Abstract
Flexible polymer linkers play an important role in various imaging and probing techniques that require surface immobilization, including atomic force microscopy (AFM). In AFM force spectroscopy, polymer linkers are necessary for the covalent attachment of molecules of interest to the AFM tip and the surface. The polymer linkers tether the molecules and provide their proper orientation in probing experiments. Additionally, the linkers separate specific interactions from nonspecific short-range adhesion and serve as a reference point for the quantitative analysis of single molecule probing events. In this report, we present our results on the synthesis and testing of a novel polymer linker and the identification of a number of potential applications for its use in AFM force spectroscopy experiments. The synthesis of the linker is based on the well-developed phosphoramidate (PA) chemistry that allows the routine synthesis of linkers with predetermined lengths and PA composition. These linkers are homogeneous in length and can be terminated with various functional groups. PA linkers with different functional groups were synthesized and tested in experimental systems utilizing different immobilization chemistries. We probed interactions between complementary DNA oligonucleotides; DNA and protein complexes formed by the site-specific binding protein SfiI; and interactions between amyloid peptide (Aβ42). The results of the AFM force spectroscopy experiments validated the feasibility of the proposed approach for the linker design and synthesis. Furthermore, the properties of the tether (length, functional groups) can be adjusted to meet the specific requirements for different force spectroscopy experiments and system characteristics, suggesting that it could be used for a large number of various applications.
1. Introduction
Single molecule AFM force spectroscopy (SMFS) is widely used to study interactions between individual molecules at the single molecule level. AFM force spectroscopy relies heavily on surface chemistry; molecules of interest can be covalently attached onto AFM probes and probed surfaces to ensure that their attachment is stronger than the force of interaction between these molecules. Therefore, the surface modification of AFM tips and probed surfaces needs to be simple, reliable and effective. When measuring interactions, the molecules immobilized on the AFM tip are approached to the molecules on the surface, allowing them to interact. To facilitate their relative proper orientation, interacting molecules are tethered via long, flexible linkers. The long, flexible tether allows the separation of specific rupture events from nonspecific shortrange adhesion that typically appears in SMFS experiments [1]. The well-defined lengths of the linkers facilitate the identification of the specific interactions on the force-distance curves.
The most common linker used for single molecule force spectroscopy is polyethylene glycol (PEG), which is available from a number of vendors. The functionalized PEG linkers are terminated with reactive groups on both ends, enabling specific coupling of a molecule to the surface. One disadvantage of a PEG linker is that it is a polyether compound of a mixture of different oligomer sizes in either broadly or narrowly defined molecular weight ranges. The molecular weight provided by a vendor is typically the average molecular weight of the mixture [2, 3]. This heterogeneity of PEG linkers results in complications in the quantitative analysis of DFS data in which multiple probing in different surface locations is typically performed [3].
In this paper, we created and tested a novel phosphoramidite (PA) linker as a convenient tether for AFM single molecule force spectroscopy experiments. This linker is synthesized by traditional phosphoramidite chemistry using automated DNA synthesizers [4, 5]. Using this approach we were able to synthesize fully homogeneously sized linkers with a predefined length as large as 72 monomer ethylene glycol units. The PA tethers were terminated with different functional groups enabling us to perform SFMS in different molecular systems.
2. Experimental design
2.1. Materials
All reagents used for the synthesis of PA linkers were purchased from Glen Research (Sterling, VA). Tris (2-carboxyethyl) phosphine (TCEP) hydrochloride was purchased from Hampton Research. All other reagents, including: dithiothreitol (DTT), diethylamine, and acetonitrile, were analytical grade from Sigma-Aldrich. Deionized water (18.2 MΩ, 0.22 µm pore size filter, APS Water Services Corp., Van Nuys, CA) was used for all experiments. SfiI was purchased from New England Biolabs (low BSA content).
2.2 Surface modification with maleimide silatrane (MAS)
This section outlines details of the surface modification technique utilizing the maleimide silatrane (MAS) methodology. The maleimide functionalization of both the mica substrate and the AFM silicon nitride tips using maleimide silatrane (MAS) has been developed previously [6], and it has been shown to provide reliable results in force spectroscopy experiments [2, 6–9]. The maleimide activated tips and the mica surfaces were used immediately for further modification to avoid unnecessary hydrolysis of the maleimide groups.
2.2.1. Tip modification
Silicon nitride (Si3N4) AFM tips (MSNL10, Veeco) were initially cleaned with ethanol for 30 min, followed by a water rinse and drying with argon flow. The tips were exposed to UV light (CL-1000 Ultraviolet Crosslinker, UVP, Upland, CA) for 30 min. After the UV treatment, the AFM tips were immersed in an aqueous solution of 167 µM maleimide silatrane (MAS) for 3 hrs followed by multiple rinses in water.
2.2.2 Mica surface modification
Mica sheets (Asheville Schoonmaker Mica Co.) were cut into 1.5 cm×1.5 cm squares and glued onto a piece of a glass slide using epoxy glue, EPO-TEK 353ND (Epoxy Technology, Inc., Billerica, MA), and cleaved. Freshly cleaved mica surfaces were treated with an aqueous solution of 167 µM maleimide silatrane (MAS) for 3 hrs. MAS modified mica squares were rinsed with water several times to remove non-bound MAS molecules.
