Abstract
Plant mitochondrial genomes are notorious for their large and variable size, nonconserved open reading frames of unknown function, and high rates of rearrangement. Paradoxically, the mutation rates are very low. However, mutation rates can only be measured in sequences that can be aligned—a very small part of plant mitochondrial genomes. Comparison of the complete mitochondrial genome sequences of two ecotypes of Arabidopsis thaliana allows the alignment of noncoding as well as coding DNA and estimation of the mutation rates in both. A recent chimeric duplication is also analyzed. A hypothesis is proposed that the mechanisms of plant mitochondrial DNA repair account for these features and includes different mechanisms in transcribed and nontranscribed regions. Within genes, a bias toward gene conversion would keep measured mutation rates low, whereas in noncoding regions, break-induced replication (BIR) explains the expansion and rearrangements. Both processes are types of double-strand break repair, but enhanced second-strand capture in transcribed regions versus BIR in nontranscribed regions can explain the two seemingly contradictory features of plant mitochondrial genome evolution—the low mutation rates in genes and the striking expansions of noncoding sequences.
Keywords: Arabidopsis, mitochondrial genome, mutation rate, DNA repair
Introduction
The mitochondrial genomes of higher plants are well known to be very different from their animal counterparts. Although presumably derived from the same endosymbiotic event (Gray 1999), the genomes of higher plant mitochondria are large, rearrange freely, and yet are reported to mutate very slowly (Palmer and Herbon 1988). The low mutation rates and the expansion of the genomes have been explained by the mutational burden hypothesis (Lynch et al. 2006; Lynch 2007), but a few species with both highly expanded genomes and high mutation rates are a difficulty for this hypothesis (Cho et al. 2004; Parkinson et al. 2005; Sloan, Muller, et al. 2012; Sloan et al. 2012). In addition, mutation rates can only be measured in sequences that can be aligned, so measurements have been made only by comparison of genes encoding proteins and rRNA but not the large quantities of DNA that do not encode known products. Complete plant mitochondrial genome sequences are becoming available, but the origin and function of the noncoding DNA is still not understood and not readily aligned between different groups of species. The recent description of complete mitochondrial sequences from two ecotypes of Arabidopsis thaliana, Columbia-0 (Col-0) and C24 (Davila et al. 2011), enables comparison of the noncoding sequences and better understanding of the evolution of plant mitochondrial genomes.
To establish a framework for comparing mitochondrial genomes, the nuclear mutation rate in the two ecotypes was estimated. Nine nuclear genes encoding mitochondrial proteins were chosen for analysis to have a set for which the role of selection will be comparable to mitochondrially encoded proteins. Table 1 presents the comparison of the coding sequences between the two ecotypes. The spectrum of the 16 differences between Col-0 and C24 can be analyzed by comparison to A. lyrata. There are 10 synonymous and 6 nonsynonymous substitutions, comprising 5 transversions and 11 transitions. The synonymous substitution rate between Col-0 and C24 is 4.20 × 10−3 per site. The synonymous substitution rate between A. lyrata and A. thaliana (Col-0) is 1.04 × 10−1 per site, similar to previous results (Koch et al. 2000; Huang et al. 2012). Using a divergence time of 5 Ma for A. thaliana and A. lyrata (Koch et al. 2000) results in a rate of 2.08 × 10−8 substitutions per site per year, similar to that previously obtained for nuclear genes in the Brassicales (Franzke et al. 2011). Using this rate to calibrate a molecular clock, the divergence time between the Col-0 and C24 ecotypes of A. thaliana is approximately 0.2 Ma.
Table 1.
