.88 | C | F | L | D | L | L | E | L | L | I | CAE89418.1 | 716 | 966 | unnamed |
| HU3 | 0.87 | A | W | A | D | L | L | E | L | I | D | BAD92985.1 | 29.3 | 75.1 | SPTBN1 |
| HU4 | 0.87 | A | W | A | D | L | L | E | L | L | D | BAA32700.2 | 4.9 | 9.4 | |
| HU5 | 0.87 | C | W | L | D | A | L | E | L | A | L | BAA96058.1 | 4.1 | 4.6 | |
| HU6 | 0.86 | L | D | V | D | L | L | E | L | V | I | BAA23700.1 | 4.9 | 4.1 | |
| HU7 | 0.86 | Y | W | L | D | L | W | L | F | I | L | CAF00026.1 | 4.1 | 3.6 | |
| HU8 | 0.85 | C | W | L | S | L | L | E | Y | L | L | AAD52694.1 | 4.6 | 5.4 | |
| HU9 | 0.85 | F | W | L | R | L | L | E | L | T | W | AAH40508.1 | 21.7 | 48.6 | PRRT3 |
| HU10 | 0.85 | C | L | A | D | L | Y | E | T | L | G | BAC02709.1 | 5.1 | 29 | |
| HU11 | 0.85 | W | W | I | D | L | L | R | R | T | G | CAE89469.1 | 3.3 | 5.6 | |
| HU12 | 0.85 | C | W | L | D | L | L | L | A | A | L | CAD33443.1 | 3.8 | 3.6 | |
| HU13 | 0.84 | L | W | I | L | L | L | E | T | S | L | BAB67803.1 | 3.6 | 4.9 | |
| HU14 | 0.84 | L | S | I | D | L | Y | E | L | I | K | AAG17028.1 | 4.4 | 6.1 | |
| HU15 | 0.84 | F | W | I | D | L | N | E | D | I | I | CAD67506.1 | 6.7 | 49.3 | unnamed |
| HU16 | 0.84 | L | W | L | D | L | W | Y | L | M | F | BAC85487.1 | 4.6 | 6.1 | |
| HU17 | 0.83 | C | T | V | D | L | L | E | F | Q | P | BAC85360.1 | 15.6 | 16.3 | |
| HU18 | 0.83 | H | W | L | D | L | F | R | L | L | G | AAB09728.1 | 4.4 | 4.4 | |
| HU19 | 0.83 | C | W | V | D | L | I | I | S | S | S | CAD38236.1 | 4.1 | 4.6 | |
| HU20 | 0.83 | F | W | A | R | L | L | E | R | L | F | CAF04283.1 | 4.6 | 4.6 | |
| HU21 | 0.83 | Y | C | I | D | L | L | E | R | L | A | CAC69378.1 | 17.4 | 48.8 | unnamed |
| HU22 | 0.83 | F | W | A | Q | L | L | E | R | V | F | BAC04745.1 | 5.1 | 4.1 | |
| HU23 | 0.83 | F | W | T | P | L | L | E | S | L | A | AAH81559.1 | 4.4 | 4.4 | |
| HU24 | 0.83 | L | C | L | D | L | L | E | L | S | T | CAI40388.1 | 5.4 | 4.6 | |
| HU25 | 0.82 | F | V | D | D | L | F | E | T | L | L | AAH09343.2 | 622 | 847 | PLXNA2 |
| HU26 | 0.82 | L | N | V | D | L | Y | E | L | S | S | BAA92381.1 | 4.9 | 7.1 | |
| HU28 | 0.82 | L | C | A | D | L | F | E | R | V | P | BAD96476.1 | 4.4 | 4.6 | |
| HU29 | 0.82 | W | W | Q | D | L | F | R | I | V | L | CAE90529.1 | 3.3 | 4.6 | |
| HU30 | 0.81 | L | K | V | D | L | L | E | Q | T | K | CAI22508.1 | 3.1 | 3.9 | |
| HU31 | 0.81 | F | V | D | D | L | F | E | T | I | F | BAB13376.3 | 973 | 1,042 | PLXNA4 |
| HU32 | 0.81 | F | E | A | D | L | L | E | M | A | E | CAF06542.1 | 5.4 | 7.9 | |
| HU33 | 0.81 | F | V | D | D | L | F | E | T | L | F | CAD35002.1 | 850 | 954 | PLXNA2 |
| HU35 | 0.81 | F | V | D | D | L | F | E | T | V | F | CAI43194.1 | 947 | 1,035 | PLXNA3 |
| HU38 | 0.81 | L | K | L | D | L | L | E | A | N | S | BAE46614.1 | 5.6 | 5.1 | |
| HU39 | 0.81 | F | W | L | P | L | L | E | K | V | Y | CAF85973.1 | 22.4 | 68.7 | unnamed |
Designations in bold indicate peptides that stimulate AV22 and AV9 in the presence of BeSO4. For Ratio to max, the score of each peptide was divided by the matrix maximum score. Peptide positions are shown in bold if peptides contain W2D4L5E7. Protein designations (if available) are provided only for peptides that gave a positive response. Hybridomas were stimulated with 0.5 µg/ml of peptide and BeSO4 at 75 µM.
Figure 5.
Characterization of candidate endogenous peptides that stimulate Be-specific T cell hybridomas. (A) Dose–response curves to stimulatory peptides identified from the biometric analysis of T cell hybridoma AV22 are shown. Equal numbers of AV22 cells and DP2.21 antigen-presenting cells were mixed with 75 µM BeSO4 and highly purified peptides. IL-2 secretion was measured by ELISA after 22 h of culture, and data are plotted as the percentage of maximum IL-2 secretion against concentration of peptide in the presence of BeSO4. EC50 values (mean ± SEM nM) for mimotope-4 and each human peptide from four independent experiments are shown. Note that error bars have been left out for viewing clarity. (B) AV22 response to antigen-presenting cells expressing HLA-DP2 with spectrin and plexin peptides covalently attached to the N terminus of the β-chain. Percentage of maximum IL-2 secretion versus the number of antigen-presenting cells added per well is shown. Results are representative of three independent experiments. (C) IL-2 response (mean ± SD pg/ml) of AV22 and dengue virus–specific hybridoma DV-13 to plexin proteins presented by an EBV-transformed B cell line derived from CBD patient 1332 is shown. Plexin proteins (A1, A2, A4, and C1, all at 100 µg/ml) were added in the presence and absence of 75 µM BeSO4. Control peptides, mimotope-4 and dengue viral peptide, were added at 100 nM and 20 µM, respectively. Representative results from three independent experiments are shown. (D) Dose–response curves of AV22 to plexin A1, A2, and A4 proteins presented by EBV-transformed B cells in the presence of 75 µM BeSO4 are shown. Representative results from three independent experiments (mean IL-2 ± SD pg/ml) performed in triplicate are shown.