2.3. Synthesis of PA linkers
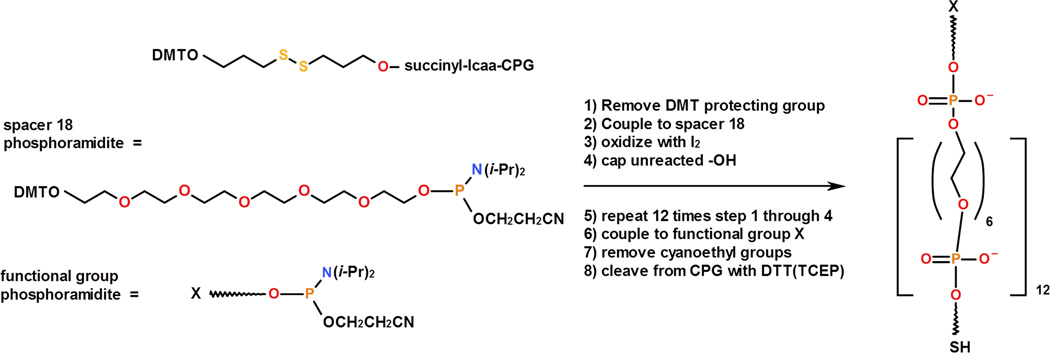
The synthetic strategy of the proposed novel PA tethers is based on the use of non-nucleoside phosphoramidite (PA) spacers. These spacers have been commonly used to place a linker arm into a defined site within synthetic oligonucleotides [4, 5]. A common procedure for PA linker synthesis is described below (Figure 1; detailed schematics are provided in Supporting information-Figure S1); however, the protocol may be modified according to experimental needs.
Figure 1.
Schematic representation of PA linker synthesis. Starting reagents are shown on the left. DMTO = dimethoxytrityl, lcaa = long chain alkyl amine, CPG = controlled porous glass support. The structure and chemical composition of the PA linker are shown on the right: the linker contains 12 units of PA spacer 18, terminal thiol (SH), and a terminal functional group (X).
The “spacer 18 phosphoramidite” (18-O-Dimethoxytritylhexaethyleneglycol,1-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite, 10–1918, Glen Research), which has six ethylene glycol units (CH2CH2O)6, was used as a monomer unit. One end of the spacer is protected with a 4, 4'-dimethoxytrityl (DMT) group and the other end is terminated with 2-cyanoethyl phosphoramidite, which is required for phosphoramidite polymerization chemistry. The synthesis process begins with the DMT-protected disulfide (3’-Thiol-Modifier C3 S-S CPG, 20–2933, Glen Research) immobilized on a controlled porous glass (CPG) support. Next, the DMT protective group is removed to form a free -OH group (step 1) which is then coupled to the phosphoramidite of “spacer 18” (step 2). Oxidation in step 3 produces a stable phosphate linkage. Capping step 4 protects the unreacted hydroxyl groups from being involved in further reactions. Steps 1–4 were repeated twelve times to yield a molecule containing 72 ethylene glycol units, followed by DMT deprotection. The resulting terminal -OH group is coupled to the phosphoramidite that contains a terminal functional group, X.
Although shorter molecules are present in addition to the desired length linker, the fraction of these molecules is small due to the high (95–99%) coupling efficiency at each step. Also, step 4 (capping) prevents these short molecules from having the terminal X functional group required for coupling to a target molecule. Therefore, these shorter molecules do not need to be removed since they cannot function as a linker. The product of the synthesis, CPG-SS-PA-X, remains bound to the CPG beads.
According to Figure 1, the synthesized linker with 72 ethylene glycol units contains 13 phosphate groups along its length in addition to the short arms connecting the terminal functional groups. The phosphate groups of the PA linker were left protected with cyanoethyl groups that were deprotected during a later step. The estimated length of this linker (~25 nm) is similar to the average length of a commercial PEG3400 tether, which we also used in this study as a control.
2.4. Immobilization of thiol-PA linkers on the maleimide-activated surface
“Thiol-Modifier C3 S-S CPG” is used in the synthesis of a PA linker terminated with thiols (thiol-PA). The disulfide bond of the modifier is cleaved by a reducing agent (DTT or TCEP) prior to immobilization onto the maleimide-activated surface. This is done during the deprotection step by adding a reducing agent to a concentrated ammonium hydroxide solution (see section 2.5.3). Alternatively, S-S cleavage can be performed separately if a gentler deprotection step is required (see sections 2.5.1, 2.5.2). The reduction of the disulfide bond results in the release of the –SH containing linker from the CPG support. The immobilization of thiol containing linkers to maleimide activated surfaces covalently attaches: 1) thiol-PA-X (X=functional group) followed by coupling of target molecules to active group X or 2) thiol-PA-molecule conjugate where molecules are coupled to the PA linker directly on CPG prior to S-S cleavage. The thiol-PA linker was immobilized on the AFM tip or MAS functionalized mica as described above. The reaction of thiol was performed in a buffer solution with neutral pH values in the range of 7.0–7.4 at room temperature for 1 hr or at 4°C overnight. This modification step was followed by blocking unreacted maleimide groups with 10 mM buffered β-mercaptoethanol for 10 min.
2.5. Reactive groups of the PA linker and coupling reactions
Table 1 shows the functional groups we have tested in the experiments with the PA linker described in section 3. These functional groups (X) were added during synthesis of the PA linker as 5’ modifiers: 1) N-hydroxysuccinimide (NHS) (5'-Carboxy-Modifier C10, 10–1935, Glen Research) = Linker-1; 2) amine modification (10–1916, Glen Research) = Linker-2; 3) trytil protected thiol group (5’-Thiol-modifier C6, 10-1926, Glen Research) = Linker-3; 4) Linker-4 containing 12 units of “spacer 18” and 20 nucleotides (5’-CCG TTG CGA TGT CAG TGG TA-3’). The same phosphoramidite chemistry was used in polymerization of both the linker and the oligonucleotide to couple the oligonucleotide to the PA linker. This was accomplished in one pot by consecutively adding “spacer 18” 12 times to the synthesized 20-mer oligonucleotide.
Table 1.
