Abstract
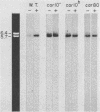
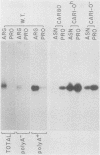
We isolated the CARI gene from Saccharomyces cerevisiae on a recombinant plasmid and localized it to a 1.58-kilobase DNA fragment. The cloned gene was used as a probe to analyze polyadenylated RNA derived from wild-type and mutant cells grown in the presence and absence of an inducer. Wild-type cells grown without the inducer contained very little polyadenylated RNA capable of hybridizing to the isolated CAR1 gene. A 1.25-kilobase CAR1-specific RNA species was markedly increased, however, in wild-type cells grown in the presence of inducer and in constitutive, regulatory mutants grown without it. No CAR1-specific RNA was observed when one class of constitutive mutant was grown in medium containing a good nitrogen source, such as asparagine. Two other mutants previously shown to be resistant to nitrogen repression contained large quantities of CAR1 RNA regardless of the nitrogen source in the medium. These data point to a qualitative correlation between the steady-state levels of CAR1-specific, polyadenylated RNA and the degree of arginase induction and repression observed in the wild type and in strains believed to carry regulatory mutations. Therefore, they remain consistent with our earlier suggestion that arginase production is probably controlled at the level of gene expression.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bossinger J., Cooper T. G. Molecular events associated with induction of arginase in Saccharomyces cerevisiae. J Bacteriol. 1977 Jul;131(1):163–173. doi: 10.1128/jb.131.1.163-173.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Atkins J. F., McGill C., Chow L. Identification and mapping of the transcriptional and translational products of the yeast plasmid, 2mu circle. Cell. 1979 Apr;16(4):827–839. doi: 10.1016/0092-8674(79)90098-9. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Magasanik B. Transcription of the lac operon of Escherichia coli. J Biol Chem. 1974 Oct 25;249(20):6556–6561. [PubMed] [Google Scholar]
- Cooper T. G., Whitney P., Magasanik B. Reaction of lac-specific ribonucleic acid from Escherichia coli with lac deoxyribonucleic acid. J Biol Chem. 1974 Oct 25;249(20):6548–6555. [PubMed] [Google Scholar]
- Dubois E. L., Wiame J. M. Catabolic synergism: a cooperation between the availability of substrate and the need for nitrogen in the regulation of arginine catabolism in Saccharomyces cerevisiae. Mol Gen Genet. 1978 Sep 8;164(3):275–283. doi: 10.1007/BF00333157. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Kindle K. L., Davidson N., Kindle K. L. The actin genes of Drosophila: a dispersed multigene family. Cell. 1980 Feb;19(2):365–378. doi: 10.1016/0092-8674(80)90511-5. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- MIDDELHOVEN W. J. THE PATHWAY OF ARGININE BREAKDOWN IN SACCHAROMYCES CEREVISIAE. Biochim Biophys Acta. 1964 Dec 9;93:650–652. doi: 10.1016/0304-4165(64)90349-6. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. Enzyme repression in the arginine pathway of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1969;35(2):215–226. doi: 10.1007/BF02219132. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. DNA rearrangements associated with a transposable element in yeast. Cell. 1980 Aug;21(1):239–249. doi: 10.1016/0092-8674(80)90131-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney P. A., Magasanik B. The induction of arginase in Saccharomyces cerevisiae. J Biol Chem. 1973 Sep 10;248(17):6197–6202. [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]