Nuclear Gene Mutations in Arabidopsis thaliana
| Gene | Nucleotide Position | Col-0 | C24 | A. lyrata | Mutation | Amino Acid Change | Classification |
|---|---|---|---|---|---|---|---|
| ATP5 | 39 | G | A | G | Transition: G:C → A:T in C24 | K → K | Synonymous |
| ATP5 | 52 | A | T | T | Transversion: A:T → T:A in Col-0 | S → T | Nonsynonymous |
| ATP5 | 375 | T | C | C | Transition: G:C → A:T in Col-0 | T → T | Synonymous |
| ATP7 | 183 | T | C | C | Transition: G:C → A:T in Col-0 | P → P | Synonymous |
| ATP7 | 504 | C | T | C | Transition: G:C → A:T in C24 | Y → Y | Synonymous |
| COX5B-1 | 157 | T | C | C | Transition: G:C → A:T in Col-0 | L → L | Synonymous |
| COX5B-1 | 158 | T | A | A | Transversion: A:T → T:A in Col-0 | Q → L | Nonsynonymous |
| COX5B-1 | 178 | G | T | G | Transversion: G:C → A:T in C24 | D → Y | Nonsynonymous |
| NDUF51 | 45 | A | T | T | Transversion: A:T → T:A in Col-0 | S → S | Synonymous |
| NDUF51 | 198 | C | G | T | Unknown | ||
| NDUF51 | 498 | C | T | T | Transition: A:T → G:C in Col-0 | H → H | Synonymous |
| NDUF51 | 1437 | T | C | C | Transition: G:C → A:T in Col-0 | N → N | Synonymous |
| NDUF51 | 1626 | T | C | C | Transition: G:C → A:T in Col-0 | V → V | Synonymous |
| NDUF51 | 1769 | A | G | A | Transition: A:T → G:C in C24 | N → S | Nonsynonymous |
| NDUF51 | 2125 | G | A | A | Transition: A:T → G:C in Col-0 | T → A | Nonsynonymous |
| NDUF51 | 2154 | G | T | G | Transversion: G:C → A:T in C24 | V → V | Synonymous |
| NDUFS8a | 95 | G | A | G | Transition: G:C → A:T in C24 | R → K | Nonsynonymous |
Note.—There were no differences between Col-0 and C24 for the genes ATP3, ATP15, ATP16, and NDUFA9.
Mitochondrial genome mutation rates were assessed by comparing the Col-0 mitochondrial genome (JF729201) to that from ecotype C24 (JF729200). These two sequences differ by rearrangement around two pairs of large repeats (6.5 and 4.3 kb), an inversion at a 205-bp repeat, a novel junction, and an insertion in Col-0 of 1.8 kb of fragmented genes (Forner et al. 2005; Davila et al. 2011). Comparison of the 364,952 bp not involved in these rearrangement polymorphisms reveals 64 single-nucleotide substitutions, 35 indels of one base, and 4 inversions of 2–4 bp (supplementary table S1, Supplementary Material online). Only 1 of these 103 mutations is in coding sequence: a synonymous substitution in the matR gene (position 230278). This gene is a maturase possibly involved in trans-splicing, although its functional role is not well understood (Keren et al. 2012). Compared with other related species, the Arabidopsis matR gene lacks an initiation codon, so it may be a pseudogene, similar to the rps14 gene in many species (Ong and Palmer 2006). If this is true, the synonymous substitution rate in genes would be 0, but even assuming matR as a bona fide gene gives a synonymous substitution rate in mitochondrial exons of 1.68 × 10−4 per site between Col-0 and C24, 25-fold lower than the rate in nuclear exons. In the A. thaliana mitochondrial genome, protein-coding gene exons comprise 30,672 bp, whereas tRNA and rRNA are 6,223 bp. Therefore, there has been one synonymous substitution (assuming matR is a functional gene) in the 36,895 bp of protein, tRNA, and rRNA genes and 102 changes (63 of which are base substitutions) in the 328,057 bp of noncoding DNA or 1.92 × 10−4 per base. Assuming a divergence time of 0.20 Ma, the synonymous substitution rate in exons is either 0 or 8.4 × 10−10 per site per year (depending on whether matR is a pseudogene), and the mutation rate in mitochondrial noncoding regions is 9.6 × 10−10 substitutions per site per year. The indel rate in the noncoding regions is roughly half the substitution rate but is 0 in coding regions.
