Abstract
Auxilin-1 is a neuron-specific membrane-binding protein involved in a late stage of clathrin-mediated endocytosis. It recruits Hsc70, thus initiating uncoating of the clathrin-coated vesicles. Interactions of auxilin-1 with the vesicle membrane are crucial for this function and are mediated via an N-terminal PTEN-like domain. We have used multiscale molecular dynamics simulations to probe the interactions of the auxilin-1 PTEN-like domain with lipid bilayers containing differing phospholipid composition, including bilayers containing phosphatidyl inositol phosphates. Our results suggest a novel, to our knowledge, model for the auxilin/membrane encounter and subsequent interactions. Negatively charged lipids (especially PIP2) enhance binding of auxilin to lipid bilayers and facilitate its correct orientation relative to the membrane. Mutations in three basic residues (R301E/R307E/K311E) of the C2 subdomain of the PTEN-like domain perturbed its interaction with the bilayer, changing its orientation. The interaction of membrane-bound auxilin-1 PTEN-like domain with negatively charged lipid headgroups results in nanoclustering of PIP2 molecules in the adjacent bilayer leaflet.
Introduction
Clathrin-mediated endocytosis enables extracellular material, including membrane receptors and ligands, to be imported into cells through the formation of clathrin-coated vesicles (1). It is involved in many plasma membrane functions and is important in the regulation of various signal transduction pathways (2). The formation of clathrin-coated vesicles starts with pit formation and the selection of cargo proteins, followed by the assembly of a clathrin lattice around the vesicle and subsequent cleavage from the plasma membrane. After disassociation from the membrane, the clathrin coat is removed and recycled (3,4), thus enabling the vesicle to travel to its intracellular target. Auxilin is a J-domain containing protein that binds to the newly budded clathrin-coated vesicles. It subsequently recruits the uncoating ATPase Hsc-70 (5,6). Thus, auxilin binding to the vesicle membrane is a key event that defines the timing of uncoating.
The neuron-specific auxilin-1 contains three main domains: an N-terminal PTEN-like domain responsible for membrane binding, a clathrin-binding domain, and a C-terminal J-domain (Fig. 1 A). X-ray diffraction studies have revealed the structure of the PTEN-like domain of auxilin-1 (7). As in the PTEN-protein itself (8), this domain is composed of phosphatase-like (PD) and C2 (sub)-domains. The C2 domain shares the same fold as many other membrane-binding C2 domains, but lacks the residues that are involved in Ca2+-mediated binding to the membrane of other C2 domains (9,10). This is also true of the PTEN C2 domain, suggesting that both C2 domains interact with membranes in a Ca2+-independent fashion, dominated by electrostatic interactions (11). The role of the PD domain of auxilin-1 in membrane binding is a little less clear. Unlike that in PTEN it is not catalytically active. However, simulation (12) and neutron reflectivity (13) studies of PTEN suggest a role of this domain in membrane binding in addition to catalysis per se. It is therefore important to understand the biophysical basis of the interactions of the PTEN domain of auxilin-1 with membranes.
Figure 1.
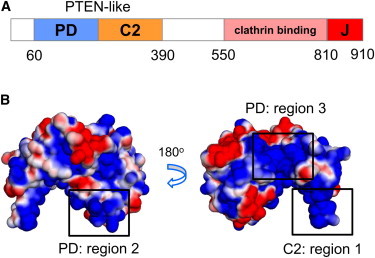
(A) Domain organization of auxilin-1 showing the location of the PD and C2 domains, which make up the PTEN-like domain, and of the C-terminal clathrin-binding and J domains. (B) Surface electrostatic potential representation of the auxilin-1 PTEN-like domain. The electrostatic calculation was performed using APBS (47) in PyMol (48). The electrostatic potential ranges from −1.5 kT/e (red) to +1.5 kT/e (blue).
Despite sharing a common fold, the sequence identity between the auxilin-1 PTEN-like domain (Protein Data Bank (PDB): 3N0A) and PTEN itself (PDB: 1DR5) is only 27%. Given the relatively low sequence identity, it is important to explore how the auxilin PTEN-like domain interacts with cell membranes, especially as it lacks the specific phosphatidyl inositol phosphate (PIP)-binding/catalytic site seen in the phosphatase-domain of PTEN. Examination of electrostatic surfaces (Fig. 1 B) reveals a basic protein surface likely to interact with the anionic bilayer that is present in both proteins (see the Supporting Material Fig. S1 A). In both proteins there is a net protein dipole, the positive end of which is directed toward the lipid-binding C2 domain. However, this dipole is much stronger for PTEN than for the auxilin PTEN-like domain. Furthermore, despite the overall conservation of fold, the lipid-binding loops of the C2 domain are substantially different between the two structures (7). In particular, loop 3 of the C2 domain of auxilin (residues 301–311; region 1 in Fig. 1 B) is longer than the corresponding loop in PTEN, and contains three positively charged residues, and two exposed hydrophobic side chains at its tip. In auxilin, loop 3 forms a twisted β-hairpin, whereas in PTEN this loop is shorter and unstructured (Fig. S1 B). These differences are likely to be significant, especially as previous simulation studies of PTEN indicate a role of this loop in interacting with the lipid bilayer (12).
