Abstract
To understand how miRNA-mediated silencing impacts on embryonic mRNAs, we conducted a functional survey of abundant maternal and zygotic miRNA families in the C. elegans embryo. We show that the miR-35-42, and the miR-51-56 miRNA families define maternal and zygotic miRNA-induced silencing complexes (miRISCs), respectively, that share a large number of components. Using a novel cell-free C. elegans embryonic extract, we demonstrate that the miRISC directs the rapid deadenylation of reporter mRNAs with natural 3’UTRs. The deadenylated targets are translationally suppressed and remarkably stable. Sampling of the predicted miR-35-42 targets reveals that roughly half are deadenylated in a miRNA-dependent manner, but with each target displaying a distinct efficiency and pattern of deadenylation. Finally, we demonstrate that functional cooperation between distinct miRISCs within 3’UTRs is required to potentiate deadenylation. With this report, we reveal the extensive and direct impact of miRNA-mediated deadenylation on embryonic mRNAs.
Introduction
Since their discovery, the small (~18–25nt) non-coding microRNAs (miRNAs) have reshaped the landscape of genetic networks in a broad variety of species. Accumulating data indicates that miRNAs directly regulate >60% of the human coding genome (Friedman et al., 2009), and leave very few (if any) genetic pathways untouched. Validated miRNA targets are now known to be implicated in a wide range of cellular functions in developmental, steady-state, and disease contexts (Bartel, 2009).
Most miRNAs are generated as primary transcripts that are sequentially matured by two RNaseIII enzymes and their associated proteins. The nuclear Drosha protein cleaves these transcripts into hairpins of ~60nt in length (pre-miRNA) (Lee et al., 2003). Pre-miRNAs are then exported to the cytoplasm and processed by Dicer (DCR-1 in C. elegans) into mature miRNAs (Grishok et al., 2001; Ketting et al., 2001). The processing of miRNAs by DCR-1 is coupled with their assembly into the miRNA-induced silencing complex (miRISC), which is composed at its core of specific members of the Argonaute family of proteins (ALG-1 and −2 in C. elegans), and additional proteins such as the GW182 homologs (AIN-1 and −2 in C. elegans). Base-pairing interactions between a miRNA and a target mRNA are important for silencing by miRISC. In canonical mRNA-miRNA interactions, the 5’ region of the miRNA (nucleotides 2–7), coined the ‘seed’, is an important determinant in the recognition of miRNA target sites, which are typically located within the 3’UTRs of target mRNAs. MiRNAs sharing the same seed sequence are said to belong in the same ‘family’ (Ibanez-Ventoso et al., 2008).
The mechanism, or the diversity of mechanisms through which miRNAs mediate gene silencing are not fully understood. Pioneering work on the mechanism of miRNA-mediated silencing in C. elegans indicated that the lin-4 miRNA represses lin-14 mRNA at the level of translation (Olsen and Ambros, 1999). Since then, several models have been proposed to explain the mode of action of miRNAs (see (Filipowicz et al., 2008) for a review). Most recently, a growing body of work indicates that miRNA targeting may often result in mRNAs degradation, which in at least some cases is preceded by decapping and/or deadenylation (Baek et al., 2008; Bagga et al., 2005; Eulalio et al., 2009; Fabian et al., 2009; Giraldez et al., 2006; Selbach et al., 2008; Wu et al., 2006). The differences between the prevailing models may stem from differences in experimental designs, but it may also be interpreted as evidence for the existence of multiple mechanisms of miRNA-mediated silencing. Resolution of these matters currently awaits systematic and comparative mechanistic studies. For example, the question of whether two different miRNA families assemble with similar molecular machineries and silence their targets through the same mechanism remains unanswered.
Here we examine the molecular function of abundant maternally- and zygotically-contributed miRNA families in C. elegans embryo. Using a novel cell-free system, we compared their mechanism of action, and surveyed their mRNA targets. We show the broad and direct impact of miRNAs on embryonic mRNA poly(A) tails, and highlight miRISC cooperation as a key feature in target deadenylation.
Results
Bulk miRISC programming by a few maternal and zygotic miRNA families in C. elegans embryos
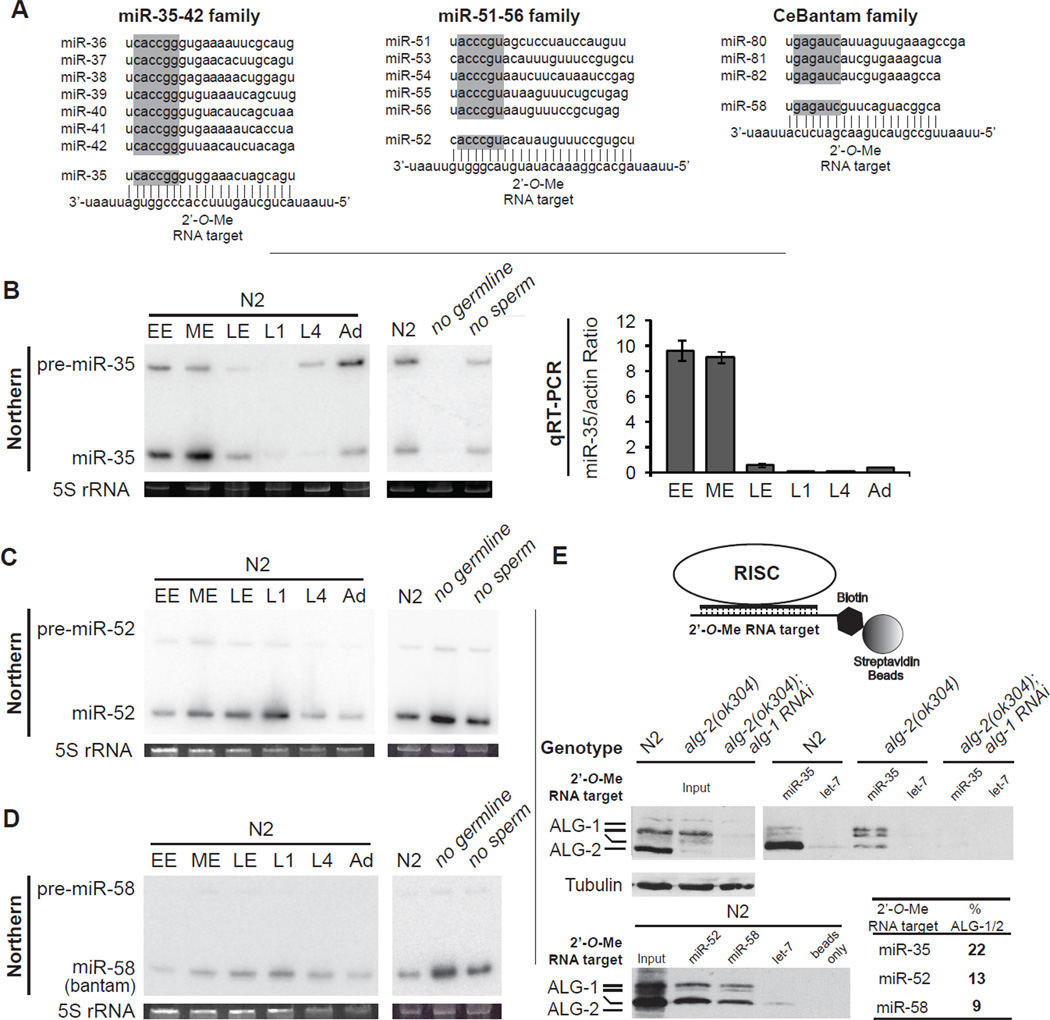
The miR-35-42 and miR-51-56 families are essential for early development (Alvarez-Saavedra and Horvitz, 2010). The miR-35-42 family is suspected to be mostly maternally-contributed, while the miR-51-56 as well as the C. elegans (Ce)Bantam families (Figure 1A) are thought to be broadly if not ubiquitously expressed (Ambros et al., 2003; Lau et al., 2001; Stoeckius et al., 2009). We refined the expression domains of these miRNAs using northern blot and qRT-PCR (Figure 1B–D). Expression of miR-35 and its precursor is very dynamic. It was strongest in early embryonic preparations (EE), but was rapidly lost at the L1 stage (Figure 1B). In contrast, miR-52, and miR-58 (Bantam) expression increased as the embryo matured, and was highest during the L1 larval stage preparations, consistent with zygotic transcription accounting for most of their expression (Figure 1C, D). Similar expression analysis of the other members of these families also indicated zygotic expression (not shown). MiR-35 is absent in germline-depleted preparations preparations, indicating a germline origin, while miR-52, and miR-58 (bantam) were enriched (Figure 1B–D no germline lane), indicating somatic expression. Deep sequencing of small RNAs confirmed that miR-35-42 family members are the most abundantly expressed miRNAs in isolated oocytes (D. Conte, personal communication), hence this family is maternally contributed.