A complete mitochondrial genome sequence from A. lyrata is not currently available as an outgroup; however, comparison to the related species Raphanus sativus (AB694744) allows inference of parts of the ancestral sequence. Alignment of the R. sativus mitochondrial genome is possible for 39 of the 103 differences between Col-0 and C24 revealing the spectrum of mutations in the Arabidopsis lineage (table 2). Twenty of the substitutions are due to mutations in Col-0 and 19 in C24. All 14 of the indels are deletions of one base pair from homopolymeric runs of 2–8 bp (including both G:C and A:T pairs). Among the substitutions, there are six transitions, all of which are G:C to A:T and can be explained by a failure of the uracil-N-glycosylase (Boesch et al. 2009) to repair cytosine deamination. Thirteen of the 16 transversions are G:C to T:A changes, which can be explained by oxidation of a guanine to 8-oxo-guanine, followed by misincorporation of an A in the opposite strand (van Loon et al. 2010; Markkanen et al. 2012). These mutations may be due to failure to remove 8-oxo-guanine from DNA before replication or failure to remove the misincorporated A after replication. This overall spectrum of mutations includes more transversions and more indels than the nuclear genome of A. thaliana (Ossowski et al. 2010).
Table 2.
Changes in Arabidopsis thaliana Compared with Raphanus sativus
| Col-0 Position | Col–0 | C24 | R. sativus | Mutation |
|---|---|---|---|---|
| Transitions | ||||
| 136702 | T | C | C | G:C → A:T in Col-0 |
| 216873 | A | G | G | G:C → A:T in Col-0 |
| 221694 | G | A | G | G:C → A:T in C24 |
| 330185 | C | T | C | G:C → A:T in C24 |
| 359929 | A | G | G | G:C → A:T in Col-0 |
| 361127 | G | A | G | G:C → A:T in C24 |
| Transversions | ||||
| 8789 | T | G | G | G:C → T:A in Col-0 |
| 18930 | A | C | C | G:C → T:A in Col-0 |
| 81609 | G | T | T | A:T → C:G in Col-0 |
| 84529 | A | C | C | G:C → T:A in Col-0 |
| 108847 | A | C | C | G:C → T:A in Col-0 |
| 112905 | A | C | C | G:C → T:A in Col-0 |
| 116312 | G | T | G | G:C → T:A in C24 |
| 119374 | G | T | G | G:C → T:A in C24 |
| 135125 | G | C | C | G:C → C:G in Col-0 |
| 167272 | A | C | C | G:C → T:A in Col-0 |
| 230278 | C | A | C | G:C → T:A in C24 |
| 243601 | C | A | C | G:C → T:A in C24 |
| 268609 | T | G | G | G:C → T:A in Col-0 |
| 297203 | T | G | G | G:C → T:A in Col-0 |
| 322252 | G | T | G | G:C → T:A in C24 |
| 324473 | G | T | T | A:T → C:G in Col-0 |
| Indels | ||||
| 28012 | CAAAAG | C–AAAG | CAAAAG | 1-bp deletion in C24 |
| 54300 | G–TTTTTA | GTTTTTTA | GTTTTTTA | 1-bp deletion in Col-0 |
| 84063 | GCCT | G–CT | GCCT | 1-bp deletion in C24 |
| 135403 | T–AAAAAAAT | TAAAAAAAAT | TAAAAAAAAT | 1-bp deletion in Col-0 |
| 156811 | AGGGC | A–GGC | AGGGC | 1-bp deletion in C24 |
| 163337 | G–AG | GAAG | GAAG | 1-bp deletion in Col-0 |
| 232507 | ACCG | A–CG | ACCG | 1-bp deletion in C24 |
| 234090 | CGGGT | C–GGT | CGGGT | 1-bp deletion in C24 |
| 236303 | CAAAT | C–AAT | CAAAT | 1-bp deletion in C24 |
| 282377 | G–AAAG | GAAAG | GAAAG | 1-bp deletion in Col-0 |
| 282384 | G–AAAT | GAAAAT | GAAAAT | 1-bp deletion in Col-0 |
| 289160 | GTTG | G–TG | GTTG | 1-bp deletion in C24 |
| 289163 | GCCG | G–CG | GCCG | 1-bp deletion in C24 |
| 340231 | A–GGA | AGGGA | AGGGA | 1-bp deletion in Col-0 |
| Inversions | ||||
| 139296 | AT | TA | AT | 2-bp inversion in C24 |
| 360174 | AA | TT | AA | 2-bp inversion in C24 |
| 292314 | TTTC | GAAA | GAAA | 4-bp inversion in Col-0 |
To characterize the noncoding DNA, the Col-0 mitochondrial genome was compared with complete mitochondrial and chloroplast genomes from several other angiosperms and one gymnosperm, not including any member of the Brassicales family (listed in supplementary table S2, Supplementary Material online). These comparisons show that most of the A. thaliana mitochondrial sequence cannot be aligned with genomes outside the Brassicales. A 221,344-bp sequence (supplementary fig. S1, Supplementary Material online) constructed by deleting the conserved regions represents 60% of the genome and shows no similarity to GenBank entries outside the Brassicales, using MegaBLAST (word size 28, no masking). Using the default parameters for BLASTn, only short regions of homology were found, and at most 2% of the mitochondrial genome was involved in any of the discovered alignments. This comparison suggests that 60% of the Arabidopsis mitochondrial genome has no function at all and could properly be called “junk” (Ohno 1972; Brenner 1998; Graur et al. 2013).