Functional groups of heterobifunctional PA linkers.
| Linker-1 | Linker-2 | Linker-3 | Linker-4 | |
|---|---|---|---|---|
| X | -NHS | -NH2 | -S-Trytil | -oligonucleotide |
| Y | -SH | -SH | -SH | -SH |
2.5.1. PA linker with N-hydroxysuccinimide (-NHS)
This functionalized PA-linker was used for immobilization of amine-terminated DNA oligonucleotides. The DNA oligonucleotides (20 mers; ssDNA1 = 5’-/5AmMC6/CCG TTG CGA TGT CAG TGG TA-3’ and ssDNA2 = 5’-/5AmMC6/TAC CAC TGA CAT CGC AAC GG-3’) with amine modification (5AmMC6) at the 5’ end were purchased from Integrated DNA Technologies, Inc. The following protocol was used for the immobilization. First, cyanoethyl protective groups were removed from the phosphates using 10 % diethylamine in acetonitrile. Then, the CPG-SS-PA-NHS linker was reacted with excess Am-DNA oligonucleotides in DMSO containing 1% N,N-diisopropylethylamine (DIPEA) overnight. The modified CPG support was thoroughly washed with DMSO to remove the unreacted oligonucleotides. The use of DMSO is necessary as rapid hydrolysis of –NHS groups occurs in aqueous environments. The amine containing DNA molecules were coupled to the immobilized PA linker in DMSO prior to cleavage of the PA linker from the CPG support. The resulting PA-DNA construct was cleaved from the CPG support with 100 mM DTT pH 8.5 potassium phosphate buffer for 30 min. DTT was extracted from the SH-PA-DNA solution using 500 µL ethylacetate, 3 times. The SH-PA-DNA construct was immobilized onto the maleimide activated surface for force spectroscopy experiments.
2.5.2 PA linker with primary amine (-NH2)
The CPG-SS-PA-NH2 (Linker-2) was synthesized and its potential application was tested in combination with the Cys-Aβ42 peptide (corresponding experiments are described in section 3.2). This peptide is a variant of the Aβ42 peptide. Its only cysteine residue was specifically introduced at the N-terminus (CDAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA) (synthesized in the lab of D. Teplow, UCLA) resulting in a single thiol group suitable for modification. The following procedure was used to convert the primary amine into maleimide in order to couple the Aβ42 peptide via the –SH group of the cysteine. First, we removed the cyanoethyl protective groups from the phosphates using 10 % diethylamine in acetonitrile. The CPG-SS-PA-NH2 was then reacted with 50-fold excess N-(γ-maleimidobutyryloxy)succinimide ester (GMBS) in a DMSO solution containing 1% DIPEA. This treatment converted the amines of the linker into maleimide groups. Again, the use of organic solvents is preferable due to the presence of hydrolysable succinimidyl ester groups. After this step, a CPG-attached maleimide functionalized linker was reacted with Cys-Aβ42 in pH 7.4 HEPES buffer (50 mM HEPES, 100 mM NaCl) for 1 hr followed by washing with buffer. The PA-peptide construct was cleaved off the CPG support with 100 mM tris (2-carboxyethyl) phosphine hydrochloride (TCEP), releasing the HS-PA-peptide construct, which was used in surface modification for force spectroscopy experiments.
2.5.3 PA linker with trytil protected thiol group (-S-Trytil)
The PA-S-Trytil linker (Linker-3) was tested using a DNA-protein complex, SfiI-DNA, previously studied using AFM force spectroscopy [8]. A thiol-maleimide coupling reaction on both sides of the linker was used in the process. One end of the linker contains thiol in a disulfide form that is cleaved by a reducing agent such as DTT or TCEP. The second end contains a trytil-protected thiol that remains protected after treatment with DTT or TCEP. The S-Trytil is deprotected by the use of silver nitrate. This strategy allows the two thiol-maleimide reactions to be decoupled. The following procedure outlines the steps for the sample preparation. Linker-3 was deprotected and cleaved off of the CPG at 55°C using a concentrated ammonium hydroxide solution containing 50 mM DTT. This step cleaved the disulfide bond and removed all of the protective groups from the linker except for the trytil protection on the 5’ thiol. Next, the linker was purified on a Sephadex G-25 column (7 inches high and 0.25 inches in diameter). The PA-linker was eluted in ~22 min with a flow rate of 0.1 mL/min and was monitored by the diode array detector (SPD-M10A, Shimadzu) at 230 nm. DTT eluted in ~50 min under these conditions. The concentration of the linker in the collected fraction was determined using 5,5’-Dithiobis(2-nitrobenzoic acid) (Ellman’s reagent; [10]) immediately after purification. The synthesized and purified Trytil-S-PA-SH linker was immobilized on a maleimide modified surface using a 100 µM solution of the linker in 0.1 M HEPES buffer (pH 7.0) at 4°C overnight. The unreacted surface maleimide groups were blocked with 10 mM β-mercaptoethanol in 0.1 M HEPES buffer (pH 7.0) for 30 min. Next, the trytil protection was removed by treatment with silver nitrate (1 volume of 0.1 M TEAA buffer and 0.15 volumes of 1 M silver nitrate) for 30 min. Excess silver nitrate was removed using the DTT solution (1 volume 0.1 M TEAA buffer and 0.2 volumes of 1 M DTT) for 5 min followed by rinsing with copious amounts of water. The free thiol groups formed during the previous steps were reacted with 1,8-bismaleimidodiethyleneglycol (BM(PEO)2, Pierce) in 0.1 M HEPES buffer (pH 7.0) for 1 hr to yield the surface bound maleimide modified PA linker. The maleimide activated surface was reacted with the 50 µM thiol modified DNA (containing a SfiI recognition sequence) in HEPES buffer containing 0.1 M NaCl. Finally, the unreacted maleimide groups were blocked with 10 mM β-mercaptoethanol in 0.1 M HEPES buffer (pH 7.0) for 30 min, followed by washing with HEPES buffer containing 0.1 M NaCl.
2.6. Sample preparation for experiments utilizing PEG3400 as a linker
The pulling experiments were performed using a bifunctional PEG (MW=3400) linker terminated with maleimide and NHS groups (MAL-PEG-SVA; Laysan Bio, Inc., Arab, AL, USA). Freshly cleaved mica surfaces were modified with a 167 µM 1-(3-aminopropyl) silatrane (APS) aqueous solution for 30 min, washed with water and dried under argon flow. Next, the surface was reacted with 167 µM MAL-PEG-SVA in DMSO for 3 hrs and washed with DMSO followed by a water rinse. The PEG modified surface was reacted with either 100 nM SH-DNA1 or 40 nM HS-PA-Aβ42 conjugate for 1 hr, followed by treatment with 10 mM β-mercaptoethanol for 10 min.