Figure 1. miRISC programming by maternal and zygotic miRNA families in C. elegans embryos.
(A) miRNAs and 2’-O-Me oligonucleotides used in this study. The seed region for each miRNA is highlighted in gray. (B) Expression profile of miR-35 by northern and real-time (qRT) PCR analysis. Total RNA from wild-type (N2) early-stage embryos (EE), middle-stage embryos (ME), late-stage embryos (LE), L1-, L4-, and adult-stage animals (Ad), or adult-stage (glp-4)bn2 (no germline) and fem-1(hc17) (no sperm) animals grown at 25°C. 5S ribosomal RNA (rRNA) is indicated as loading control. qRT-PCR results are presented as the mean from triplicate samples and error bars represent standard deviation. (C,D) Northern analysis of miR-52 and miR-58 (bantam) expression. (E) (Top) Schematic representation of the miRISC 2’-O-Me pulldown strategy. (Middle and bottom) Extracts prepared from wild-type (N2), alg-2(ok304), or alg-2(ok304); alg-1 RNAi embryos were incubated with the indicated 2’-O-Me matrices. Bound proteins were probed for ALG-1 and ALG-2, and average percentage pulled down of two independent experiments is indicated in bold. Related data in Figures S1.
Based on miRNA-specific qRT-PCR, we estimated the concentration of miR-35 in ME fractions to be approximately 3–8 nM with little batch-to-batch variation, a concentration confirmed using northern blots (Figure S1). To further address the abundance of these miRNAs in embryos, we used biotinylated, non-hydrolyzable 2’-O-Methylated (2’-O-Me) oligonucleotides that mimic miRNA target sites as baits to capture programmed miRISC complexes from embryonic lysates (Figure 1E, upper panel) (Hutvagner et al., 2004). The pool of miR-35-42 miRNAs, even the most divergent family members, was strongly depleted from the lysate using this strategy (Figure S1D). Pull down of miR-35-42 miRISC in embryonic lysates was effective as indicated by the presence of the Argonautes ALG-1 and ALG-2 (Figure 1E middle panel: note that ALG-1 migrates as multiple species). Quantification indicates that approximately 22% of the entire endogenous embryonic ALG-1/ALG-2 pool is programmed by the miR-35-42 family alone (Figure 1E). In contrast a let-7 affinity matrix, which is at most very weakly expressed during embryogenesis, did not pull down significant amounts of miRISC from embryo extracts (Figure 1E middle and lower panel, let-7 lanes). Using similar capture experiments, we estimate that miR-51-56 and the CeBantam families program 13% and 9%, respectively, of the ALG-1/2 pool in ME preparations (Figure 1E, lower panel & Table). We conclude that a few abundant miRNA families occupy a large fraction of miRISC in C. elegans embryos.
Table 1. Comparative proteomic analysis of embryonic miRISCs.
MudPit analysis of proteins interacting with α-miR-35 and α-miR-52 2’-O-Meoligonucleotides in wild-type (N2) C. elegans embryonic extracts. Identified genes are listed along with their protein description and corresponding peptide coverage (%). The number of times the protein was detected in independent pull downs is indicated in parentheses. Interactions were confirmed by western blot for those proteins with available antibodies (check-marks). ND: not detected. Related data in Figure S2.
| Gene | Protein Description | % Peptide coverage (# Independent detection) |
Western | |
|---|---|---|---|---|
| α-miR-35 | α-miR-52 | |||
| C06G1.4 | AIN-1 (GW182 homolog) | 25.7 (5/5) | 14.0 (4/4) | √ |
| B0041.2 | AIN-2 (GW182 homolog) | 18.5 (5/5) | 11.2 (4/4) | √ |
| K12H4.8 | DCR-1, Dead box helicase/RNaseIII | 16.9 (5/5) | 2.0 (4/4) | √ |
| F48F7.1 | ALG-1, Piwi/PAZ domain | 9.7 (5/5) | 5.2 (4/4) | √ |
| T07D3.7 | ALG-2, Piwi/PAZ domain | 20.1 (5/5) | 6.8 (4/4) | √ |
| R10E4.2b | Tag-310, RRM domain | 24.7 (3/5) | 10.3 (1/4) | |
| W07B3.2 | GEI-4, Coiled-coil domain | 11.0 (3/5) | 5.4 (1/4) | |
| T20G5.11 | RDE-4, dsRBD | 14.0 (2/5) | 22.1 (1/4) | √ |
| R09B3.3 | Rna15 subunit homolog | 32.9 (2/5) | 32.9 (1/4) | |
| F58B3.7 | G patch/RRM domain | 10.5 (2/5) | 7.0 (1/4) | |
| Y23H5A.3 | Novel | 7.8 (4/5) | ND | |
| EEED8.1 | MEL-47, RRM domain | 8.4 (3/5) | ND | |
| Y73B6BL.6 | SQD-1 (HRP-1 subunit homolog) | 19.3 (3/5) | ND | |
| R10E9.1 | MSI-1 (HRP-1 subunit homolog) | 12.8 (3/5) | ND | |
| R74.5a | ASD-1, RRM domain | 6.4 (3/5) | ND | |
Comparative Proteomic analysis of embryonic miRISCs
To investigate whether abundant maternal and zygotic miRISC complexes are composed of similar machineries, we used Multi-Dimensional Protein Identification Technology (MuDPIT) (Wu and MacCoss, 2002) to identify proteins that copurify with miR-35-42 and miR-51-56-miRISC. A set of 15 proteins were identified in at least 3 independent capture experiments, but were never detected in either mock purifications (beads alone) or using a matrix directed at a non-specific miRNA (hsamiR-16) (Table 1). Ten of the interacting proteins were detected in at least 1 capture experiment for both the miR-35-42 and miR-51-56-miRISC affinity matrices. Known miRISC components (ALG-1, ALG-2, AIN-1, AIN-2) were detected in all affinity purifications for both miR-35-42- and miR-51-56-directed matrices (5 out of 5 miR-35 & 4 out of 4 miR-52 captures). Interestingly, DCR-1 was detected in all fractions recovered with both matrices, and its interacting partner RDE-4 (Tabara et al., 2002) was also detected, although less consistently (2 out of 5 miR-35 & 1 out of 4 miR-52 captures). This observation suggests that, as in mammalian and Drosophila, C. elegans DCR-1 not only associates with the pre-miRNA maturation machinery, but is also a component or the holo-RISC complex (Pham et al., 2004). The capture of these six proteins was further confirmed by western blot (Figure S2).