The low mutation rates in plant mitochondria have been a challenge to explain (Lynch et al. 2006; Lynch 2007). Selection on gene function would presumably be similar for mitochondrial proteins encoded by either the nuclear or mitochondrial genome, yet the rate of synonymous substitutions between Col-0 and C24 is 4.2 × 10−3 in the nucleus and 1.7 × 10−4 in the mitochondrion, whereas the nonsynonymous rate is 2.7 × 10−4 in the nucleus and 0 in mitochondria (<10−5). The substitution rates calculated for noncoding DNA using Col-0 and C24 may be an underestimate of the massive changes that occur in plant mitochondrial DNA that prevent alignment of more than 200 kb of the Arabidopsis mitochondrial genome with any organism that diverged more than about 20 Ma or that over evolutionary time scales the noncoding DNA is subject to very error-prone processes. Still, the low mutation rates in genes must be contrasted with the high rates of rearrangement and mutation in noncoding sequences.
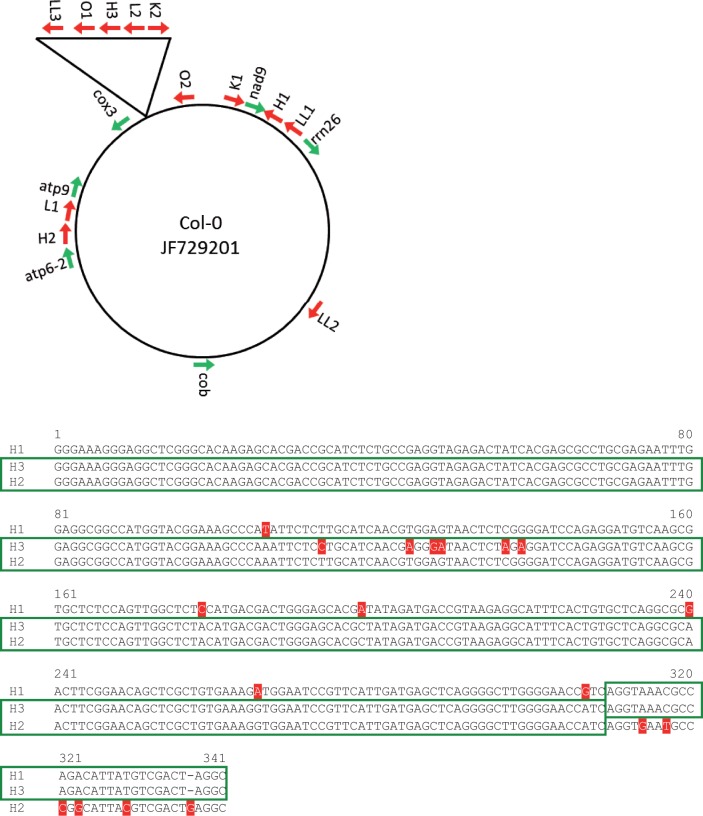
Additional information may be gleaned from a segment of the genome that is novel in Col-0 compared with C24. This segment is a cluster of at least five fragments from other locations in the genome assembled into a 1.8 kb chimera inserted upstream of the cox3 gene (Forner et al. 2005). Although these five fragments are found elsewhere in the genome, this combination of fragments is not found in any other plant species, thus it is likely that this insertion occurred in the lineage leading to Col-0 after its reproductive isolation from C24. The age of this insertion must therefore be no older than 0.2 Ma, making it the most recent plant mitochondrial genome expansion event that can be characterized. The repeats within this cluster have been identified (Arrieta-Montiel et al. 2009; Davila et al. 2011) and can be compared with the original sequences. Repeat H is particularly informative because there were already two copies of this 340 bp repeat within the Col-0 and C24 genomes, and the fragment inserted as part of the 1.8 kb chimera upstream of cox3 introduces a third copy in Col-0 (see fig. 1). Repeat H-1, located between nad9 and rrn26, is identical between Col-0 and C24. Repeat H2, located between atp6-2 and atp9, is also identical between Col-0 and C24, but repeats H1 and H2 are distinguished by 11 substitutions and 1 indel. This suggests H1 and H2 duplicated and diverged in the common ancestor of Col-0 and C24, whereas repeat H3 originated after the ecotypes diverged. The substitution