2.9. Force measurements
The single molecule force spectroscopy measurements were performed with a Molecular Force Probe 3D AFM system (MFP-3D, Asylum Research) and a Force Robot 300 (FR-300, JPK, Germany). The spring constants of each tip were calibrated by the thermal noise analysis method. The spring constants of the cantilevers were ~0.03 N/m. A low trigger force (100 pN) was applied on the experimental system. The approach velocity was fixed at 500 nm/s. All measurements were performed at room temperature in an appropriate buffer solution: 1) DNA-DNA interactions were measured in 10 mM HEPES, 50 mM NaCl, pH 7.0; 2) interactions between Aβ42 peptides were measured in pH 7.4 HEPES buffer (50 mM HEPES, 100 mM NaCl); 3) DNA-SfiI interactions were measured in 10 mM HEPES, 50 mM NaCl, 2 mM CaCl2, pH 7.0.
2.10. Data analysis
The rupture event analysis in the force distance curves was conducted using Igor Pro software for experiments performed on the MFP-3D, or JPK Data processing software for experiments performed on the FR-300. Rupture events were fitted with the worm like chain (WLC) polymer model [6, 9]. Two parameters were determined from the fit of the force-distance curves: 1) contour length, LC and 2) persistence length, Lp.
3. Single molecule AFM force spectroscopy experiments with the use of PA linkers
3.1. DNA-DNA interactions
3.1.1 DNA-DNA interactions using a PA linker in a single tether approach
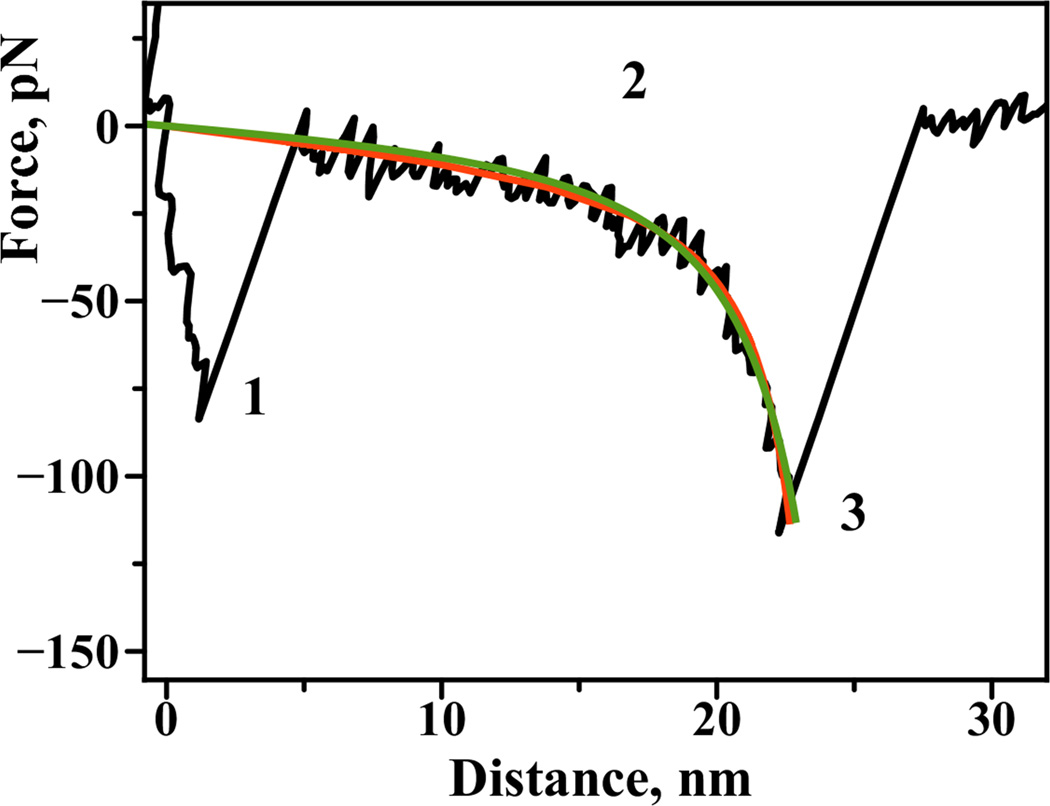
We first probed interactions between the two complementary 20 base long single strands of DNA tethered via PA-linkers. Figure 3A schematically shows our experimental approach. Complementary single stranded oligonucleotides were immobilized on the substrate (ssDNA1) and on the tip (ssDNA2) by either thiol or amine groups placed at the 5’ ends of the oligos. The sequence of these oligonucleotides does not contain self-complementary regions and thus favors the formation of a fully hybridized duplex [11]. The duplex melting curves showed no unexpected intramolecular secondary structures (Figure S2 in Supporting information). Upon approach of the tip to the surface (Figure 3, A), the DNA is hybridized to form a duplex. The tip retraction pulls the opposite 5’ end, leading to the rupture of the duplex resulting in a specific rupture signature on the force curve. Figure 2 shows a typical force-distance curve. An adhesive peak at the beginning of the force curve (section 1 of the force curve) is accompanied by a force extension curve (section 2). This section corresponds to the stretching of the polymer linkers followed by a rupture event (3). The details of the analysis of the specific rupture peaks have been published elsewhere [8, 9]. Importantly, the rupture peak appears at a certain distance along the force curve corresponding to the extension of all of the linkers in the system. We have fitted this part of the force curves with the worm like chain polymer model (WLC), thereby determining the contour length value of the linker [6, 9]. For our experimental systems WLC model consistently provided a good quality fit to the measured force-distance curves similar to the one obtained with Freely Jointed Chain model (see Fig. 2).
Figure 3.