Interestingly, among the detected interactions, TAG-310, SQD-1, and MSI-1, all encode tandem RRM domain proteins, and were also previously detected in AIN-1/2 immunoprecipitates (Ding et al., 2005). This raises the possible implication of a new family of proteins in the miRISC. For five of the interacting proteins (Y23H5A.3, MEL-47, SQD-1, MSI-1, and ASD-1), an interaction was only detectable when using the miR-35-42 capture matrix. Although this may reflect differences in the composition of the maternal and zygotic miRISCs, it may also be a consequence of different sensitivities for capture with the two matrices, or a consequence of the less-than-quantitative detection using MuDPIT. Nevertheless, as 10 out of 15 of the consistently detected interactions are common between the two capture matrices, our analysis suggests that the maternal and zygotic miRISCs are composed of similar components. This similarity is further supported by the functional analyses provided below.
Cell-free silencing by maternal and zygotic miRNAs
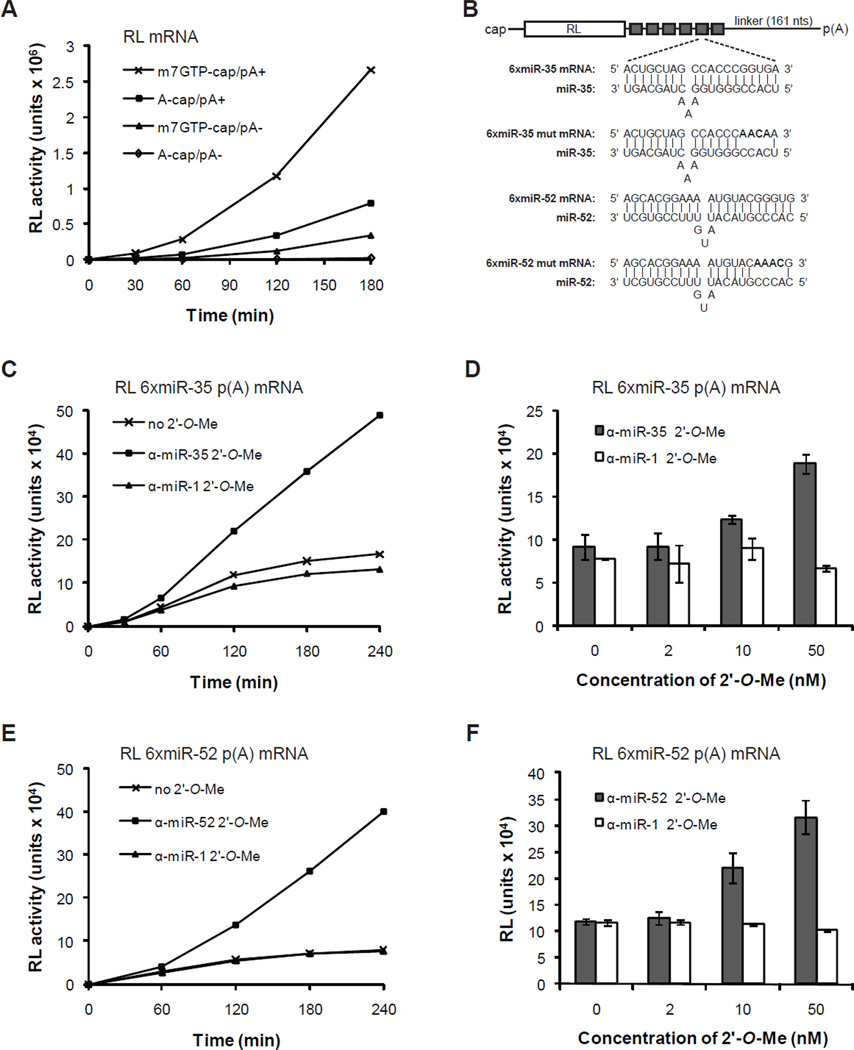
To investigate the mechanism of silencing employed by the miR-35-42 and miR-51-56 families, we developed a cell-free translation system from C. elegans embryos (see Materials and Methods). Using a Renilla reniformis luciferase (RL) reporter mRNA, translation in our system was heavily dependent on 3’ poly(A) tail and 5’-m7GpppG-cap structures (Figure 2A). Translation was most efficient for mRNAs bearing both a m7GpppG-cap and a poly(A) tail and was greater than the additive contributions of either a poly(A) tail or m7GpppG-cap alone (Figure 2A). Hence, this C. elegans cell-free translation system recapitulates functional synergy between the 5’ m7GpppG-cap and the 3’-poly(A) tail (Gallie, 1991).
Figure 2. Cell-free miRNA-mediated translational repression by maternal and zygotic miRNAs.
(A) Cap and poly(A) tail synergy in C. elegans embryonic extracts. The translation of 10 nM RL reporters bearing a physiological 5’ m7GpppG-cap, a 5’ ApppG-cap, and/or 3’ poly(A) tail was monitored over a 3-hour time-course. (B) Schematic representation of the RL reporter mRNAs used. Sequences of the miR-35- and miR-52-binding sites (6xmiR-35 and 6xmiR-52) and mutated binding sites (6xmiR-35 mut and 6xmiR-52 mut) are shown. (C,E) Translation time-course of RL 6xmiR-35 (C) and 6xmiR-52 mRNAs (E) with or without 50 nM specific (α-miR-35 (C), α-miR-52 (E)) or non-specific α-miR-1 2’-O-Me. (D,F) Dose-response translation de-repression by α-miR-35 (D) and α-miR-52 (F) 2’-O-Me for a 3h reaction. Each bar represents the mean from triplicate independent experiments and error bars indicate standard deviation.
To assay for miRNA-mediated silencing activity, we first examined the translation of RL mRNA fused to a synthetic 3’UTR encoding six copies of a miR-35-42 binding site (Figure 2B, 6xmiR-35 mRNA). Reporters were added to the translation system at a concentration of 1nM mRNA, which corresponds to 1/3rd-1/8th of the measured miR-35 concentration. Translation of 6xmiR-35 was dramatically impaired in comparison to RL mRNA (compare Figure 2A with Figure 2C), with activity slowing down and reaching a near-plateau at about 2 hours of incubation (Figure 2C). This repression was dependent on miR-35, since addition of increasing concentrations of 2’-O-Me antisense to miR-35 (α-miR-35) released the translation inhibition of 6xmiR-35 (Figure 2C, D). De-repression reached threefold when using 50nM, for a 3-hour (180 min) translation reaction (Figure 2C, D). Addition of the same concentrations of a 2’-O-Me oligonucleotide complementary to the non-related miR-1 did not affect the translation of 6xmiR-35 (Figure 2C, D). This concentration of 2’-O-Me oligonucleotide was therefore used for the additional experiments. Similar results were obtained using a miR-51-56 family reporter, and the corresponding 2’-O-Me inhibitor (compare Figure 2C with 2E). Thus, miRNA-mediated silencing by the C. elegans miR-35-42 and miR-51-56 families can be recapitulated in vitro.