We next expressed HLA-DP2 with plexin A2 (PLXNA2), plexin A4 (PLXNA4), erythrocytic spectrin (SPTB), or spectrin β-chain (SPTBN1) peptides covalently attached to the N terminus of the HLA-DPB1*0201 gene on mouse fibroblasts. Titering the number of fibroblasts as antigen-presenting cells in the presence of BeSO4, both AV22 and AV9 showed equivalent responses to plexin A2, plexin A4, and erythrocytic spectrin peptides, as well as mimotopes 2 and 4 (Fig. 5 B). The peptide derived from spectrin β-chain (SPTBN1; HU3) induced a significantly lower IL-2 response with both purified peptide and the covalently linked version, likely because of poor HLA-DP2 binding as a result of an A at the p1 position (Fig. 5, A and B). Based on these data, we focused on the plexin family of proteins to ascertain whether these molecules might be relevant in vivo in CBD.
Plexins are transmembrane proteins encoded by nine genes (PLXNA1-4, B1-3, C1, and D1). Only the plexin A family contains the stimulatory epitope for our Be-responsive TCRs. However, the presence of this epitope in multiple plexin A proteins meant that an shRNA knockdown approach was not feasible. Therefore, we first determined whether plexin proteins can be processed by antigen-presenting cells to yield the stimulatory, Be-dependent HLA-DP2–binding epitope. Purified recombinant proteins encoding the cytoplasmic domains of several mouse plexin molecules were generated (He et al., 2009; Wang et al., 2012). All of these proteins are >600 aa in length and have >98% identity with human plexins. Each of the plexin A proteins (PLXNA1, PLXNA2, and PLXNA4) was processed by HLA-DP2+ EBV-transformed B cells and stimulated IL-2 secretion from AV22 only in the presence of Be (Fig. 5 C). As expected, PLXNC1 protein, which does not contain the stimulatory epitope, did not induce IL-2 secretion by AV22, and none of the plexin proteins stimulated the dengue virus–specific hybridoma (Fig. 5 C). Furthermore, the natural Be-specific response observed using HLA-DP2–transfected fibroblasts (DP8302) as antigen-presenting cells for AV22 was significantly enhanced with the addition of all of the plexin A proteins (not depicted). Titering the concentration of plexin A proteins to as low as 6 µg/ml still resulted in robust stimulation of the AV22 hybridoma for PLXNA1 and PLXNA4 (Fig. 5 D). Conversely, PLXNA2 was ∼15-fold less potent at stimulating Be-dependent IL-2 secretion and could be detected beginning at 50 µg/ml.
Be-loaded HLA-DP2–plexin A4 tetramer staining of CD4+ T cells in BAL
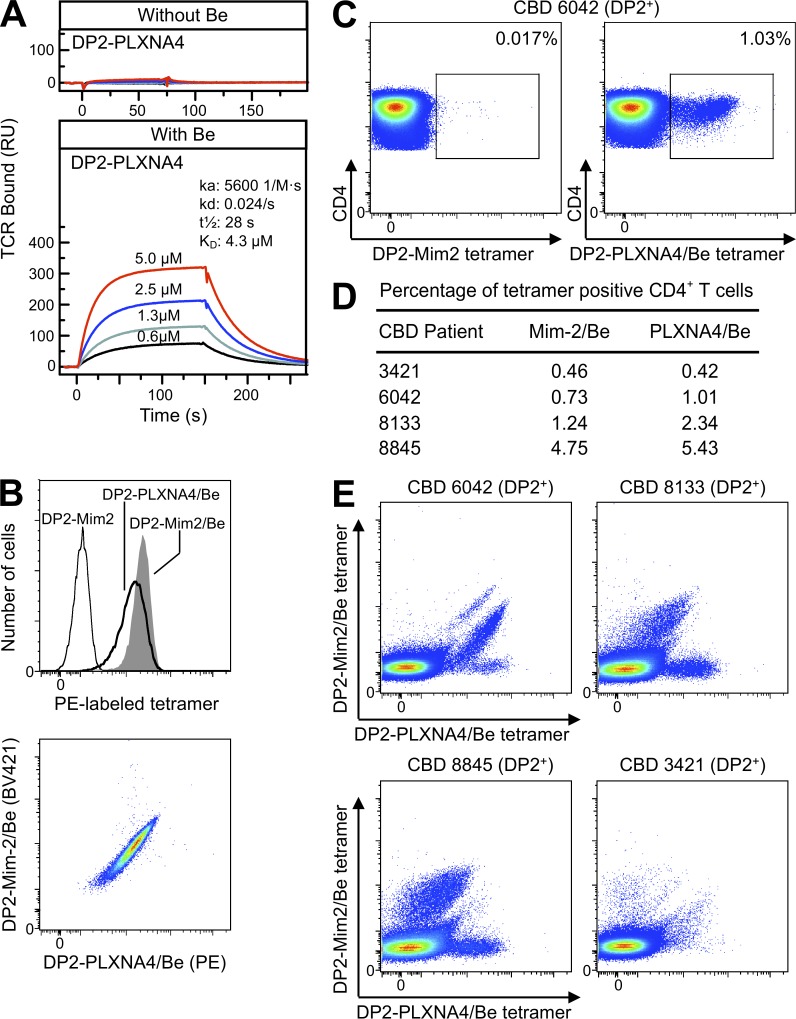
Similar to our analysis of mimotope-2, we generated soluble recombinant HLA-DP2–PLXNA4 and assessed the binding affinity of the AV22 TCR for this complex in the presence of BeSO4. As shown in Fig. 6 A, the AV22 TCR binds to HLA-DP2–PLXNA4 only in the presence of Be, with identical binding kinetics for each of the four concentrations of TCR injected. The measured TCR affinity (4.3 µM) for this MHCII is very similar to that of the HLA-DP2–mimotope-2/Be complex (compare with Fig. 4 A). An HLA-DP2–PLXNA4 tetramer, saturated in BeSO4 and labeled with PE, stained the AV22 hybridoma with slightly lower fluorescence intensity than the PE-labeled HLA-DP2–mimotope-2/Be tetramer (Fig. 6 B, top). Using a mimotope-2/Be tetramer labeled with brilliant violet 421 (BV421), hybridoma cells stained with both tetramers showed a uniform pattern of costaining (Fig. 6 B, bottom). Using ex vivo BAL cells from four of the HLA-DP2+ CBD patients previously stained with the HLA-DP2–mimotope-2/Be tetramer in Fig. 4 E, we identified PLXNA4/Be tetramer–staining CD4+ T cells in all subjects (Fig. 6, C and D). No PLXNA4/Be tetramer–staining T cells were seen using BAL cells from an HLA-DP2− CBD patient (not depicted). In three of the HLA-DP2+ CBD patients, the frequency of PLXNA4/Be tetramer–staining T cells was higher than that seen with the mimotope-2/Be tetramer (Fig. 6 D).
Figure 6.