A) Schematic representation of the experimental design for force spectroscopy probing of DNA-DNA interactions in a single tether approach. The expected contour length value is 35 nm. Superposition of the force distance curves for the experiments with the single tether approach B) PA linker and D) PEG3400 linker. Corresponding contour length distributions C) PA linker (HWHM = 1.5 nm) and E) PEG3400 linker (HWHM = 3.5 nm).
Figure 2.
A typical force–distance curve obtained for the interactions between complementary DNA strands in a single tether approach: 1) Short-range adhesion peak due to nonspecific interactions between the surface and the tip of the probe, 2) this region is indicative of polymer linker stretching, 3) specific rupture peak. Region 2 of the curve is fitted with a) worm-like chain polymer model (WLC, continuous green line) and b) freely-jointed chain polymer model for comparison (FJC, continuous red line). Both models provide a good fit with the following residual RMS values: 6.9 pN for WLC model and 6.7 pN for FJC model.
For the single tether studies (Figure 3, A), only the oligonucleotide on the surface was coupled via the PA linker while the complementary strand was immobilized onto the tip via a shorter MAS linker. Figure 3B shows a superposition of the force distance curves collected for multiple probing experiments with the PA linker. The curves overlay well, suggesting that the interactions detected in the multiple approach-retraction cycle are reproducible. We interpret the peaks appearing at 20–30 nm to be the cooperative unbinding of the DNA duplex. This single step duplex rupturing with no plateaus was expected for pulling DNA in a shearing geometry [12].
The contour length measurements set is shown as a histogram in Figure 3C. The maximum of the Gaussian fit (a continuous line) was 26 ± 1 nm. This contour length value is shorter than expected for a fully extended linker (Figure 3A suggests that the expected value is 35 nm, taking into account the contribution of the 20 bp DNA duplex that would contribute 6.8 nm (0.34 nm per bp)). It has been previously reported [13, 14] that a water-mediated suprastructure of PEG based linkers shortens the length of the polymer by ~80% in the range of the rupture forces we observed. Therefore, the lower than expected LC values may be a result of incompletely stretched linkers in our experiments.
3.1.2 DNA-DNA interactions using a PEG linker in the single tether approach
The pulling experiments for the same DNA oligonucleotides were performed with the use of a PEG linker instead of a PA linker. Figure 3D shows the superposition of the force distance curves collected in the experiment with the single tether PEG approach. Qualitatively both approaches were similar, but there were quantitative differences. The contour length distribution for the PEG linker is shown in Figure 3E with the maximum of the Gaussian fit being 30 ± 1 nm. Notably, the LC histogram for the PA experiment under these conditions is narrower with the width (HWHM) being 1.5 nm in contrast to the PEG experiment wherein the HWHM is 3.5 nm. Thus, the use of the PEG3400 linker resulted in a broader LC histogram than the histogram for the PA tether. The effect can be explained by the heterogeneity of the PEG linker, resulting in a broad distribution of contour lengths.
The stiffness of the linker can be estimated from the persistence length values measured from the WLC analysis. Figure S4 shows the results of a WLC analysis for both linkers, demonstrating that the persistence length values are very close to each other, 0.27 nm (PA) vs. 0.28 nm (PEG). The PA linker is primarily composed of ethylene glycol units with phosphate groups evenly spaced along the length of the linker. This design does not change inertness of the linker towards biomolecules – a property that resulted in a wide use of PEG. The phosphate groups of the PA linker are negatively charged at neutral pH values and the electrostatic repulsion between the charges can increase polymer flexibility [15]. The analysis performed in the supplement shows that the contribution of the phosphate charges is not high, supporting the observation of the small difference in the persistence lengths obtained from the WLC analysis.
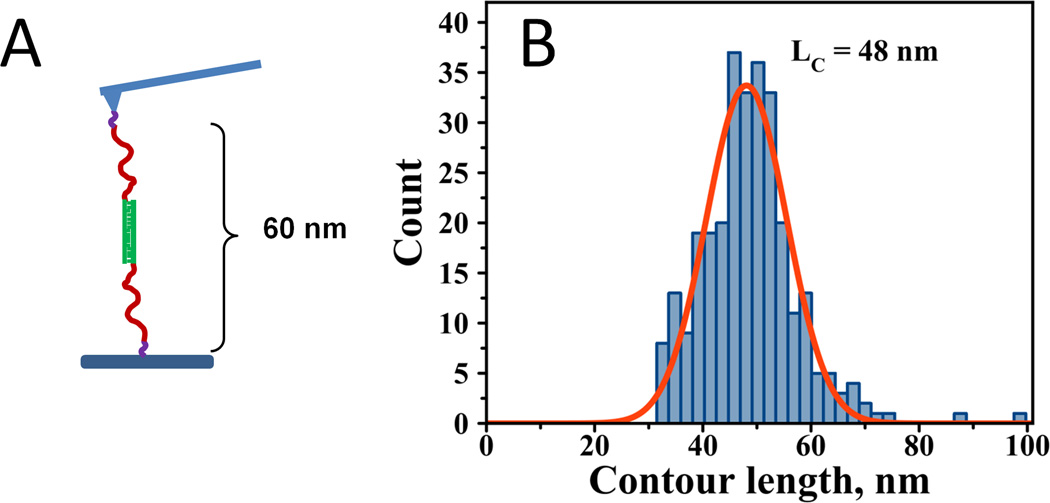
3.1.3 DNA-DNA interactions using a PA linker in the double tether approach
We have performed experiments in a double tether approach in which both oligonucleotides on the tip and the surface were coupled via a PA linker. Figure 4A shows the experimental design of this analysis and the expected contour lengths for fully stretched linkers. The histogram of the contour length values in Figure 4B shows that the maximum of the distribution is 48 ± 2 nm. It is evident that the rupture peaks have shifted to longer distances as expected for specific interactions after the addition of extra linker to the system. The maximum contour length distribution is 22 nm longer for the double tether approach than the single tether approach. In each type of experiment, the contour length is the combination of the PA contour length and the length of MAS linker. The data for the single tether and double tether experiments allowed us to determine the contribution of the PA linker itself to the total value of the contour length. The difference in the mean contour length values between the double tether and single tether approaches is 22 ± 3 nm, corresponding to the contour length of the PA linker under these experimental conditions.