Zygotic and maternal miRNAs direct deadenylation
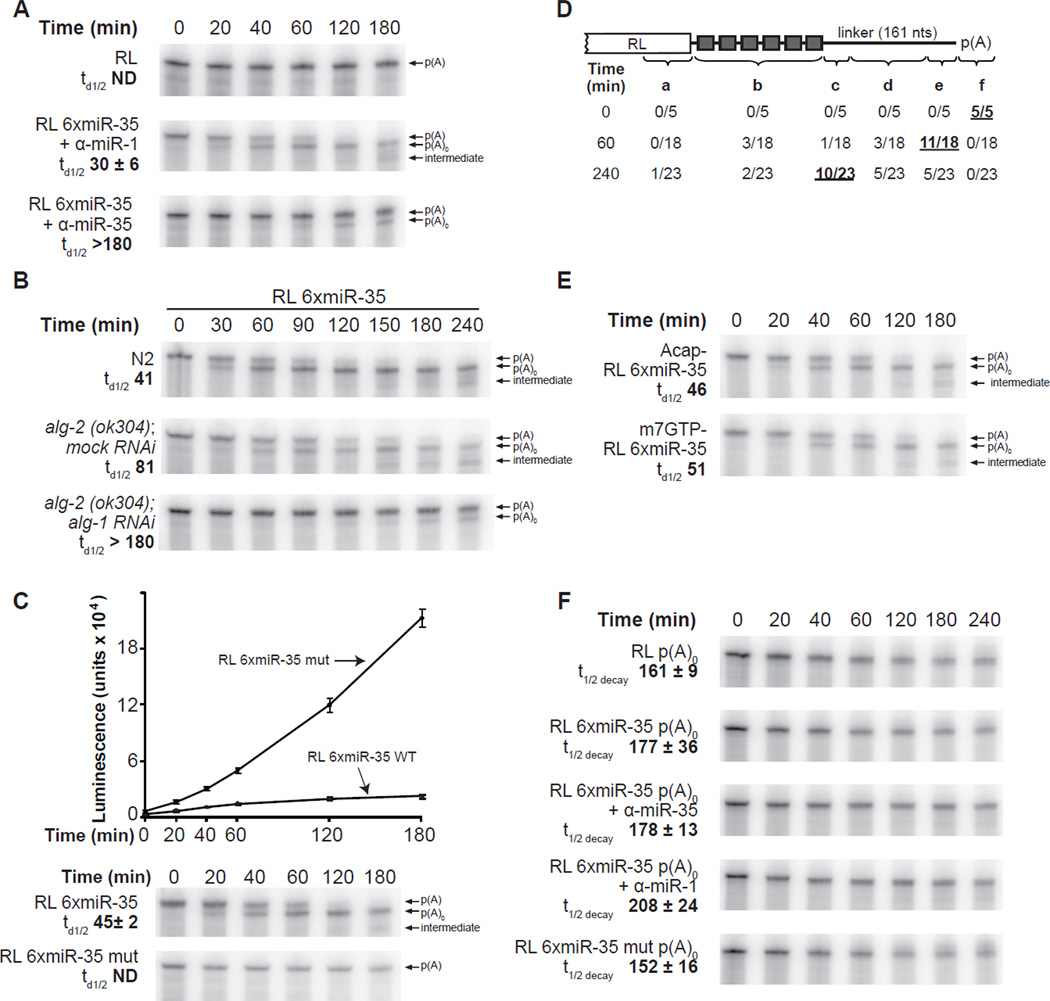
To determine the mechanism of miRNA-mediated silencing in our translation system, we examined the integrity of 32P-radiolabeled reporter mRNAs by polyacrylamide gel electrophoresis (Figure 3). The RL mRNA reporter was remarkably stable over the 3 hours of incubation (Figure 3A, RL panel). In contrast, the RL 6xmiR-35 reporter was completely converted to a second, shorter RNA species within 120 minutes (Figure 3A, B, C). Cloning and sequencing revealed that this RNA species corresponds to the deadenylated RL 6xmiR-35 reporter (see below). Quantification of multiple independent experiments, indicates that deadenylation reached half completion (td1/2) within the first 45 minutes of incubation, with slight variations between the extract preparations (for example, compare Figure 3A: td1/2 30 +/− 6 min, and Figure 3C: td1/2 45 +/− 2 min). Three series of control experiments indicate that the deadenylation of RL 6xmiR-35 mRNA is dependent on targeting by miR-35-RISC. Firstly, deadenylation was specifically blocked by the addition of α-miR-35 2’-O-Me (Figure 3A + α-miR-35 panel) but was insensitive to the addition of α-miR-1, α-miR-52, or α-let-7 2’-O-Me (+α-miR-1 panel, and not shown). Secondly, the deadenylation of RL 6xmiR-35 mRNA was substantially delayed in the alg-2(ok304); alg-1 RNAi extract, with less than half of the RL 6xmiR-35 reporter mRNA deadenylated after 4 hours (Figure 3B alg-2(ok304); alg-1 RNAi). Thirdly, RL 6xmiR-35 mut reporters, where miR-35 complementary sites have been altered (see Figure 2B for mutation design), were not deadenylated in the extract (Figure 3C RL 6xmiR-35 mut panel).
Figure 3. Embryonic miRISCs direct deadenylation but do not promote target decay in vitro.
(A) Deadenylation time-course of RL and RL 6xmiR-35 with the indicated 50 nM 2’-O- Me, and (B) of RL 6xmiR-35 in wild-type (N2), alg-2(ok304); mock (gfp) RNAi, or alg-2(ok304); alg-1 RNAi embryonic extracts. (C) Time-course of RL 6xmiR-35 WT and mutant translation and deadenylation. The same samples from each time-points were examined in translation (upper panel) and PAGE-autoradiography (lower panels). (D) Schematic representation of 3’RACE products from RL 6xmiR-35 at the indicated time-points. The indicated number of reads terminated a. within the RL open reading frame, b. between the miR-35 binding sites, c. within the first 40 nts 3’ of the miR-35 binding sites, d. within the middle region of the 3’UTR, e. within less than 25 nts 5’ of the poly(A) tail, f. within the poly(A) tail. (E) Deadenylation time-course of RL 6xmiR-35 mRNA bearing a m7GpppG cap or a ApppG cap. (F) Decay time-course of unadenylated reporters. Panels B and E are representative of two independent experiment, panels A, C and F are representative of triplicate experiments conducted using the same extract preparation. Half-deadenylation (td1/2) and half-life (t1/2decay) were quantified using ImageJ. +/− indicates standard deviation. Related data in Figure S3.
The RL 6xmiR-52 reporter was deadenylated with similar kinetics, and again processing was specifically prevented by a 2’-O-Me inhibitor (Figure S3 + α-miR-52 panel), or by mutation of the seed-complementary site (Figure S3, RL 6xmiR-52 mut panel, see Figure 2B for mutation design). We conclude that both miR-35-42 and miR-51-56 families direct potent and sequence-specific deadenylation in C. elegans embryonic extracts.
To precisely match the timing of translation repression with the fate of the reporter mRNAs, radiolabeled and polyadenylated RL 6xmiR-35 and RL 6xmiR-35mut reporters were subjected to a time-course of miRNA-mediated translation repression, and the same samples were examined for translation and PAGE-autoradiography (Figure 3C). Strikingly, the progression of deadenylation paralleled the course of translation repression of the reporters. Considered with the important contribution of the poly(A) tail for translation in our system (see Figure 2A), this observation suggests that deadenylation accounts for a major part of the translation repression observed in our system. It does not rule out, however, a minor contribution for additional mechanisms in the early phases of the target recognition by miRISC.
Embryonic miRISC does not mediate target decay in vitro
MiRNAs often direct the destabilization of target mRNAs. In our system, miRNA target reporters proved remarkably stable, even after being fully deadenylated (see Figure 3A–C p(A)0 labeled band, average t1/2 decay 183 min). This observation prompted us to ask whether targeting by embryonic miRISC results in target degradation in addition to deadenylation. Upon close examination of miR-35-42 and miR-51-56-deadenylated reporter autoradiograms, we noticed the appearance of shorter RNA species at or around 3 hours of incubation (Figure 3A, B & C, and Figure S3, see intermediate arrows). These intermediates accumulated in a miRNA- and/or deadenylationdependent manner, as cognate miR-35-42 and miR-51-56 2’-O-Me inhibitors, or genetic depletion of alg-1/2 prevented their accumulation (See intermediate arrow in Figure 3A, B & C). Sequencing of the recovered reporter mRNA population indicated that while the vast majority of reads terminated at, or very near the polyadenylation site at the 60 minutes time point (region e in Figure 3D), reads from clones recovered after 240 minutes clustered closely in the 3’ region bordering the miRNA-binding site repeats (Figure 3D region c). This indicates that the embryonic extract is capable of mRNA decay. The continuous removal of sequences further upstream of the poly(A) tail over time suggests the involvement of a 3’→5’ exonuclease activity.