rate between H1 and H2 is 3.3 × 10−2 per site (alignments shown in fig. 1). Using the rate of 2.08 × 10−8 substitutions per site per year gives a divergence time for H1 and H2 of 1.57 Ma. Repeat H3 can be aligned with either H1 or H2 but appears to consist of two different domains: The first 310 bp appear to be most similar to H2, whereas the next 30 bp are most similar to H1. There are also five substitutions near the center of the H2-like domain of H3. If the 310-bp H2-like region of H3 is compared with H2, the substitution rate is 1.6 × 10−2 substitutions per site. The maximum possible divergence time of these two repeats is 0.2 Ma, corresponding to a rate of 8.1 × 10−8 substitutions per site per year—two orders of magnitude higher than the synonymous substitution rate of the mitochondrial genes. The junctions between the repeats in the 1.8 kb insertion are also impossible to align, suggesting a high mutation rate associated with duplication and chimera formation that makes alignment and measurement of the rate impossible. It is also possible that template switching accompanies the formation of chimeras and duplications by BIR, making this process mutagenic (Llorente et al. 2008).
Fig. 1.—
Repeats H1, H2, and H3. (A) Diagram of the Col-0 genome with a few genes indicated as landmarks and showing the 1.8 kb insertion and other copies of its repeats. (B) Alignment of repeats H1, H2, and H3. Mutations are indicated in red, and boxes surround the likely origins of the H3 sequence.
In contrast, two of the other repeats within this insertion, repeats L-2 and K-2, are fragments of atp9 and rps3/rpl16, respectively. When these repeats are compared with the parent genes, the substitution rate is 0. These two repeats have also been noted to be quite active in recombination processes (Martinez-Zapater et al. 1992; Sakamoto et al. 1996; Abdelnoor et al. 2003; Shedge et al. 2007). The much lower mutation rate of these two regions cannot be explained by selection, because they are merely gene fragments of 249 and 251 bp and must therefore be due to the mechanism of repair.
Together, these results suggest a very accurate mechanism of repair in coding regions, a mechanism for duplication of sequences, and a less accurate mechanism of repair in noncoding sequences. These phenomena, including the expansions of plant mitochondrial genomes and the occasional exceptional species with high mutation rates accompanied by still more genome expansion can all be explained by a model of DNA repair and recombination.