Figure 4.
A) Schematic representation of the experimental design for force spectroscopy probing of DNA-DNA interactions in a double PA tether approach. The expected contour length is 60 nm. B) Contour length distribution; LC = 48 ± 1 nm with HWHM = 8.7 nm.
3.1.4 DNA-DNA interactions using the PA-DNA construct
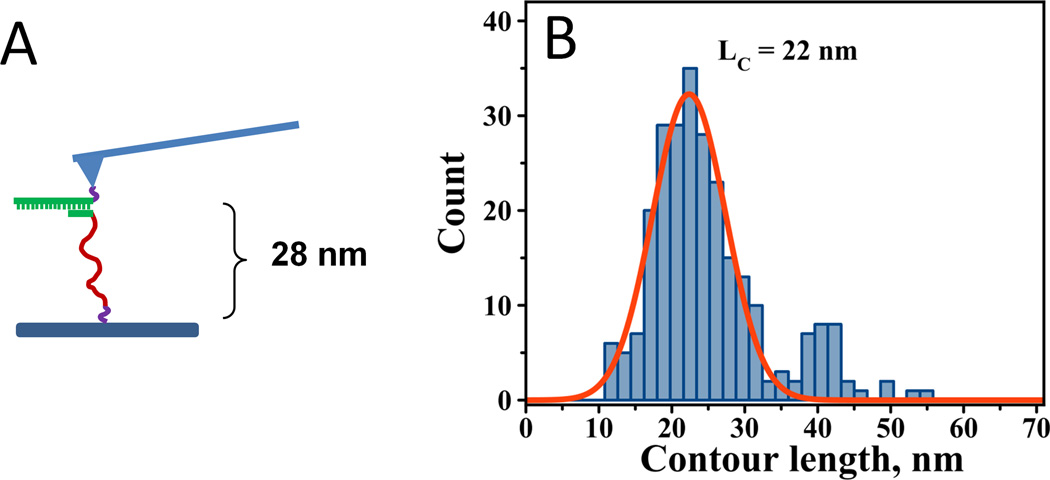
As mentioned previously, in experiments involving DNA, the PA linker can be incorporated into the DNA, allowing the synthesis of both the DNA and the PA linker as one polymer. We tested this approach by synthesizing a 20-mer-PA linker hybrid (Linker-4). This construct was immobilized on the AFM tip and a 5-nucleotide complement was immobilized on the MAS-mica surface (Figure 5A). The interactions were probed in the multiple approach-retraction cycle resulting in force distance curves similar to the ones shown in Figure 3B. Contour length analysis by curve fitting with WLC revealed a narrow distribution of LC values (Figure 5B). Fitting the distribution with one Gaussian function resulted in an LC_MAX of 22 ± 1 nm. Importantly, in these experiments, the duplex is ruptured via an unzipping mechanism rather than the shearing mechanism described in the experiments above. In addition to main peak at 22 nm, Fig. 5B shows a small fraction of LC values centered at ~41 nm. We have not observed the second peak for fully complementary 20 bp duplexes (Fig. 3). Thus, we attribute these values to the formation of a less stable complex between 5 nucleotides and 20 nucleotides in addition to the complementary duplex at the beginning of 20 oligomer (as illustrated in Fig. 5). Rupturing of such a complex located farther in the sequence from the point of attachment results in bigger values of LC.
Figure 5.
A) Schematic showing the experimental design for force spectroscopy probing of DNA-DNA interactions in a single tether approach using the 20-mer-PA linker hybrid. The expected contour length is 28 nm. B) Contour length distribution for the corresponding experiments (LC = 22 ± 1 nm with HWHM = 5.9 nm).
3.2. Amyloid beta (Aβ42) interactions
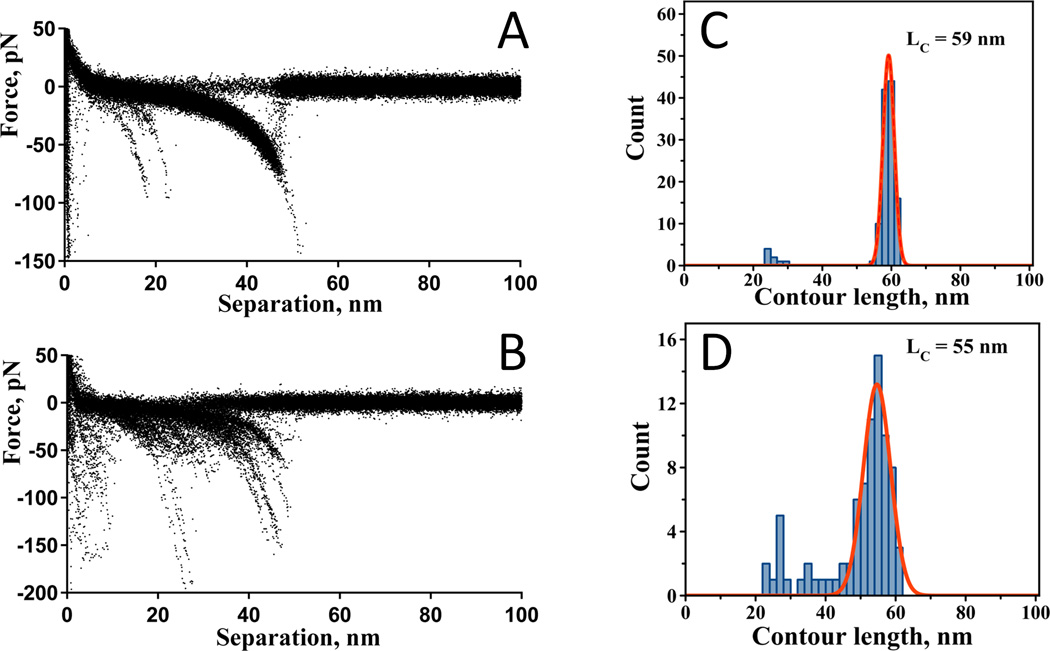
We also tested whether PA linkers can be an appropriate tether for probing protein-protein interactions. Aβ42 peptide was selected as a test system for these experiments. Aggregation of Aβ42 is critically involved in the development of Alzheimer’s disease; therefore, by studying the interaction mechanisms of this peptide, we will increase our understanding of the self-assembly process. We have shown previously that AFM force spectroscopy is capable of characterizing Aβ42 peptide misfolding and its propensity to form stable dimeric complexes [2, 16]. Cys-Aβ42 was immobilized by covalent attachment via the thiol of the only cysteine at the N-terminus to the tip with MAS and to the mica surface with a PA linker. The N-terminus was chosen because this part of the peptide is not involved in fibril formation. The interactions between individual molecules were measured in the multiple approach-retraction cycle. Figure 6A shows a superposition of the force distance curves collected at pH 7.4. Similar to prior studies [2], the peaks due to the rupture of a single Aβ-Aβ pair appear at a specific distance that is primarily defined by the length of the flexible linkers and the unstructured segments of the peptide. WLC analysis of the distances for all of the rupture events is shown as a statistical histogram in Figure 6C. The maximum of the distribution is 59 nm. The use of a PEG linker instead of a PA linker resulted in similar results, but with a wider LC distribution (Figure 6, B and D) that we attribute to the inherent length heterogeneity of the PEG linker [3]. The most probable LC is 55 nm, as determined by the Gaussian fit of the distribution.