A number of studies have suggested that miRNA-promoted decay involves a decapping step (Bagga et al., 2005; Behm-Ansmant et al., 2006). To address whether de-capping is involved in the slow turnover of the reporters, we examined the fate of ApppG-capped mRNAs that are not recognized by cellular de-capping enzymes (Grudzien-Nogalska et al., 2007; Wang et al., 2002). The time-course of deadenylation and decay for the ApppG-capped transcript closely mirrored the profile of the m7GpppG-capped reporters (Figure 3E), indicating that reporter decay does not require decapping in the extract. It also further supports the notion that mRNA decay occurs via a 3’→5’ activity in the embryonic extracts.
The observed decay could be due to a non-specific 3’→5’ activity acting on every reporter in the extract, or it could be the result of the miRISC actively promoting decay of the deadenylated reporters. Hence, we examined the stability of RL reporters lacking a poly(A) tail, but bearing functional (6xmiR-35) or non-functional (6xmiR-35mut) miRISC-binding sites (Figure 3F). RL 6xmiR-35 p(A)0 (t1/2 decay 177 +/− 36 min) was at least as stable as RL 6xmiR-35mut p(A)0 (t1/2 decay 152min +/− 16 min), or RL p(A)0 (t1/2 decay 161 +/− 9 min). Addition 2’-O-Me oligonucleotides slightly increased the stability of RL 6xmiR-35 p(A)0 but not in a sequence-specific manner, presumably due to competition for non-specific RNases in the extract. Similarly, the RL6xmiR-52-p(A)0 reporter was at least as stable as the RL6xmiR-52mut-p(A)0 (Figure S3B). Overall, these results indicate that the miRISC does not directly mediate the destabilization of the target mRNA but rather directs the generation of a stable deadenylated mRNA in the embryonic extract.
Pervasive deadenylation of embryonic miRNA targets
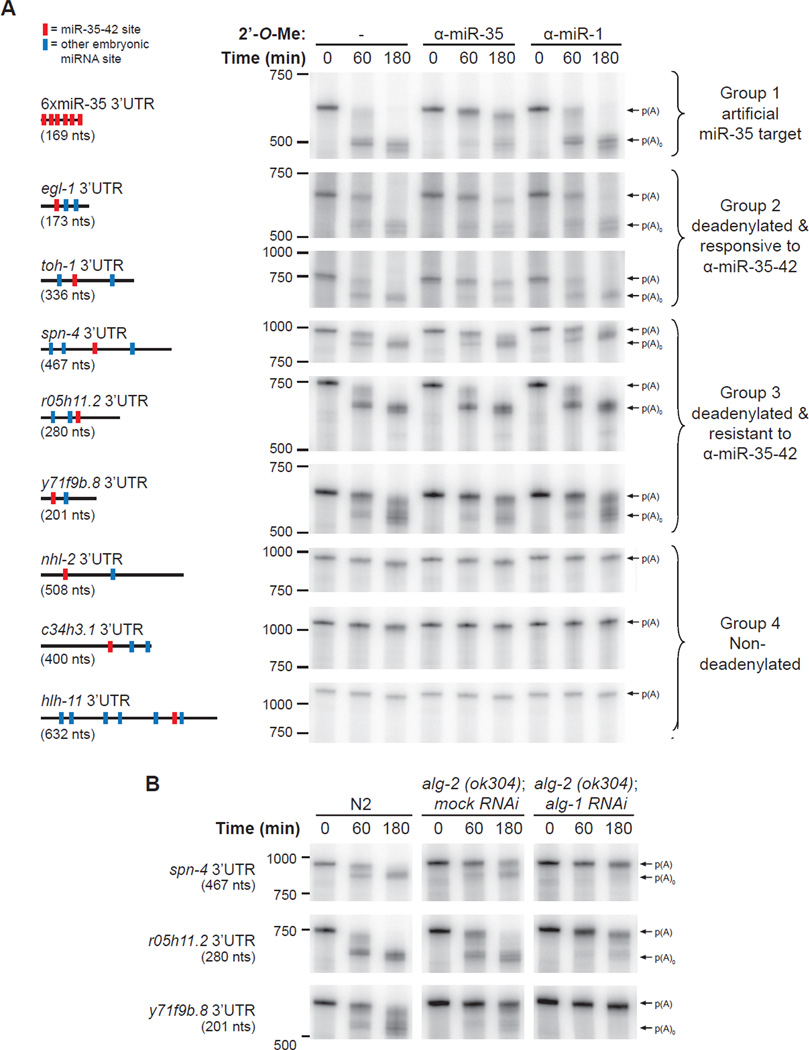
To obtain a measure of if and how natural 3’UTRs would undergo miRNA-mediated silencing in this cell-free system, we undertook a survey of mRNA deadenylation and decay by sampling the predicted miR-35-42 3’UTR targets. 3’UTRs of miR-35-42 targets (as per TargetScan (Friedman et al., 2009) and mirWIP (Hammell et al., 2008) predictions) were cloned as fusions to a truncated fragment of RL mRNA sequence to improve gel resolution in the deadenylation assay. Transcripts were then incubated in embryonic extracts, recovered and resolved by denaturing PAGE, as presented above. A control experiment with the 6xmiR-35 3’UTR, as well as a representative sample of the natural 3’UTRs surveyed is presented in Figure 4A.
Figure 4. miRNA-mediated deadenylation is prevalent in embryos.
(A) Deadenylation of natural 3’UTR reporters in embryonic extracts. 3’UTRs were fused to a truncated RL fragment (nts 764-936 (172-nt long)), for all UTRs screened except c34h3.1 where nts 491–936 where included. Reporters also encoded a 161-nt linker and a poly(A) tail of 87 nts. Schematic representation of the each 3’UTRs is depicted on the left (size in parentheses). Red bars denote miR-35-42 sites, blue bars denote sites for miRNAs that are known to be expressed in embryos (Stoeckius et al., 2009). Courses were realized with or without 50 nM 2’-O-Me (either α-miR-35 or α-miR-1(C-)). 3’UTRs are divided into four groups: 1. deadenylated artificial miR-35 target (6xmiR-35, (control)), 2. deadenylated 3’UTR targets that are responsive to α-miR-35, 3. deadenylated 3’UTR targets that are resistant to α-miR-35, 4. 3’UTRs not subjected to detectable deadenylation. (B) Time-course of group 3 in N2, alg-2(ok304); mock (gfp) RNAi and alg-2(ok304); alg-1 RNAi embryonic extracts. Experiments were reproduced at least twice in independent extract preparations. Related data in Figure S4.
Roughly half of the 3’UTRs examined were rapidly deadenylated in the extract, highlighting the prevalence of deadenylation as an embryonic mRNA regulation mechanism (Figure 4A, groups 2 and 3). The rate of deadenylation (compare spn-4 to r05h11.2 3’UTR for example) as well as the pattern (compare spn-4, and r05h11.2 to y71f9b.8 3’UTRs) varied broadly, indicating the 3’UTR-specific properties of the deadenylation process. Deadenylation of a subset of these targets was substantially blocked by incubation with the α-miR-35 2’-O-Me inhibitors but not the α-miR-1 2’-O-Me inhibitor (group 2), indicating that deadenylation was dependent on miR-35. This subset includes the 3’UTR of the proapoptotic BH3-only homolog egl-1, and the toll-ish homolog toh-1 (Figure 4A, group 2). Since all of the natural 3’UTRs were also predicted to be targeted by additional embryonic miRNAs (see 3’UTR legend on left, blue crossbars), deadenylated target 3’UTRs were incubated in extracts depleted of ALG-2 or both ALG-1 and −2 (Figure 4B for examples, also see Figure S4 for a control of the extract translation activity). Strikingly, depletion of both ALG-1 and ALG-2 together prevented deadenylation for all deadenylated targets screened thus far, including group 3 targets that were resistant to α-miR-35. These data indicate the involvement of embryonic miRISCs in the deadenylation of an important variety of natural 3’UTR targets.