HLA-DP2–PLXNA4/Be tetramer staining of ex vivo BAL cells from CBD patients. (A) The binding kinetics of soluble AV22 TCR to biotinylated HLA-DP2–PLXNA4 with and without Be are shown. Four concentrations of soluble AV22 TCR were injected through the flow cells before and after loading with 200 µM BeSO4, and the surface plasmon resonance signal was obtained. Data shown are representative of three independent experiments. (B) Staining of the AV22 T cell hybridoma with HLA-DP2–mimotope-2, HLA-DP2–mimotope-2/Be, and HLA-DP2–PLXNA4/Be tetramers individually (top) and costaining with Be-saturated mimotope-2 (BV421 labeled) and PLXNA4 (PE labeled) tetramers (bottom). The fluorescence intensity of cells was evaluated after staining with 20 µg/ml of tetramers for 2 h at 37°C. Representative results from three independent experiments are shown. (C) Density plots showing control HLA-DP2–mimotope-2 without Be (left) and HLA-DP2–PLXNA4/Be (right) tetramer staining of CD4+ T cells from ex vivo BAL cells of a HLA-DP2+ CBD patient. Results are representative of four HLA-DP2+ CBD subjects stained. (D) Summary of the frequency of tetramer staining of ex vivo BAL CD4+ T cells from four HLA-DP2+ CBD patients individually stained with the HLA-DP2–mimotope-2/Be and HLA-DP2–PLXNA4/Be tetramers. (E) Density plots of ex vivo BAL cells from four DP2+ CBD patients costained with HLA-DP2–PLXNA4/Be and HLA-DP2–mimotope-2/Be tetramers are shown.
Having tetramers labeled with different fluorophores enabled us to simultaneously stain ex vivo BAL cells from CBD patients with the HLA-DP2–mimotope-2/Be and HLA-DP2–PLXNA4/Be tetramers. As shown in Fig. 6 E, distinct populations of CD4+ T cells emerged that differentially stain with the two tetramers. Although the relative affinity of the BAL CD4+ T cells for each tetramer varies, the majority of CD4+ T cells that bind the mimotope-2/Be tetramer also bind the PLXNA4/Be tetramer, as indicated by the diagonal staining distribution (Fig. 6 E). For CBD patient 6042, the dual staining cells are shifted to the right and bind to the PLXNA4/Be tetramer more strongly than in patients 8133 and 8845 (Fig. 6 E). Importantly, a discrete population of BAL T cells that only bind the PLXNA4/Be tetramer was seen in these CBD patients (Fig. 6 E). Thus, these data highlight the heterogeneity of the BAL CD4+ T cells that bind these ligands and demonstrate a unique population of cells that specifically recognizes only the HLA-DP2–PLXNA4/Be complex, strongly supporting a role of plexin A as an endogenous antigen for a set of Be-specific TCRs.
Plexin A protein expression in antigen-presenting cells, BAL fluid, and lung tissue
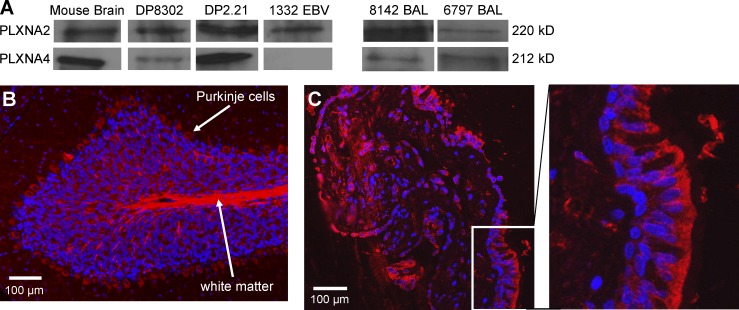
To address whether plexins are present at the site of inflammation in CBD, we used Western blot analysis to assess expression of plexin A2 and A4 proteins in the antigen-presenting cells routinely used in Be-specific T cell hybridoma activation assays. Using mouse brain as a positive control, plexin A2 was present in both HLA-DP2–expressing fibroblasts (DP8302 and DP2.21) and EBV-transformed B cells, whereas plexin A4 was expressed in DP8302 and DP2.21 (Fig. 7 A). Using concentrated BAL fluid from CBD patients, we documented expression of plexin A2 in 6/6 and plexin A4 in 5/6 samples analyzed (Fig. 7 A). Using the same plexin A2 mAb and mouse cerebellum as a positive control, Fig. 7 B shows plexin A2 staining in Purkinje cells and white matter. Staining of a transbronchial lung biopsy from a CBD patient showed plexin A2 expression predominantly within the bronchial epithelium (Fig. 7 C). Similar findings were observed in lung tissue obtained from two other CBD patients (not depicted), and isotype control antibody staining was negative. Thus, we demonstrate that plexin A proteins are readily available as a source of antigen in lung tissue and BAL fluid of CBD patients.
Figure 7.
Distribution of plexin A proteins in antigen-presenting cells, BAL fluid, and lung tissue derived from CBD patients. (A) Western blot analysis of cell extracts from the mouse fibroblast cell lines (DP8302 and DP2.21) and EBV-transformed B cells from CBD patient 1332 and BAL fluid from two representative CBD patients is shown. Anti-plexin antibodies cross-react to mouse and human plexin A2 and A4. BAL fluid samples were concentrated 300-fold and resolved on a 7.5% polyacrylamide gel. Results are representative of a minimum of three independent experiments for cell extracts and BAL fluids. (B and C) Immunofluorescence staining of plexin A2 (red) and cell nuclei (blue) in mouse cerebellum (B) and transbronchial lung biopsy tissue from a CBD patient (C) is shown. An enlarged view of the area within the white box in C showing plexin A2 expression in bronchial epithelial cells is shown in the right panel. Results are representative of the three transbronchial lung biopsy specimens analyzed.
DISCUSSION
Understanding the pathogenesis of CD4+ T cell–mediated diseases requires an elucidation of the ligands responsible for driving T cell activation. In the case of CBD, βGlu69-containing HLA-DP molecules and Be are known requirements for activation of Be-specific T cells. Until now, the identity of the peptides that complete this ligand has remained unknown. Using positional scanning libraries and biometric data analysis, we delineated a spectrum of mimotopes and endogenously derived human peptides that stimulate Be-specific TCRs isolated from the lung of a patient with active CBD. Be-loaded HLA-DP2 tetramers with mimotope-2 and plexin A4 peptides in the binding groove identified antigen-specific CD4+ T cells in the target organ of all HLA-DP2–expressing CBD patients evaluated, confirming the importance of these related ligands in driving recruitment of these pathogenic T cells to the lung.