Formation of duplications and chimeric genes are relatively rare, occurring at evolutionary time scales, and can readily be explained by nonhomologous end joining and microhomology-mediated break-induced replication (BIR; initiated at ectopic sites). One such event has been captured in the Col-0 lineage and clearly originated by an error-prone process such as these. Importantly, these processes do not provide a precise mechanism for removing DNA. BIR at short regions of homology occurs frequently in various mutants, leading to half-crossovers (Zaegel et al. 2006; Shedge et al. 2007; Cappadocia et al. 2010; Davila et al. 2011; Janicka et al. 2012; Miller-Messmer et al. 2012). This process presumably occurs in wild type, but at a lower frequency, and is a mechanism for genome expansion but not accurate contraction. Duplications followed by mutation, rearrangement, and drift are the likely source of the genome expansions found in plant mitochondria, with the 1.8-kb chimera in Col-0 being a particularly recent example. If duplications of noncoding sequences undergo substitutions at the rate seen in repeat H3, they will become unalignable and unrecognizable in relatively short times, leading to large regions of plant mitochondrial genomes (221 kb in the case of A. thaliana) having unrecognizable origins.
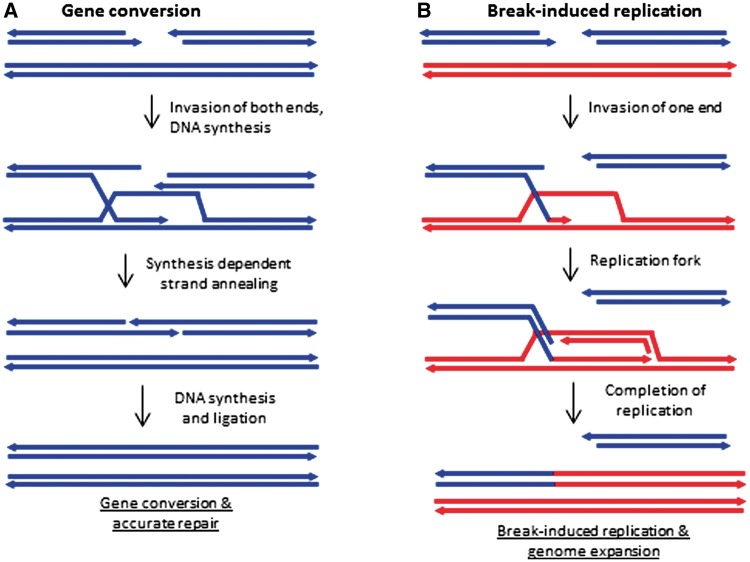
If BIR and a high mutation rate explain the expansions and divergence of the noncoding regions, what explains the low mutation rates in coding regions? Selection on function cannot explain the low synonymous substitution rate. One possibility is a biased repair process (Birky and Walsh 1992; Sloan et al. 2012). The lack of synonymous substitutions is suggestive of accurate template-directed repair. I suggest that gene conversion is the mechanism of repair in coding regions. Accurate double-strand break repair, accompanied by crossovers, is frequent at the very large repeats (Klein et al. 1994) and likely also occurs between sister molecules within mitochondria, but symmetric events involving both ends of a double-strand break are not frequent elsewhere in the genome (Davila et al. 2011). Double-strand break repair or synthesis-dependent strand annealing (SDSA) involving both ends of a break would result in gene conversion and repair that does not depend on recognition of which base in which strand is mismatched or damaged because both are removed and replaced. Although in the nucleus gene conversion is associated with a higher mutation rate than replication that is due to the involvement of DNA polymerases Polζ, Polη, and Pol32 (Hicks et al. 2010). In A. thaliana mitochondria, it appears that both replication and repair are carried out by two very similar gamma-type DNA polymerases (Parent et al. 2011; Cupp and Nielsen 2013), which are more accurate. I propose that transcribed regions direct repair to these pathways, which must involve degradation of both strands at the site of a lesion or mismatch, and the use of both ends in template-directed repair (see fig. 2). The difference could be due to enhanced breakage of the second strand, enhanced second-strand capture by coordination of the two ends, or promotion of SDSA in transcribed regions compared with nontranscribed. It is also possible that mismatches and base excision events lead to double-strand breaks; if this is more likely in transcribed regions, this would account for the low mutation rates in genes and might also be a mechanism for homoplasmy in mitochondrial genomes. BIR in noncoding regions and gene conversion in transcribed regions would explain both the phenomena of genome expansion and the low mutation rate in genes, two peculiar features of plant mitochondrial genome evolution. Occasional repair by error-prone processes in noncoding regions, perhaps associated with the duplications and rearrangements mediated by BIR and nonhomologous end-joining can then explain the rapid divergence of noncoding regions.