Target deadenylation requires miRISC cooperation
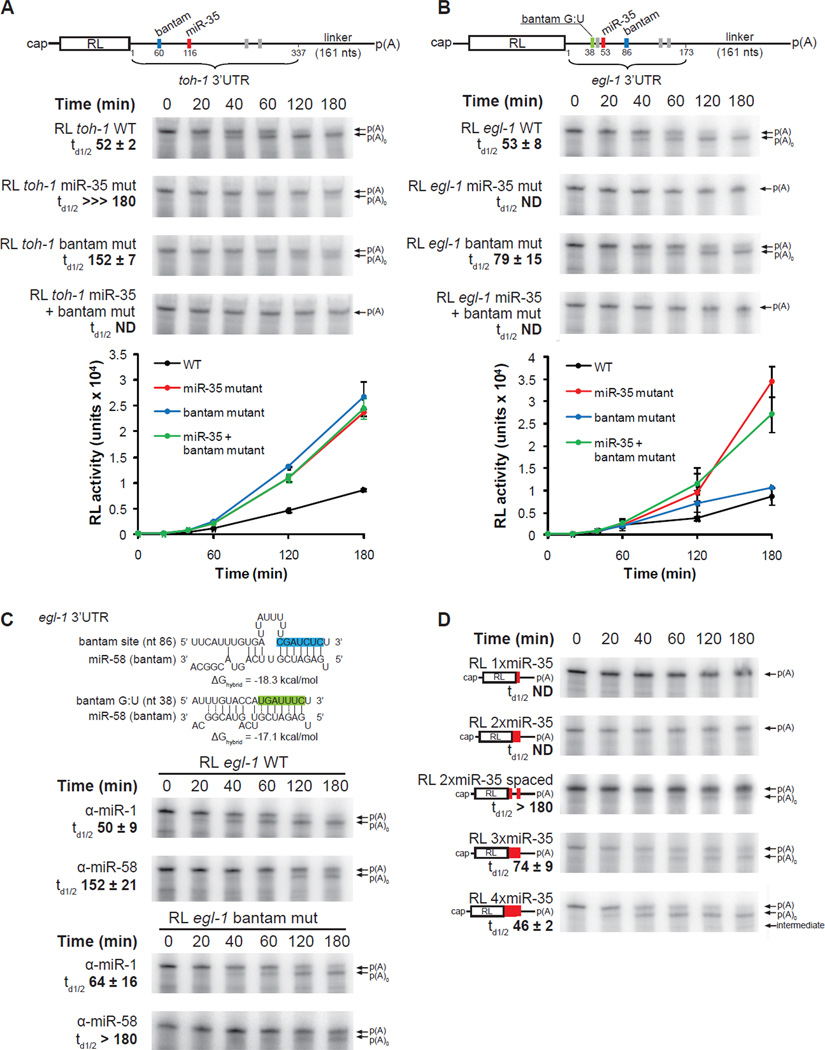
To better understand how miR-35-42 miRNAs direct deadenylation and repress translation, we further characterized the properties of the toh-1 and egl-1 3’UTRs (Figure 5A–C). According to bioinformatic predictions using the TargetScanWorm program (release 5.1) the 3’UTRs of toh-1 and egl-1 encode four and five miRNA-binding sites, respectively. Among those, the sites for the miR-35-42 and CeBantam families (Figure 5A, B, colored boxes on UTR legends) match miRNAs that are detectable in the early embryo. The remaining sites (grey boxes) match miRNAs that are undetectable in our system by northern blotting (Figure S5A), or that did not have any detectable functional implications when inhibited using 2’-O-Me (Figure S5B). Strikingly, deadenylation of reporters encoding these 3’UTRs was slowed by negating a single one of these two miRNA families (miR-35-42 or CeBantam) using sequence-specific 2’-O-Me inhibitors (Figure 4, group 2, and Figure S5B, C), suggesting that both miRNA families are required to initiate efficient deadenylation on these 3’UTRs. To assess the precise contribution of each miRNA-binding site, we mutated the predicted miR-35-42 and bantam-binding sites within the toh-1, and egl-1 3’UTRs and examined their effects on deadenylation, and translation repression assays (Figure 5A, B). For reporters containing the toh-1 3’UTR (RL toh-1 WT, td1/2 52 +/− 2 min), mutating either the miR-35-42 or the bantam site alone effectively impaired deadenylation (RL toh-1 miR-35 mut, td1/2 ⋙180 min; RL toh-1 bantam mut, td1/2 152 +/− 7 min), wereas no deadenylation could be detected when using the double mutant 3’UTR (RL toh-1 miR-35 + bantam mut). These reporters were also de-repressed to the same extent in translation assays (Figure 5A, bottom panel). While these data cannot rule out a weak and residual activity for the miR-35-42 site on its own, they indicate that the miR-35-42 and the CeBantam miRNA families cooperate synergistically in promoting the deadenylation and silencing on the toh-1 3’UTR.
Figure 5. Target deadenylation requires miRISC cooperation.
(A,B) Deadenylation and translation time-courses of RL toh-1 WT (A) and RL egl-1 WT (B) 3’UTR reporters in wild-type (N2) embryo extract. Detailed schematic representation of 3’UTR reporter mRNAs is shown. Red bars indicate miR-35-42 sites, blue and green bars indicate sites for CeBantam family members, and gray bars indicate sites for miRNAs that were not detected and/or had no detectable functional implications in our system (See also Figure S5). (C) (Top) Pairing of the the egl-1 3’UTR miR-58 (bantam) sites; the site with canonical base-pairing in blue, and the non-canonical site containing G:U wobble base-pairing in green. (Middle and bottom) Deadenylation time-course of the RL egl-1 WT, and the RL egl-1 bantam mut mRNA (encodes mutations within the canonical bantam site) in the presence of 50 nM α-miR-58, or the negative control α-miR-1. (D) Deadenylation time-course of RL reporter mRNAs encoding 1 to 4 copies of miR-35 binding sites. The 2xmiR-35 spaced reporter contains two miR-35 separated by 29 nts. Translation and deadenylation assays were conducted as triplicate of independent experiments. Quantifications of time of half-deadenylation (td1/2) were realized using ImageJ. Error bars, and +/− indicate standard deviation. Related data in Figure S5.
In similar reporter assays, the 3’UTR of the egl-1 mRNA also mediated a potent translation repression, and a rapid deadenylation (Figure 5B, RL egl-1 WT, td1/2 53 +/− 8 min). Mutation of the miR-35-42 binding site on its own, or in combination with an additional mutation in the predicted bantam site at position 86, completely abrogated reporter deadenylation and translation repression (RL egl-1 miR-35 mut, and RL egl-1 miR-35 + bantam mut). Mutation of this bantam target site on its own, however, had only a mild effect on the course of deadenylation, and on translation repression (RL egl-1 bantam mut, td1/2 79 +/− 15 min). This observation first appeared surprising, as bantam-specific 2’-O-Me inhibitors efficiently inhibited the deadenylation, and derepressed the translation of the RL egl-1 WT reporter (Figure 5C upper panels, S5B & S5C). Further analysis of the egl-1 3’UTR using the mirWIP algorithm (Hammell et al., 2008) revealed a second, atypical bantam-binding site in the 5’ vicinity of the miR-35-42 binding site, starting at position 38 (Figure 5C, upper panel). This second site (named bantam G:U) base-pairs extensively with miR-58 (ΔGhybrid −17.1 kcal/mol, in comparison to −18.3 kcal/mol for the first bantam site at nt 38), includes four G:U wobble base pairs, two of which are located within the seed sequence region, and also features an extensive base-pairing (6 bp) with the 3’ sequence of miR-58. Interestingly, deadenylation of the RL egl-1 bantam mut reporter was specifically, and potently impaired by the presence of α-miR-58 2’-O-Me inhibitor (Figure 5C bottom panel). This suggests that the bantam G:U site accounts for a major part of the impact of bantam miRNAs on the egl-1 3’UTR. These observations also suggest that non-canonical miRNA-binding sites can contribute to the cooperativity between multiple miRNA-binding sites that is required for miRNA-mediated deadenylation.