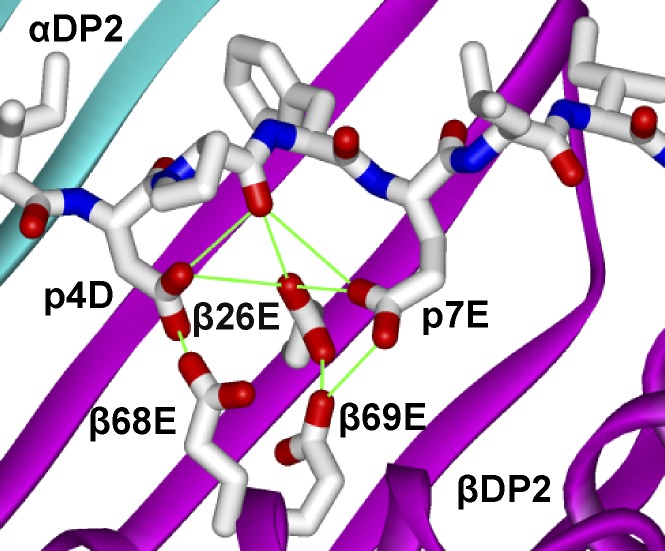
Because of the inability of effector memory CD4+ T cells obtained from a target organ to vigorously undergo repeated rounds of stimulation (Fontenot et al., 2005), delineating antigen specificity of human CD4+ T cells has been difficult. We overcame this obstacle by using the immortalized T cell hybridoma line 54ζ expressing the TCRs of interest and an unbiased positional scanning library. From the deconvolution of the positional scanning library data, important characteristics of Be-dependent mimotopes were identified, including (a) preference for bulky hydrophobic or nonpolar amino acids at the p1 and p6 anchor positions that match the known HLA-DP2–binding motif (Díaz et al., 2005; Sidney et al., 2010), (b) independence of Be-dependent TCR recognition on the C terminus of the peptide, (c) distinct profile of allowed amino acids at the TCR contact sites p2, p3, and p5, and (d) negatively charged aspartic and glutamic acid residues at p4 and p7 that represent potential Be coordination sites. Modeling of mimotope-2 in the peptide-binding groove of the solved HLA-DP2 structure reveals the acidic pocket composed of glutamic acid residues at positions 26, 68, and 69 of the HLA-DP2 β-chain with an additional two acidic amino acids contributed by the peptide in the Be-binding pocket (Fig. 8). The positions of the p4 and p7 amino acids of the peptide strongly suggest their role in capturing Be for T cell recognition. Thus, this acidic cluster provides multiple electron donors for the coordination of a Be moiety within the αβTCR footprint. Our present and previously published data (Bill et al., 2005; Dai et al., 2010) show that mutagenesis of any of these five acidic residues abolishes T cell activation, suggesting that this acidic pocket is the Be-binding site responsible for the induction of an adaptive immune response. In the absence of Be, this solvent-exposed collection of acidic amino acids is likely stabilized by other divalent cations (e.g., magnesium) or a water molecule. In retrospect, our inability to identify a Be-dependent peptide from the list of peptides commonly bound to HLA-DP2 is likely caused by the preference of those peptides to possess a positively charged arginine and lysine at the p4 position (Díaz et al., 2005). As further evidence of the critical contribution of peptide in coordinating Be, the response of the HLA-DP2–restricted dengue virus–specific hybridoma to its ligand (NS3, p254-265) is not affected by the presence of Be (unpublished data). Because the NS3 peptide does not contain acidic amino acids at p4 and p7 (L and histidine [H], respectively), it is thus incapable of coordinating Be, providing an explanation of why Be does not inhibit TCR interaction with the HLA-DP2–dengue viral complex.
Figure 8.
Model of mimotope-2 in the peptide-binding groove of HLA-DP2. The amino acids of mimotope-2 were introduced into the peptide of the HLA-DP2–pDRa structure (Protein Data Bank accession no. 3LQZ) using Swiss PDB Viewer 4.0. Rotamers of the amino acids of mimotope-2 that avoided conflict with the structure were selected. Ribbon representations of DP2 α (cyan)- and DP2 β (magenta)-chains are shown. Wireframe representations of the side chains of mimotope-2, βGlu26, βGlu68, and βGlu69 with CPK coloring are shown.
The identification of Be-dependent mimotopes enabled us to answer several critical questions regarding TCR recognition of this unconventional antigen. Using surface plasmon resonance, the AV22 TCR only interacted with the HLA-DP2–peptide complex if Be was present, and this interaction occurred with high affinity (Kd = 4.6 µM), with the majority of TCR/pMHCII affinity constants (Kd) ranging between 10 and 100 µM (Davis et al., 1998; van der Merwe and Davis, 2003). In addition, we predict that Be binding to HLA-DP2 is nearly irreversible, based on the stability of the complex on the BIAcore chip as well as the stability of the staining intensity of Be-specific T cells for the BeSO4-soaked HLA-DP2–mimotope tetramer over an 8-mo period.
The ability to track antigen-specific T cells using tetramers of MHC molecules has been a significant advance over the last decade (Nepom, 2012). However, the application of this technology to CD4+ T cells has proven difficult, predominantly because of a low frequency of antigen-specific CD4+ T cells that expand in response to antigen (Homann et al., 2001). Consequently, in vitro expansion or magnetic bead capture of antigen-specific CD4+ T cells is typically necessary before staining (Novak et al., 1999; Day et al., 2003; Moon et al., 2009; Wambre et al., 2012). Initially, we used an HLA-DP2–mimotope-2/Be tetramer to stain ex vivo CD4+ T cells from the BAL of CBD patients and detected large numbers of CD4+ T cells specific for this ligand in all DP2-expressing CBD patients tested, with frequencies ranging from 0.1 to 4.8%. In addition, lungs of HLA-DP2 transgenic mice exposed to BeO contain CD4+ T cells that share the same TCR specificity and bind to the DP2-mimotope/Be tetramer (unpublished data), reinforcing the importance of this ligand in the generation of Be-specific adaptive immunity in humans and mice.
Our data also suggest that the frequency of Be-responsive T cells as determined by intracellular cytokine staining underestimates the number of Be-specific T cells in the lung because two thirds of Be-specific, tetramer-binding T cells fail to express IFN-γ and/or IL-2. We have previously shown that some Be-specific CD4+ T cells proliferate poorly after antigen exposure, while maintaining the ability to secrete Th1-type cytokines (Fontenot et al., 2002, 2005). The current study extends those findings by showing that a portion of Be-responsive T cells in BAL have lost their Th1 cytokine–secreting ability, suggesting a terminally differentiated T cell phenotype and consistent with up-regulated programmed death-1 expression on Be-specific CD4+ T cells in CBD (Palmer et al., 2008). Although it is possible that the tetramer-positive cells may be secreting cytokines other than IFN-γ and IL-2, we have never detected IL-4, IL-10, or IL-17 in BAL-derived CD4+ T cells in response to either BeSO4 or staphylococcal enterotoxin B (unpublished data). Overall, addition of HLA-DP2 tetramer staining to Be-induced cytokine staining may allow a more accurate assessment of the frequency of Be-specific T cells in the lung and could potentially be developed into a biomarker of disease development and progression.
To identify endogenous peptides for the Be-specific TCRs, we used the biased W2D4L5 positional scanning library data to rank all overlapping peptides in a human protein database for their stimulatory potential. We focused on plexin A peptides based on screening of a limited number of these peptides and show that plexin A proteins are efficiently processed by HLA-DP2–expressing EBV-transformed B cells and fibroblasts to present the core TCR recognition motif in association with Be to our T cell hybridomas. Furthermore, these proteins are present in lung tissue and BAL fluid from CBD patients and likely are accessible to antigen-presenting cells during Be-induced lung inflammation. Using an HLA-DP2–PLXNA4/Be tetramer to stain ex vivo BAL cells from CBD patients, CD4+ T cells that bind to this complex were present in all subjects. Furthermore, simultaneous staining with mimotope-2/Be and PLXNA4/Be tetramers revealed distinct populations of T cells that bind these tetramers with varying affinities. Although the amino acid sequence of mimotope-2 was dictated based on the preferences of a single TCR, the binding patterns of ex vivo BAL CD4+ T cells reflect a heterogeneous collection of T cells that display a range of affinities for both ligands. Our findings of a distinct population of BAL CD4+ T cells that only bind to the PLXNA4/Be tetramer provide strong support that plexin A proteins are a source of endogenous self-peptides in vivo that are required for T cell recognition of Be.