Fig. 2.—
Model of two types of DNA repair. The blue and red lines indicate different sequences. (A) The consequences of coordination of both ends following a break. (B) The consequences of invasion of a single DNA end, which ultimately leads to genome expansions. Invasion occurs at an ectopic site due to a small region of homology.
An additional peculiar finding in plant mitochondrial genomes is that in rare lineages there are both high mutation rates in genes and even more dramatic expansions of the genome (Parkinson et al. 2005; Mower et al. 2007; Sloan et al. 2012). These observations can also be explained as follows. In addition to double-strand break repair mechanisms, there are base excision repair mechanisms involved in removal of uracil (Boesch et al. 2009), 8-oxo-guanine, and adenines mispaired with 8-oxo-guanine (Macovei et al. 2011). These mechanisms are highly accurate because they are specific for a damaged base. If one of the mechanisms of base-excision or mismatch repair is missing, more mismatched or damaged bases would accumulate, with the following consequences. First, more of these changes will become fixed in the population, leading to a higher measured mutation rate. Second, unrepaired bases will more frequently lead to double-strand breaks, BIR, and genome expansion. Third, gene conversion in coding regions will fix mutations due to the repair templates having more errors in them. A prediction of this model is that in these rare lineages, the spectrum of mutations will be biased, revealing which repair pathway is defective, and the bias will be different in different lineages depending on which repair pathway is/was impaired. It would be expected that these losses of repair will be rare across groups of taxa and may have been transient losses in an ancestor, with the current genome structures being evolutionary remnants of that event due to the absence of a mechanism for deletions.
Materials and Methods
Sequence manipulation was done using the VectorNTI 11.5.0 package from Invitrogen. Alignments were done using MUSCLE (Edgar 2004) as implemented in MEGA5 (Tamura et al. 2011). Synonymous substitution rates were calculated by MEGA5, using the Kumar model. Substitution rates in noncoding regions were calculated with MEGA5, using the Tamura–Nei model.
Nuclear genes encoding mitochondrial proteins from A. thaliana Col-0 that were used for comparison to C24 and A. lyrata were ATP3 = NM_128864.3; ATP5 = NM_121348.3; ATP7 = NM_115090.3; ATP15 = NM_104043.2; ATP16 = NM_124074.3; COX5B-1 = NM_106672.4; NDUF51 = NM_180772.1; NDUFA9 = NM_180772.1; and NDUFS8a = NM_106551.3. Noncoding sequences at the 5′- and 3′-ends were removed and compared with C24 using the Basic Local Alignment Search Tool (BLAST) tool at the 1001 Genomes site, http://1001genomes.org/cgi-bin/blast/blast.cgi, last accessed May 17, 2013 (Schneeberger et al. 2011), and compared with A. lyrata using the BLAST server at (http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed May 17, 2013) with the default MegaBLAST settings (Zhang et al. 2000).
Comparison of the mitochondrial genomes of Col-0 and C24 was also done using MegaBLAST but with no masking for low complexity sequences. To avoid rearrangement polymorphisms, this comparison was done using segments of the Col-0 genome as follows: 1–48,895; 48,896–112,984; 112,985–129,990; 129,991–197,428; 197,429–257,567; 257,568–268,497; 268,498–276,591; 276,592–276,625; 276,626–330,317; and 332,108–366,750.
To identify “junk” DNA, the Col-0 mitochondrial genome sequence JF729201 was compared using MegaBLAST (no masking) to the complete mitochondrial and chloroplast genome sequences listed in supplementary table S1, Supplementary Material online. The sequence that results from deletion of all identified conserved regions is presented in supplementary figure S1, Supplementary Material online, as a FASTA format file.
Supplementary Material
Supplementary tables S1 and S2 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Conversations about plant mitochondrial genome evolution with Jeff Mower and his laboratory were helpful in shaping my thinking on these issues. I am also grateful to Cynthia Haseltine, Etsuko Moriyama, and two anonymous reviewers for comments on the manuscript. The Root Marm Chicken Farm Jug Band was inspirational. This work was supported in part by an EAGER award from the National Science Foundation (MCB-1104677).