Figure 6.
Superposition of the force distance curves for the experiments with Aβ42 in a single tether approach: A) PA linker and B) PEG3400 linker. Corresponding contour length distributions: C) PA linker and D) PEG3400 linker.
3.3. Protein – DNA interactions: synaptic SfiI – DNA complex
We have also used the PA based linker (Linker-3) to monitor interactions within an even more complex system utilizing a different approach for the preparation of the PA linker. We probed the interaction of SfiI protein that is capable of binding two DNA duplexes containing a specific recognition site [17]. Figure 7 schematically illustrates the experimental setup used for studying synaptic SfiI-DNA complexes. Two identical oligonucleotides containing the 13 bp SfiI recognition site were attached to the AFM tip and the MAS functionalized mica surface, by using either one PA linker on the tip in a single tether approach (Figure 7, A) or a PA linker on both the tip and the surface in a double tether approach (Figure 7, B). The following procedure was used for the PA linker preparation. The PA linker (Linker-3) for these experiments was cleaved from the CPG support and covalently attached to a maleimide-activated surface before DNA coupling. The DNA was then coupled via bis-maleimide to the thiol groups of the PA linker. This procedure validated that multi-step modification of the functional group, X, can be effectively carried out after the linker is attached to the surface.
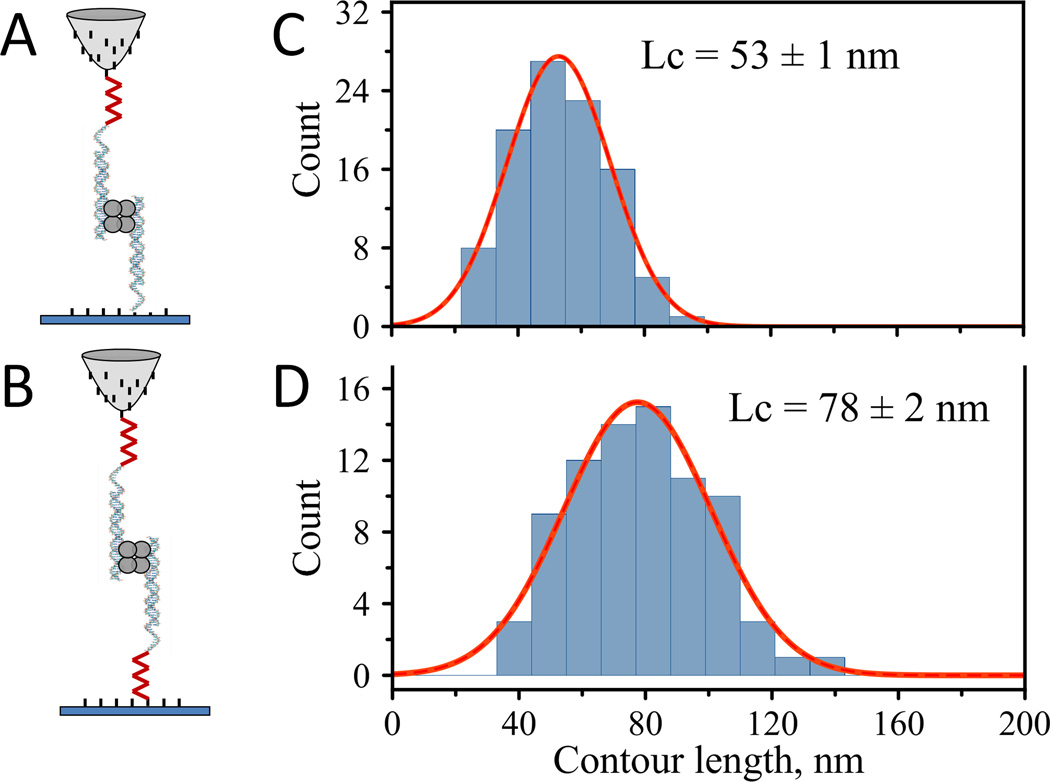
Figure 7.
Schematic representations of the experimental design for the probing experiments with SfiI-DNA: A) single tether approach and B) double tether approach. Contour length distributions: C) single tether, LC = 53 nm (HWHM = 19 nm) and D) double tether LC = 78 nm (HWHM = 27 nm).