Although we have identified mimotopes and endogenous plexin A peptides that stimulate Be-specific CD4+ T cells in the presence of HLA-DP2 and Be, our data indicate that there are other Be-responsive T cells that do not bind these tetramers and must recognize other peptides. It is unknown whether the number of additional peptides is large or small. Furthermore, although the source of these stimulating peptides is likely from self-proteins, they may also be derived from exogenous sources such as viruses or bacteria. However, based on our library screen, we predict that the common theme of Be-dependent peptides will be the presence of acidic, Be-coordinating amino acids at p4 and p7, with amino acids at p2, p3, and p5 being dictated by the TCR contact requirements of particular Be-specific T cells.
In addition to the role of conventional MHC-bound peptides in anchoring to MHC and interacting with TCR, our data establish a novel function for peptide in metal hypersensitivity, that of metal ion capture. Importantly, this process can result in the conversion of a self-peptide into a neoantigen. The creation of neoantigens that are absent in the thymus and arise in target organs plays a key role in the genesis of autoimmunity (Marrack and Kappler, 2012). For example, in celiac disease, transglutaminase enzymatically modifies gliadin, resulting in deamidated gliadin peptides that are recognized as nonself by CD4+ T cells and lead to the generation of a T cell–mediated response in genetically susceptible hosts (Molberg et al., 1998). The abacavir hypersensitivity syndrome, which occurs in HLA-B*57:01 patients treated with abacavir, results from binding of the drug to the peptide-binding groove of the MHCI molecule and generation of altered peptides that activate antigen-specific CD8+ T cells (Illing et al., 2012). In the case of CBD, self-peptides bound in the HLA-DP2–binding groove, such as those derived from plexin A and spectrin proteins, may be altered in the presence of Be, resulting in the conversion of endogenous peptides into neoantigens and culminating in Be-specific adaptive immunity. Alternatively, Be binding to HLA-DP2 may partially neutralize the acidity of the p4 pocket and subtly alter the peptide repertoire that binds to this MHCII, contributing to the generation of de novo antigen-specific responses. Although Be is an absolute requirement for T cell recognition, it is unknown whether the CDR loops of the AV22 TCR directly contact a solvent-exposed Be moiety as suggested by our model of the HLA-DP2–mimotope-2/Be complex or recognize an altered self-peptide with no direct contacts between Be and the TCR. Collectively, our findings further blur the distinction between hypersensitivity and autoimmunity, showing how a modified self-peptide can create a neoantigen as the target in these inflammatory diseases.
In conclusion, our findings identify the first complete ligand for a metal-specific TCR and demonstrate the requirement for additional acidic amino acids contributed by the peptide for Be capture and presentation. This additional function of the peptide in metal hypersensitivity provides an explanation for the generation of a human disease in a genetically susceptible host exposed to an environmental antigen.
MATERIALS AND METHODS
Study population and Be-specific T cell lines.
17 patients with CBD were enrolled in this study. CBD was diagnosed using previously defined criteria, including a history of Be exposure, the presence of granulomatous inflammation on lung biopsy, and a positive proliferative response of blood and/or BAL T cells to BeSO4 in vitro (Rossman et al., 1988; Newman et al., 1989). For select subjects, BAL-derived, Be-specific T cell lines were generated as previously described (Fontenot et al., 2000; Bill et al., 2005). HLA-DP typing was performed by ClinImmune Labs. Informed consent was obtained from each patient, and the Human Subject Institutional Review Boards at the University of Colorado Denver and National Jewish Health approved the protocol.
Generation of mouse hybridomas expressing TCRs from human T cell clones.
TCRs expressed by two Be-specific T cell clones derived from the lung of CBD patient 1332 (Bowerman et al., 2011) and a dengue virus–specific T cell clone were introduced into the TCR-negative mouse recipient hybridoma cell line 54ζ, which expresses human CD4 (Boen et al., 2000). In brief, PCR fragments encoding the extracellular variable domains of the TCR α- and β-chains of each T cell clone were introduced into separate expression vectors that encode either a mouse Cα or Cβ domain. The full-length chimeric TCRA and TCRB gene constructs were cotransfected into 54ζ recipient cells by electroporation, and transfectants were selected in 1 mg/ml G418-containing medium (Cellgro; Bowerman et al., 2011). Transfectants were screened for TCR expression using a PE-labeled H57-597 mAb (eBioscience). Subcloning by limiting dilution was performed, and the subclones with the highest TCR expression, as determined by staining with the H57-597 mAb, were chosen for stimulation experiments.
EBV-transformed B cells and fibroblast cell lines.
The EBV-transformed B lymphoblastoid cell line from CBD patient 1332 was generated from PBMCs as previously described (Fontenot et al., 2000). Mouse fibroblasts transfected with genes encoding wild-type HLA-DP2 or DP2 with peptides covalently attached were used as antigen-presenting cells to present Be and peptides. DP8302 expresses the DPA*0103 and DPB*0201 (HLA-DP2) genes (Bill et al., 2005). DP2.21 cells were derived from kidney fibroblasts of IAb−/−, Ii−/− C57BL/6 mice (Huseby et al., 2003) and express DP2 (DPA*0103/DPB1*0201) on their surface. For T cell hybridoma activation experiments, DP2.21 cells were adapted to grow in the protein-free medium OptiPro SFM (Invitrogen) supplemented with 4 mM GlutaMAX (Invitrogen).
Positional scanning libraries, mixtures, peptides, and plexin proteins.
A decapeptide, N-acetylated, C-terminal–amidated, L–amino acid positional scanning library was synthesized. This library consists of 200 mixtures prepared in an OX9 format, where O represents a specific amino acid at a defined position and X represents an equal molar mixture of 20 natural amino acids (except cysteine) in each of the remaining 9 positions. Each OX9 mixture consists of 3.2 × 1011 different decapeptides, and the total number of peptides in the library is 6.4 × 1012. Mixtures containing multiple fixed positions and a biased W2D4L5 positional scanning library were synthesized as previously described (Pinilla et al., 1994; Hemmer et al., 1999).
Individual candidate peptides were initially synthesized using the PEPScreen 96-well array (Sigma-Aldrich). Peptides chosen for further study were synthesized on a larger scale and tested at 95% purity (CPC Scientific). All peptides were first dissolved in DMSO at high concentration before making a working stock in PBS. DNA encoding the cytoplasmic domains of mouse plexins (A1, residues 1269–1894; A2, 1264–1894; A4, 1263–1893; C1, 975–1571) was cloned into a modified pET28 vector (EMD Millipore) and expressed in ArticExpress bacteria (Agilent Technologies). Proteins were purified to >95% with a HisTrap column (GE Healthcare) followed by ion exchange chromatography (1 ml resource Q; GE Healthcare) and resuspended in 10 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, and 2 mM DTT (He et al., 2009; Wang et al., 2012).
T cell hybridoma activation assays.