Literature Cited
- Abdelnoor RV, et al. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc Natl Acad Sci U S A. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009;183:1261–1268. doi: 10.1534/genetics.109.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW, Jr, Walsh JB. Biased gene conversion, copy number, and apparent mutation rate differences within chloroplast and bacterial genomes. Genetics. 1992;130:677–683. doi: 10.1093/genetics/130.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesch P, et al. Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 2009;37:5690–5700. doi: 10.1093/nar/gkp606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. Refuge of spandrels. Curr Biol. 1998;8:R669. doi: 10.1016/s0960-9822(98)70427-0. [DOI] [PubMed] [Google Scholar]
- Cappadocia L, et al. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell. 2010;22:1849–1867. doi: 10.1105/tpc.109.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci U S A. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp JD, Nielsen BL. Arabidopsis thaliana organellar DNA polymerase IB mutants exhibit reduced mtDNA levels with a decrease in mitochondrial area density. Physiol Plant. Forthcoming 2013 doi: 10.1111/ppl.12009. http://onlinelibrary.wiley.com/doi/10.1111/ppl.12009/abstract#. [DOI] [PubMed] [Google Scholar]
- Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J, Weber B, Wietholter C, Meyer RC, Binder S. Distant sequences determine 5' end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res. 2005;33:4673–4682. doi: 10.1093/nar/gki774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 2011;16:108–116. doi: 10.1016/j.tplants.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Graur D, et al. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5:578–590. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. Evolution of organellar genomes. Curr Opin Genet Dev. 1999;9:678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, et al. Evolutionary rates of commonly used nuclear and organelle markers of Arabidopsis relatives (Brassicaceae) Gene. 2012;499:194–201. doi: 10.1016/j.gene.2012.02.037. [DOI] [PubMed] [Google Scholar]
- Janicka S, et al. A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J. 2012;72:423–435. doi: 10.1111/j.1365-313X.2012.05097.x. [DOI] [PubMed] [Google Scholar]
- Keren I, et al. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 2012;71:413–426. doi: 10.1111/j.1365-313X.2012.04998.x. [DOI] [PubMed] [Google Scholar]
- Klein M, et al. Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J. 1994;6:447–455. doi: 10.1046/j.1365-313x.1994.06030447.x. [DOI] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome achitecture. Sunderland (MA): Sinauer Associates, Inc; 2007. [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Macovei A, Balestrazzi A, Confalonieri M, Fae M, Carbonera D. New insights on the barrel medic MtOGG1 and MtFPG functions in relation to oxidative stress response in planta and during seed imbibition. Plant Physiol Biochem. 2011;49:1040–1050. doi: 10.1016/j.plaphy.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Markkanen E, Hubscher U, van Loon B. Regulation of oxidative DNA damage repair: the adenine:8-oxo-guanine problem. Cell Cycle. 2012;11:1070–1075. doi: 10.4161/cc.11.6.19448. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Gil P, Capel J, Somerville CR. Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell. 1992;4:889–899. doi: 10.1105/tpc.4.8.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Messmer M, et al. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 2012;159:211–226. doi: 10.1104/pp.112.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- Ong HC, Palmer JD. Pervasive survival of expressed mitochondrial rps14 pseudogenes in grasses and their relatives for 80 million years following three functional transfers to the nucleus. BMC Evol Biol. 2006;6:55. doi: 10.1186/1471-2148-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Parent JS, Lepage E, Brisson N. Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 2011;156:254–262. doi: 10.1104/pp.111.173849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CL, et al. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 2005;5:73. doi: 10.1186/1471-2148-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Kondo H, Murata M, Motoyoshi F. Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell. 1996;8:1377–1390. doi: 10.1105/tpc.8.8.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci U S A. 2011;108:10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Muller K, McCauley DE, Taylor DR, Storchova H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 2012;196:1228–1239. doi: 10.1111/j.1469-8137.2012.04340.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Zaegel V, et al. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006;18:3548–3563. doi: 10.1105/tpc.106.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.