Thus far, our results indicate that cooperation between at least two separate miRISC-binding sites in a natural 3’UTR is required to potentiate miRNA-mediated deadenylation. To better define this cooperation, we engineered reporter mRNAs bearing 1, 2, 3 or 4 miR-35-42 binding sites, and examined their fate in deadenylation assays (Figure 5D). Interestingly, deadenylation was not observed for the artificial reporters bearing one or two miR-35 target sites. However, increasing the distance between the miR-35 target sites from 5 to 29 nts in the 2xmiR-35 reporter resulted in a detectable, but modest deadenylation (see 2xmiR-35 spaced). Deadenylation was dramatically accelerated by additional miR-35-42 binding sites, with td1/2 74 +/− 9 min and 46 /− 2 min for 3xmiR-35 and 4xmiR-35, respectively (Figure 5D). A similar effect was observed when analogous (1x-4x) miR-51-56 family reporters were examined (Figure S5D). This effect was not the result of varying distances between the sites and the poly(A) tail, as all the reporters encode the same sequence between the last miRNA-binding site and the poly(A) tail, and shortening or doubling the distance to the poly(A) tail had, by comparison, only a minor effect on the course of deadenylation (Figure S5E). Altogether, these results demonstrate that miRISC cooperation is required to potentiate miRNA target deadenylation.
Discussion
Impact of embryonic miRNAs on mRNA polyadenylation and stability
Previous work indicates that miRNA-mediated deadenylation correlates with miRNA-directed destabilization. This has been particularly well supported in zebrafish and Drosophila embryos where a few abundant zygotic miRNA families drive deadenylation and rapid turnover of maternal mRNA targets, in a process required for a timely maternal-to-zygotic gene expression transition (MZT) (Giraldez et al., 2006). On this particular aspect, the in vitro properties of the C. elegans maternal and zygotic embryonic miRISCs appear to contrast. Even though the miR-35-42, miR-51-56, and CeBantam miRISCs directed rapid deadenylation of artificial and natural targets, the deadenylated mRNAs remained surprisingly stable. The slow 3’→5’ destabilization of mRNA targets in this cell-free embryonic system seems remained unaffected by alteration of the m7GpppG-cap structure, and was not directly promoted by miRISC recruitment. Consistently with miRNAs not promoting the destabilization of certain target mRNAs in vivo, neither toh-1 nor egl-1 mRNA levels were significantly increased in alg-2(ok304); alg-1 RNAi embryo (Figure S6). Transcriptional compensation for rapid miRNA-mediated decay appears unlikely, in particular for maternal miRNA targets, as gene expression in the early embryo is largely governed by maternally provided mRNAs and is under extreme transcriptional restriction (Seydoux and Fire, 1994). We hypothesize, instead, that miRNA-mediated deadenylation in the early C. elegans embryo is either completely uncoupled, or only conditionally coupled with target destabilization.
Uncoupling between deadenylation, decapping and decay in the maturing oocyte and in the early embryo may be essential to prevent the premature degradation of maternal mRNA targets that are co-inherited with highly abundant miRNAs. Such a biochemical condition might be a feature of P-bodies (a structure involved in miRNA-mediated silencing (Ding et al., 2005)) in the germline (Boag et al., 2008) and in the earliest phases of embryonic development (Gallo et al., 2008). A recent study, which revealed that P-bodies are inherited with -but are distinct from- germ granules and lack essential decapping activators in the early embryo, lends credence to this model. This property may, under certain conditions, allow for the de-repression and mRNA expression in a temporal manner via readenylation (See model in Figure 6, and Figure legends). Interestingly, somatic P-bodies ‘mature’ biochemically, and later acquire the LSM-1 & LSM-3 decapping activators (Gallo et al., 2008). In time, this maturation, and possibly other means of miRISC regulation could be key events to couple deadenylation with further decay, hence accelerating the degradation of miRNA targets.
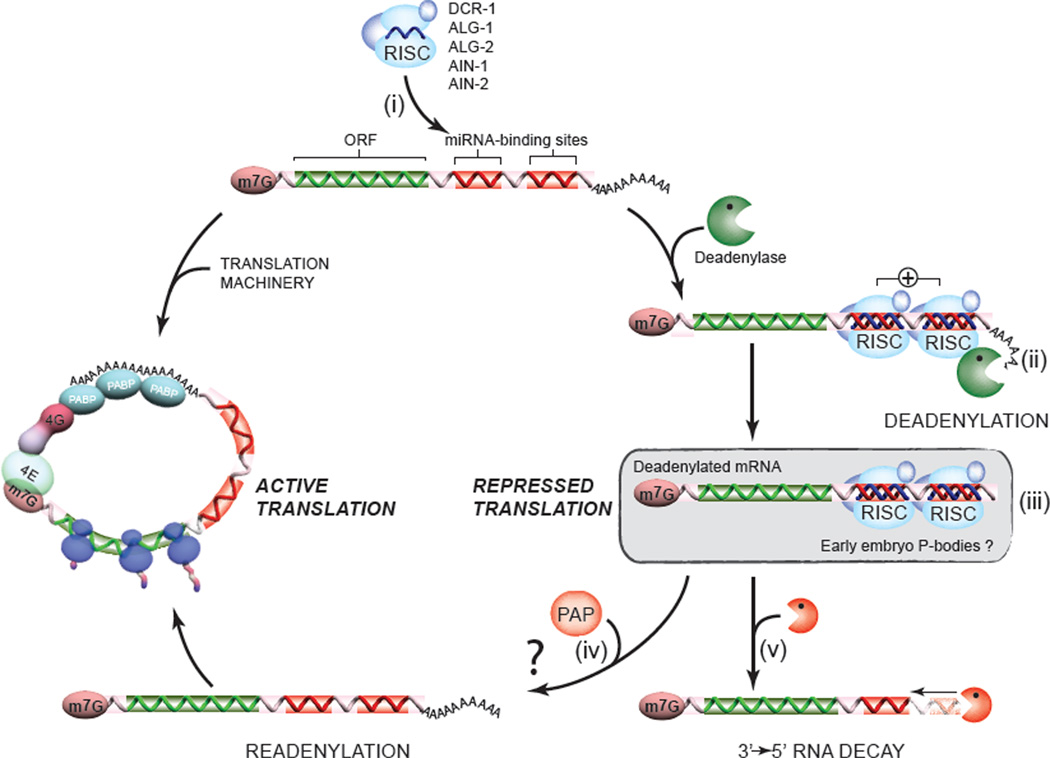
Figure 6. A model for the deadenylation and decay of early embryo miRNA targets.