T cell hybridomas (2.5 × 104–5 cells per well) were cultured in protein-free medium in 96-well flat-bottomed tissue culture plates (Falcon) in the presence of HLA-DP2–transfected mouse fibroblasts (105 cells) and 75 µM BeSO4. Initial experiments involving the decapeptide positional scanning library were performed using mixtures at 200 µg/ml. Subsequent mixtures were tested multiple times in duplicate at concentrations ranging from 20 to 200 µg/ml. Concentrations of crude peptides were 0.5, 5, and 50 µg/ml. For dose–response curves to purified peptides in the presence of BeSO4, peptide was added at concentrations ranging from 0.1 nM to 10 µM. For testing plexin proteins, assays were completed in IMDM/GlutaMAX tissue culture medium (Invitrogen) supplemented with 10% FBS (Hyclone), 1 mM Na pyruvate, 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Invitrogen). 6–400 µg/ml of protein was added to wells in triplicate containing autologous EBV-transformed B cells (2 × 105 cells) or DP8302 fibroblasts (105 cells) in either the absence or presence of 75 µM BeSO4.
After 22–24 h, supernatants were harvested, and mouse IL-2 was measured by ELISA (eBioscience). In some experiments, the concentration of peptide that provided 50% of the maximum IL-2 response (EC50) for each hybridoma was determined using nonlinear regression (sigmoidal-fit; Prism; GraphPad Software).
Scoring matrix and database searches.
A scoring matrix was generated by assigning numerical values to the stimulatory potency of defined amino acids at each position in the W2D4L5 decapeptide positional library. The values were calculated as the logarithm of IL-2 (pg/ml) in the presence of peptide mixtures/Be. For the three defined positions (W2D4L5), a value of 0 was assigned to all the amino acids except for the amino acid that was fixed at that position (i.e., W in position 2, D in position 4, and L in position 5). The value for these amino acids was assigned the maximum stimulatory potency measured among all the mixtures at all positions (E7). The predicted stimulatory potential of a peptide, or score, was calculated by summing the values associated with each amino acid in each position of the peptide. The sum of the maximum values at each position is defined as the maximum matrix score. Using a web-based search tool, the scoring matrix was applied to rank, according to their stimulatory score, all of the overlapping peptides within each protein sequence of a human GenPept (version 156) protein database, as previously described (Hemmer et al., 1999; Zhao et al., 2001; Pinilla et al., 2003).
BIAcore measurement of TCR affinity and kinetics.
Approximately 2,000 resonance units of HLA-DP2–mimotope-2, HLA-DP2–PLXNA4, or an irrelevant MHCII molecule, HLA-DR52c–WIR (Yin et al., 2012), were captured in separate flow cells of a BIAcore streptavidin biosensor chip. Various concentrations of soluble AV22 TCR were injected through the flow cells before and after loading with 200 µM BeSO4, and the surface plasmon resonance signal was recorded during and after the injections. The resonance signal in the control flow cell was subtracted from those in the other flow cells to correct for the fluid phase signal. The kinetics of AV22 binding to HLA-DP2–mimotope-2 and HLA-DP2–PLXNA4 after loading with Be2+ were calculated using BIAeval software (version 4.1).
Preparation of soluble MHCII–covalent peptide tetramers.
Gene fragments encoding the extracellular domains of the α- and β-chains of HLA-DP2 were cloned into separate baculovirus transfer vectors. Sequence encoding mimotope-2 (FWIDLFETIG) or PLXNA4 peptide (FVDDLFETIF) and a flexible linker were inserted between the signal peptide and the N terminus of the mature β-chain to tether the peptide to MHCII. The complete constructions were incorporated into Sapphire baculovirus DNA (Orbigen) to produce high-titer virus stock. Soluble HLA-DP2–mimotope-2 and HLA-DP2–PLXNA4 proteins were purified from supernatants of infected High5 insect cells (Invitrogen) by immunoaffinity chromatography and size-exclusion chromatography using Superdex200 10/300GL (GE Healthcare). Both proteins were biotinylated with the BirA enzyme (Avidity) and incorporated into saturated complexes with PE-streptavidin (Prozyme) or BV421-streptavidin (BioLegend; Crawford et al., 1998), with and without 200 µM BeSO4. Tetramers were separated from monomeric protein by FPLC size-exclusion chromatography. An IAg7-insulin B:9-23 tetramer and a T cell hybridoma specific for this ligand (8-1.1; Crawford et al., 2011) were used as staining controls.
Tetramer and dual intracellular cytokine/tetramer assay.
Hybridoma cells were incubated in 25 µl of culture medium containing the HLA-DP2–mimotope-2, HLA-DP2–mimotope-2/Be, DP2-PLXNA4/Be, or IAg7-insulin tetramer (20 µg/ml) for 2 h at 37°C in a humidified 10% CO2 incubator, with gentle mixing every 20 min. In separate tubes, cells were stained with PE-labeled H57-597 mAb (eBioscience) at 4°C to confirm TCR expression.
For stimulation of cytokine expression, 5–20 × 106 cells were exposed to either medium or 100 µM BeSO4 for 6 h at 37°C, with 10 µg/ml brefeldin A (BD) added after the first hour. For T cell lines, an equal number of autologous EBV-transformed B cells was added to the assay as antigen-presenting cells. Cells were washed and stained with 20 µg/ml PE-labeled DP2–mimotope-2/Be tetramer or IAg7-insulin tetramer. After 2 h, cells were stained for 30 min at 4°C with the following mAbs: anti-CD3–Texas Red, anti–CD4-PerCP-Cy5.5, anti–CD8-v500, anti–CD14-FITC, and anti–CD19-FITC (all from eBioscience). Cells were fixed, permeabilized, and stained with anti–IFN-γ–PE-Cy7 (BD) and anti–IL-2–AF700 (BioLegend) mAb for 30 min. The viable mononuclear cell population was evaluated for fluorescence intensity on an LSR-II flow cytometer (BD). Data were analyzed with FlowJo software (Tree Star). Cells expressing CD8, CD14, and CD19 were excluded from the analysis. T cells positively staining for CD3 and CD4 were gated and analyzed for tetramer binding and cytokine expression.
For tetramer costaining of ex vivo BAL cells from CBD patients, cells were incubated in 25 µl of staining medium containing the HLA-DP2–mimotope-2 (control), HLA-DP2–mimotope-2/Be, or HLA-DP2–PLXNA4/Be tetramer for 2 h at 37°C. Cells were surface stained and analyzed as described above.
Western blot analysis and immunofluorescence microscopy.
Cells from fibroblast lines DP8302 and DP2.21 and EBV-transformed B cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1% Triton X-100, 0.1% SDS glycerol, 1 mM PMSF, 0.5% Na deoxycholate, and protease inhibitor cocktail [Sigma-Aldrich]), incubated for 30 min at 4°C with continuous agitation, and centrifuged at 16,000 g for 20 min at 4°C. Total protein was determined using a Bradford protein assay (Bio-Rad Laboratories). Equivalent amounts of protein (50 µg) were resolved by SDS-PAGE on a 7.5% polyacrylamide gel (Bio-Rad Laboratories) with mouse brain extract (Santa Cruz Biotechnology, Inc.) as a positive control and transferred to polyvinylidene fluoride membranes (EMD Millipore). After blocking with TBS–5% nonfat powdered milk, membranes were incubated with polyclonal rabbit anti–human plexin A2 (H-70; Santa Cruz Biotechnology, Inc.) or a rabbit anti–plexin A4 (C5D1; Cell Signaling Technology) mAb at 4°C overnight. Membranes were incubated with goat anti–rabbit IgG-HRP secondary antibody (Santa Cruz Biotechnology, Inc.), and immunoreactive proteins were visualized by an ECL system (Thermo Fisher Scientific).