With no SfiI protein present in the solution, the probing of interactions between the tip and the surface did not produce any specific rupture events beyond the short-range adhesion peak. We continued to probe interactions using the same tip and the same substrate with the addition of 2 nM SfiI to the cell. Adding SfiI protein resulted in the formation of a complex between the protein and the DNA duplexes after the tip approached the surface (Fig. 7, A or B). Note that only synaptic complexes can be probed in our experimental setup, in which SfiI binds both DNA molecules: one from the tip and another from the surface. Any other binding arrangement would not result in the observation of specific rupture peaks. Interactions between covalently attached DNA molecules and SfiI protein were probed in the multiple approach-retraction cycle on various spots of the surface. Thousands of force distance curves were collected and filtered for specific rupture events which were then fitted with the WLC polymer model. The DNA duplexes were immobilized using a PA linker along with single-stranded dT10. Therefore, in the pulling experiments we needed to include the length of this unit to calculate the expected contour length. The expected rupture length was estimated by adding together the length of the maleimide silatrane linker containing five polyethylene glycol monomeric units in an all-trans conformation (3 nm), the size of SfiI (6.8 nm) [18], the extension of the (dT)10 spacer, the part of the oligonucleotide that is not complexed with the protein (assuming 0.34 nm per base-pair), and the length of the synthesized PA linker (~25 nm). Therefore, the overall stretching of the system in these experiments should be ~ 60 nm.
Figure 7C shows a histogram of contour length values obtained from the single tether approach. Fitting of the histogram with a single Gaussian function resulted in a LC_MAX of 53 ± 2 nm. This value is in agreement with the expected full extension of the linkers for the system (~60 nm). The experiments with the double tether approach, shown in Figure 7D, resulted in a contour length value of 78 ± 3 nm. The difference in these values is 25 ± 5 nm. Given the contribution of the (dT)10 extension (~ 3 nm) to this value, the estimated contour length of the PA linker is in agreement with the contour length value, 22 ± 3 nm, determined in the DNA pulling experiment described above (Figures 3 and 4).
4. Conclusions
The results presented in this paper revealed a number of features of the PA linker that make it valuable for AFM force spectroscopy studies: 1) the length homogeneity of the linker, 2) the possibility to control and vary the length of the linker, and 3) the ability to functionalize the ends of the linker. The PA spacer 18 used in this paper contains 6 ethylene glycol units and 12 spacer units made up of the PA linker. Given the fact that its synthesis involves DNA synthetic chemistry, the synthesis of the linker used in this study is equivalent to the synthesis of only the 12-mer oligonucleotide. Therefore, the procedure can be extended to a 50-mer, thereby increasing the length of the linker without sacrificing length homogeneity. We successfully tested several different applications of the end-functionalized linker. Future potential development in this area includes the use of click chemistry approaches, such as those utilizing alkyne terminal groups. Click chemistry is a superior alternative to the widely used succinimidyl ester functional groups that are hydrolysable in aqueous environments. Additionally, the assortment of PA spacers is large, including non-PEG based spacers with hydrophobic characteristics that can be used for linker synthesis. These hydrophobic linkers can be useful in probing very hydrophilic surfaces to minimize the non-specific interactions of hydrophilic PEG moieties. Finally, although AFM force spectroscopy applications are outlined here, PA tethers can be used in other single molecule studies that require molecule tethering, such as optical trapping, particle tethering and single molecule fluorescence studies.
Supplementary Material
Acknowledgements
We thank A. Portillo, Yuliang Zhang and other group members for insightful suggestions and discussions.
Funding
The work was supported by grants from the National Institutes of Health (5P01GM091743-02 and 5R01 GM096039-02), U.S. Department of Energy Grant DE-FG02-08ER64579, National Science Foundation (EPS – 1004094).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ray C, Akhremitchev BB. Journal of the American Chemical Society. 2005;127:14739–14744. doi: 10.1021/ja052932e. [DOI] [PubMed] [Google Scholar]
- 2.Kim BH, Palermo NY, Lovas S, Zaikova T, Keana JF, Lyubchenko YL. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratto TV, Langry KC, Rudd RE, Balhorn RL, Allen MJ, McElfresh MW. Biophysical journal. 2004;86:2430–2437. doi: 10.1016/S0006-3495(04)74299-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolinnaya NG, Blumenfeld M, Merenkova IN, Oretskaya TS, Krynetskaya NF, Ivanovskaya MG, Vasseur M, Shabarova ZA. Nucleic Acids Res. 1993;21:5403–5407. doi: 10.1093/nar/21.23.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shchepinov MS, Udalova IA, Bridgman AJ, Southern EM. Nucleic Acids Res. 1997;25:4447–4454. doi: 10.1093/nar/25.22.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kransnoslobodtsev AV, Shlyakhtenko LS, Ukraintsev E, Zaikova TO, Keana JF, Lyubchenko YL. Nanomedicine. 2005;1:300–305. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krasnoslobodtsev AV, Peng J, Asiago JM, Hindupur J, Rochet JC, Lyubchenko YL. PLoS One. 2012;7:e38099. doi: 10.1371/journal.pone.0038099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krasnoslobodtsev AV, Shlyakhtenko LS, Lyubchenko YL. Journal of molecular biology. 2007;365:1407–1416. doi: 10.1016/j.jmb.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Malkova S, Lyubchenko YL. J Mol Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Ellman GL. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 11.Strunz T, Oroszlan K, Schafer R, Guntherodt HJ. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11277–11282. doi: 10.1073/pnas.96.20.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kufer SK, Puchner EM, Gumpp H, Liedl T, Gaub HE. Science. 2008;319:594–596. doi: 10.1126/science.1151424. [DOI] [PubMed] [Google Scholar]
- 13.Oesterhelt F, Rief M, Gaub HE. New Journal of Physics. 1999;1:6. [Google Scholar]
- 14.Kreuzer HJ, Wang RLC, Grunze M. New Journal of Physics. 1999;1:21. [Google Scholar]
- 15.Dobrynin AV, Rubinstein M. Progress in Polymer Science. 2005;30:1049–1118. [Google Scholar]
- 16.Lyubchenko YL, Kim BH, Krasnoslobodtsev AV, Yu J. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2:526–543. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 17.Wentzell LM, Nobbs TJ, Halford SE. Journal of molecular biology. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 18.Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, Aggarwal AK. Embo J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.