The miRISC complex (ALG-1/2, AIN-1/2, DCR-1 and other accessory proteins), programmed by the abundant maternal and zygotic miRNA families, scans and recognizes mRNA targets (i). Through functional cooperation (indicated by a + sign), embryonic miRISC recruit and/or activate the deadenylase complex (CCR4/NOT was previously identified in a number of studies, including our own), and direct the rapid deadenylation of the target (ii). The stability of deadenylated mRNAs, and the association with GW182 proteins AIN-1 and AIN-2 on our target site baits in proteomics suggest that deadenylated targets may be protected and/or stored within the miRISC, or possibly within P-body-like structures (iii). One consequence of this stability is the possibility that deadenylation may be reverted, or outcompeted by poly(A) polymerase activities (PAP) (iv). Although this last hypothesis remains to be tested, evidence for competing deadenylation and polyadenylation activities exists in paradigms such as the germline and in the early embryo (Goldstrohm and Wickens, 2008). Finally, a fraction of the deadenylated mRNA pool may be decayed through a slow 3’→5’ route (v). This destabilization could be accelerated by the recruitment of decapping machinery by the miRISC, for example (See Discussion).
3’UTR-specific modulation of miRNA-mediated silencing outcomes
The survey of 3’UTR targets of the miR-35-42 family unveiled the direct, and potentially broad impact of miRNAs on the deadenylation of embryonic mRNAs. The synergistic contribution of neighboring RISC-binding sites on silencing had been noticed through the early studies of artificial reporters in transfection assays, and through genome-wide bio-informatic studies (Grimson et al., 2007; Saetrom et al., 2007). The Grimson study even identified the distance between RISC-binding sites and the poly(A) tail as a significant parameter for the potency of silencing, but how these determinants altered the mechanism of miRNA-mediated silencing was unknown. Our embryonic system allowed a direct perspective on the mechanistic impact of this cooperation: we show that synergy between distinct miRNA-binding sites can drastically potentiate deadenylation.
Potentiation of deadenylation through miRISC cooperation appears to be a common feature of the two targets studied in details here: the tollish family member toh-1, and the BH3-only protein encoding egl-1. In this latter case, the biological implications of the collaborative regulation by multiple miRNA families are potentially immense for embryonic development. A finely tuned level of EGL-1 protein is thought to be the key to trigger apoptosis in a large number of cell lineages in C. elegans (Nehme and Conradt, 2008). Our observations also point to a striking evolutionary conservation of the role for miRNA in the regulation of apoptosis: CeBantam miRNAs, just like the Drosophila Bantam miRNA which down regulates hid (Brennecke et al., 2003), antagonize apoptosis. Curiously, regulation of egl-1 homologs by miRNAs also occurs in human and is often altered in cancer. Mammalian egl-1 homolog and pro-apoptotic Bim is a known target of the oncogenic miR-17-92 polycistron (Inomata et al., 2009; Ventura et al., 2008), and its protein partner BCL-2 is also heavily regulated by miRNAs including miR-15a, miR-16-1 (Calin et al., 2008; Cimmino et al., 2005) and miR-34 (Ji et al., 2009). Hence, coordinated regulation of the egl-1 transcript by maternal and zygotic miRNAs represents yet another aspect in the tight control of the BH-3 family of proteins in apoptotic cellular decisions.
Some observations in the 3’UTR functional survey may suggest that cooperation between cis elements in promoting deadenylation is not restricted to miRISC-binding sites. One example, the y71f9b.8 3’UTR encodes two miRISC-binding sites which match known embryonically-expressed miRNA families (i.e. the miR-35-42 & miR-72- 74). Yet, this 3’UTR drives efficient deadenylation, even when miR-35-42 is inhibited, although with a distinct, non-processive pattern (Figure 4). At this point, we cannot rule out that non-canonical miRISC-binding sites may have been missed in the predictions on y71f9b.8 3’UTR sequences. An attractive, and alternative possibility however is that miRISC-binding sites may cooperate with additional cis-acting sequences within the y71f9b.8 3’UTR to promote deadenylation. Such a possibility finds echoes in recent findings by the Ambros group indicating that RNA-binding proteins (Hammell et al., 2009) can be required to potentiate miRISC action on specific targets.
In closing, our survey suggests that an accurate assessment of miRNA-mediated silencing mechanisms requires a careful consideration of context- and 3’UTR-specific outcomes. The modulation of miRNA-mediated silencing mechanisms through miRISC cooperation, or through interactions with additional elements within UTRs could provide flexibility in adapting the function of miRNAs to different genetic environments such as the transcriptionally silent embryo and fully differentiated somatic cells.
MATERIALS AND METHODS
C. elegans Strains and RNAi
N2 was used as the wild-type strain. Alleles used: glp-4(bn2), fem-1(hc17), and alg-2(ok304) were cultured as in (Brenner, 1974). alg-2(ok304) animals were exposed to alg-1 RNAi or gfp RNAi (mock), starting with L3 larvae. RNAi was carried out as in (Fire et al., 1998; Timmons et al., 2001).
Construction of Plasmids
For the backbone of the reporters, the Renilla luciferase (RL) ORF was cloned in NheI-XbaI sites of pCI neo vector (Promega) and a poly(A) tail of 87 nucleotides was cloned into NotI/MfeI. See Supplemental Methods and Table S1 sections for details on the additional reporters.
Northern Analysis
Total RNA from animals taken at different stages was prepared using the TRIZOL (Invitrogen) method. Embryos from adults bearing 1–3 embryos per animal (EE) were harvested, and allowed to further develop for 6 hours at 17°C (ME), and 12 hours (LE) in M9 saline suspensions. Animals were also harvested as synchronous populations of L1, L4 and adult stages. 10 ug of total RNA were analyzed by northern as in (Duchaine et al., 2006).
Real-Time PCR
miR-35 real-time PCR analysis throughout C. elegans development was performed using methods described in (Raymond et al., 2005).
2’-O-Methyl (2’-O-Me) Pulldown
2’-O-Me pull down was done as described in (Hutvagner et al., 2004). 10 ^L of the beads were loaded on gel for western blot analysis with a polyclonal antibody against peptides in the C-terminal region of ALG-1 and ALG-2, rabbit polyclonal antibody against DCR-1, rabbit polyclonal antibody against RDE-4, and GFP as in (Duchaine et al., 2006).
Multidimensional Protein Identification (MudPIT) performed as described in (Duchaine et al., 2006).
Preparation of Embryonic Extracts & In vitro Translation Assays were performed as described in details in the Supplemental Materials and Methods section.
Deadenylation Assays were performed in the same condition as translation (See Supp. Methods section) using 1 ng radiolabeled RNA. Autoradiography was realized as in Fabian et al. 2009.
Supplementary Material
Acknowledgments
We thank Darryl Conte Jr., Maxime Bouchard, and Ahilya N. Sawh for their comments on the manuscript. We thank Noriko Uetani for her artistic assistance with the model. We thank Min Han for providing ain-2::gfp transgenic animals/worms and rat polyclonal AIN-1 antibody; Richard E. Davis for providing rat polyclonal DCP-2 antibody. This work was supported by Canadian Institute of Health Research (grants MOP 86577 and 93607), the National Sciences and Engineering Council of Canada (grant RGPIN 86577), the Canada Foundation for Innovation (CFI), and the Fonds de la Rercherche en Santé du Québec (Chercheur-Boursier Salary Award J.1) to T.F.D.
References
- Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and Other Tiny Endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol. 2008;182:543–557. doi: 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. Rna. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Stepinski J, Jemielity J, Zuberek J, Stolarski R, Rhoads RE, Darzynkiewicz E. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enzymol. 2007;431:203–227. doi: 10.1016/S0076-6879(07)31011-2. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Nehme R, Conradt B. egl-1: a key activator of apoptotic cell death in C. elegans. Oncogene. 2008;27(Suppl 1):S30–40. doi: 10.1038/onc.2009.41. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. Rna. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P, Heale BS, Snove O, Jr., Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Maaskola J, Colombo T, Rahn HP, Friedlander MR, Li N, Chen W, Piano F, Rajewsky N. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat Methods. 2009;6:745–751. doi: 10.1038/nmeth.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C.elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr Opin Mol Ther. 2002;4:242–250. [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.