For microscopic demonstration of plexin A2 expression in human lung tissue, formalin-fixed, paraffin-embedded transbronchial lung biopsies from three CBD patients and mouse cerebellum (positive control) were deparaffinized, followed by epitope retrieval (heat induced, Tris/EDTA, pH 9.0) and blocking with Fc receptor block (Miltenyi Biotec) in 1% BSA/PBS. Slides were incubated for 3 h with 2 µg/ml of the anti–plexin A2 mAb or rabbit polyclonal IgG antibody (negative control), followed by 2 µg/ml of highly cross-adsorbed Alexa Fluor 555 goat anti–rabbit IgG (Invitrogen) for 1 h. Hoechst 33342 was used to stain the nuclei of cells. Slides were visualized on an Axio microscope (Carl Zeiss).
Concentration of BAL fluid.
Previously frozen (−80°C) BAL fluids from CBD patients were filtered (0.22-µm filter; Corning), and 15-ml aliquots were concentrated in an Ultra-15 Centrifugal Filter Unit (50,000 MWCO; Amicon). Retentates were further concentrated in an Ultra-0.5 Centrifugal Filter Unit (100,000 MWCO; Amicon) to a final volume of 50 µl. The integrity of BAL fluid proteins was assessed by SDS-PAGE, and equal amounts of protein were resolved by Western blot.
Acknowledgments
We thank Tina Gibbins for the synthesis of peptide mixtures.
This work was supported by National Institutes of Health grants HL062410, ES011810, HL102245 (to A.P. Fontenot), and GM088197 (to X. Zhang) and Clinical and Translational Sciences Institute grant UL1 TR000154 from the National Center for Advancing Translational Sciences. The work was also supported by the Multiple Sclerosis Research Institute (grant to C. Pinilla) and the Welch Foundation (grant I-1702 to X. Zhang).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAL
- bronchoalveolar lavage
- Be
- beryllium
- CBD
- chronic beryllium disease
References
- Bill J.R., Mack D.G., Falta M.T., Maier L.A., Sullivan A.K., Joslin F.G., Martin A.K., Freed B.M., Kotzin B.L., Fontenot A.P. 2005. Beryllium presentation to CD4+ T cells is dependent on a single amino acid residue of the MHC class II β-chain. J. Immunol. 175:7029–7037 [DOI] [PubMed] [Google Scholar]
- Boen E., Crownover A.R., McIlhaney M., Korman A.J., Bill J. 2000. Identification of T cell ligands in a library of peptides covalently attached to HLA-DR4. J. Immunol. 165:2040–2047 [DOI] [PubMed] [Google Scholar]
- Bowerman N.A., Falta M.T., Mack D.G., Kappler J.W., Fontenot A.P. 2011. Mutagenesis of beryllium-specific TCRs suggests an unusual binding topology for antigen recognition. J. Immunol. 187:3694–3703 10.4049/jimmunol.1101872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford F., Kozono H., White J., Marrack P., Kappler J. 1998. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 8:675–682 10.1016/S1074-7613(00)80572-5 [DOI] [PubMed] [Google Scholar]
- Crawford F., Stadinski B., Jin N., Michels A., Nakayama M., Pratt P., Marrack P., Eisenbarth G., Kappler J.W. 2011. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc. Natl. Acad. Sci. USA. 108:16729–16734 10.1073/pnas.1113954108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Murphy G.A., Crawford F., Mack D.G., Falta M.T., Marrack P., Kappler J.W., Fontenot A.P. 2010. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc. Natl. Acad. Sci. USA. 107:7425–7430 10.1073/pnas.1001772107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M., Boniface J.J., Reich Z., Lyons D., Hampl J., Arden B., Chien Y. 1998. Ligand recognition by α β T cell receptors. Annu. Rev. Immunol. 16:523–544 10.1146/annurev.immunol.16.1.523 [DOI] [PubMed] [Google Scholar]
- Day C.L., Seth N.P., Lucas M., Appel H., Gauthier L., Lauer G.M., Robbins G.K., Szczepiorkowski Z.M., Casson D.R., Chung R.T., et al. 2003. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest. 112:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz G., Cañas B., Vazquez J., Nombela C., Arroyo J. 2005. Characterization of natural peptide ligands from HLA-DP2: new insights into HLA-DP peptide-binding motifs. Immunogenetics. 56:754–759 10.1007/s00251-004-0735-5 [DOI] [PubMed] [Google Scholar]
- Fontenot A.P., Maier L.A. 2005. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 26:543–549 10.1016/j.it.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Fontenot A.P., Falta M.T., Freed B.M., Newman L.S., Kotzin B.L. 1999. Identification of pathogenic T cells in patients with beryllium-induced lung disease. J. Immunol. 163:1019–1026 [PubMed] [Google Scholar]
- Fontenot A.P., Torres M., Marshall W.H., Newman L.S., Kotzin B.L. 2000. Beryllium presentation to CD4+ T cells underlies disease-susceptibility HLA-DP alleles in chronic beryllium disease. Proc. Natl. Acad. Sci. USA. 97:12717–12722 10.1073/pnas.220430797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot A.P., Canavera S.J., Gharavi L., Newman L.S., Kotzin B.L. 2002. Target organ localization of memory CD4+ T cells in patients with chronic beryllium disease. J. Clin. Invest. 110:1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot A.P., Palmer B.E., Sullivan A.K., Joslin F.G., Wilson C.C., Maier L.A., Newman L.S., Kotzin B.L. 2005. Frequency of beryllium-specific, central memory CD4+ T cells in blood determines proliferative response. J. Clin. Invest. 115:2886–2893 10.1172/JCI24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Yang T., Terman J.R., Zhang X. 2009. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc. Natl. Acad. Sci. USA. 106:15610–15615 10.1073/pnas.0906923106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmer B., Pinilla C., Appel J., Pascal J., Houghten R., Martin R. 1998. The use of soluble synthetic peptide combinatorial libraries to determine antigen recognition of T cells. J. Pept. Res. 52:338–345 10.1111/j.1399-3011.1998.tb00658.x [DOI] [PubMed] [Google Scholar]
- Hemmer B., Gran B., Zhao Y., Marques A., Pascal J., Tzou A., Kondo T., Cortese I., Bielekova B., Straus S.E., et al. 1999. Identification of candidate T-cell epitopes and molecular mimics in chronic Lyme disease. Nat. Med. 5:1375–1382 10.1038/70946 [DOI] [PubMed] [Google Scholar]
- Homann D., Teyton L., Oldstone M.B. 2001. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat. Med. 7:913–919 10.1038/90950 [DOI] [PubMed] [Google Scholar]
- Huseby E.S., Crawford F., White J., Kappler J., Marrack P. 2003. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc. Natl. Acad. Sci. USA. 100:11565–11570 10.1073/pnas.1934636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby E.S., White J., Crawford F., Vass T., Becker D., Pinilla C., Marrack P., Kappler J.W. 2005. How the T cell repertoire becomes peptide and MHC specific. Cell. 122:247–260 10.1016/j.cell.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Illing P.T., Vivian J.P., Dudek N.L., Kostenko L., Chen Z., Bharadwaj M., Miles J.J., Kjer-Nielsen L., Gras S., Williamson N.A., et al. 2012. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 486:554–558 [DOI] [PubMed] [Google Scholar]
- Kreiss K., Mroz M.M., Zhen B., Martyny J.W., Newman L.S. 1993a. Epidemiology of beryllium sensitization and disease in nuclear workers. Am. Rev. Respir. Dis. 148:985–991 10.1164/ajrccm/148.4_Pt_1.985 [DOI] [PubMed] [Google Scholar]
- Kreiss K., Wasserman S., Mroz M.M., Newman L.S. 1993b. Beryllium disease screening in the ceramics industry. Blood lymphocyte test performance and exposure-disease relations. J. Occup. Med. 35:267–274 [PubMed] [Google Scholar]
- Kreiss K., Mroz M.M., Newman L.S., Martyny J., Zhen B. 1996. Machining risk of beryllium disease and sensitization with median exposures below 2 µg/m3. Am. J. Ind. Med. 30:16–25 [DOI] [PubMed] [Google Scholar]
- Lombardi G., Germain C., Uren J., Fiorillo M.T., du Bois R.M., Jones-Williams W., Saltini C., Sorrentino R., Lechler R. 2001. HLA-DP allele-specific T cell responses to beryllium account for DP-associated susceptibility to chronic beryllium disease. J. Immunol. 166:3549–3555 [DOI] [PubMed] [Google Scholar]
- Marrack P., Kappler J.W. 2012. Do MHCII-presented neoantigens drive type 1 diabetes and other autoimmune diseases? Cold Spring Harb. Perspect. Med. 2:a007765 10.1101/cshperspect.a007765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molberg O., Mcadam S.N., Körner R., Quarsten H., Kristiansen C., Madsen L., Fugger L., Scott H., Norén O., Roepstorff P., et al. 1998. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4:713–717 10.1038/nm0698-713 [DOI] [PubMed] [Google Scholar]
- Moon J.J., Chu H.H., Hataye J., Pagán A.J., Pepper M., McLachlan J.B., Zell T., Jenkins M.K. 2009. Tracking epitope-specific T cells. Nat. Protoc. 4:565–581 10.1038/nprot.2009.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz M.M., Kreiss K., Lezotte D.C., Campbell P.A., Newman L.S. 1991. Reexamination of the blood lymphocyte transformation test in the diagnosis of chronic beryllium disease. J. Allergy Clin. Immunol. 88:54–60 10.1016/0091-6749(91)90300-D [DOI] [PubMed] [Google Scholar]
- Nepom G.T. 2012. MHC class II tetramers. J. Immunol. 188:2477–2482 10.4049/jimmunol.1102398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L.S., Kreiss K., King T.E., Jr, Seay S., Campbell P.A. 1989. Pathologic and immunologic alterations in early stages of beryllium disease. Re-examination of disease definition and natural history. Am. Rev. Respir. Dis. 139:1479–1486 10.1164/ajrccm/139.6.1479 [DOI] [PubMed] [Google Scholar]
- Novak E.J., Liu A.W., Nepom G.T., Kwok W.W. 1999. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J. Clin. Invest. 104:R63–R67 10.1172/JCI8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto Y., Kurane I., Leporati A.M., Ennis F.A. 1998. Definition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4+ T cell clones. J. Gen. Virol. 79:697–704 [DOI] [PubMed] [Google Scholar]
- Palmer B.E., Mack D.G., Martin A.K., Gillespie M., Mroz M.M., Maier L.A., Fontenot A.P. 2008. Up-regulation of programmed death-1 expression on beryllium-specific CD4+ T cells in chronic beryllium disease. J. Immunol. 180:2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla C., Appel J.R., Blanc P., Houghten R.A. 1992. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 13:901–905 [PubMed] [Google Scholar]
- Pinilla C., Appel J.R., Houghten R.A. 1994. Investigation of antigen-antibody interactions using a soluble, non-support-bound synthetic decapeptide library composed of four trillion (4 x 1012) sequences. Biochem. J. 301:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla C., Appel J.R., Borràs E., Houghten R.A. 2003. Advances in the use of synthetic combinatorial chemistry: mixture-based libraries. Nat. Med. 9:118–122 10.1038/nm0103-118 [DOI] [PubMed] [Google Scholar]
- Richeldi L., Sorrentino R., Saltini C. 1993. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 262:242–244 10.1126/science.8105536 [DOI] [PubMed] [Google Scholar]
- Rossman M.D., Kern J.A., Elias J.A., Cullen M.R., Epstein P.E., Preuss O.P., Markham T.N., Daniele R.P. 1988. Proliferative response of bronchoalveolar lymphocytes to beryllium. A test for chronic beryllium disease. Ann. Intern. Med. 108:687–693 10.7326/0003-4819-108-5-687 [DOI] [PubMed] [Google Scholar]
- Sidney J., Steen A., Moore C., Ngo S., Chung J., Peters B., Sette A. 2010. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J. Immunol. 184:2492–2503 10.4049/jimmunol.0903655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkle S.S., Kittle L.A., Schumacher B.A., Newman L.S. 1997. Beryllium induces IL-2 and IFN-γ in berylliosis. J. Immunol. 158:518–526 [PubMed] [Google Scholar]
- van der Merwe P.A., Davis S.J. 2003. Molecular interactions mediating T cell antigen recognition. Annu. Rev. Immunol. 21:659–684 10.1146/annurev.immunol.21.120601.141036 [DOI] [PubMed] [Google Scholar]
- Wambre E., DeLong J.H., James E.A., LaFond R.E., Robinson D., Kwok W.W. 2012. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J. Allergy Clin. Immunol. 129:544–551 10.1016/j.jaci.2011.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., He H., Srivastava N., Vikarunnessa S., Chen Y.B., Jiang J., Cowan C.W., Zhang X. 2012. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal. 5:ra6 10.1126/scisignal.2002636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Crawford F., Marrack P., Kappler J.W., Dai S. 2012. T-cell receptor (TCR) interaction with peptides that mimic nickel offers insight into nickel contact allergy. Proc. Natl. Acad. Sci. USA. 109:18517–18522 10.1073/pnas.1215928109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Gran B., Pinilla C., Markovic-Plese S., Hemmer B., Tzou A., Whitney L.W., Biddison W.E., Martin R., Simon R. 2001. Combinatorial peptide libraries and biometric score matrices permit the quantitative analysis of specific and degenerate interactions between clonotypic TCR and MHC peptide ligands. J. Immunol. 167:2130–2141 [DOI] [PubMed] [Google Scholar]