Auxilin binding to clathrin-coated vesicles is thought to occur via interactions of the PTEN-like domain with PIP lipids in the vesicle membrane (7). However, the molecular details of auxilin/lipid interactions, including the binding mechanism and orientation of the protein, relative to the bilayer remain uncertain. In particular, we wish to know whether both the PD and C2 domains bind to PIPs, and how this may be related to the lipid composition of the vesicle membrane.
Molecular dynamics (MD) simulations provide a computational tool for probing membrane proteins and their interactions with the lipid bilayer (14). There have been a number of MD simulations of the interactions of (e.g., C2 (15)), and other (16–18) lipid-recognizing domains with membrane surfaces. Comparisons with experiment for (e.g., PH (19)) and talin (20) domains have shown that MD simulations can reveal specific protein/lipid interactions, and this approach has been used to explore a model of the PTEN/PIP3 complex in a bilayer (12).
Here, we use a multiscale approach, combining coarse-grained (CG) and atomistic (AT) MD simulations (21), to define and analyze the interactions of the PTEN-like domain of auxilin-1 with PIP-containing lipid bilayers. Our results suggest a novel, to our knowledge, mechanism that explains how the auxilin-1 PTEN-like domain binds to membranes, and in addition reveal the formation of PIP nanoclusters in the bilayer leaflet adjacent to a bound auxilin-1 PTEN-like domain.
Methods
CG-MD simulations
The crystal structure of the PTEN-like domain of auxilin-1 (PDB: 3N0A) was converted to a CG representation providing the initial structure for the simulations. In the MARTINI CG model used (22,23) four heavy atoms (i.e., nonhydrogen) are combined as one CG particle. Four CG particle types exist: charged (Q), nonpolar (N), apolar (I), and polar (P). P particles represent water-soluble neutral groups, C represents hydrophobic atom groups, Q represents charged atom groups, and N represents a mixed (polar/apolar) group of atoms. Additional particle subtypes (0, d, α, and dα for N and Q particles) exist for assigning H-bond capabilities. An elastic network was applied to all backbone particles using a cutoff distance of 7 Å to model secondary and tertiary structure. The missing region on the PD domain (residues 76–87) was added and/or mutations were made using Modeler (http://salilab.org/modeler/) (24). During the mutations or the modeling the structure of the protein was kept restrained. The CG protein was displaced 120 Å away from a preformed lipid bilayer, the lipid compositions of which are described in Table 1. Bilayers were constructed by performing separate CG-MD self-assembly simulations. In these simulations the lipids were randomly added to a simulation box and a simulation of duration 200 ns was performed for the bilayer to be formed. All the bilayers contained ∼350 lipids. The CG-MD simulations were performed using the MARTINI force field (23). All systems were solvated with CG water and subsequently counter ions were added to neutralize the system. After solvation, the systems were energy minimized for 200 steps, and equilibrated for 5 ns with the protein backbone particle restrained (force constant = 10 kJ/mol/Å2). An ensemble of 10 production simulations was run for each system with all simulations starting from the same configuration but with different initial velocities. All simulations were run for 1.5 μs.
Table 1.
Summary of simulations
| Simulation | Bilayer | Protein | Duration (ns) |
|---|---|---|---|
| PC | POPC | 3N0A | 10 × 1500 |
| PS15 | POPC(85%) + POPS(15%) | 3N0A | 10 × 1500 |
| PS30 | POPC(70%) + POPS(30%) | 3N0A | 10 × 1500 |
| PS60 | POPC(40%) + POPS(60%) | 3N0A | 10 × 1500 |
| PIP2 | POPC(85%) + PIP2(15%) | 3N0A | 10 × 1500 |
| loopPIP2 | POPC(85%) + PIP2(15%) | 3N0A + loop (resid:76–87) | 5 × 1500 |
| mutPIP2 | POPC(85%) + PIP2(15%) | 3N0A; R301E/R307E/K311E | 10 × 1500 |
| mutPIP2-2 | POPC(85%) + PIP2(15%) | 3N0A; R190E/R206E/R207E | 10 × 1500 |
| PIP2-AT | POPC(85%) + PIP2(15%) | 3N0A | 4 × 60 |
| PIP3 | POPC(85%) + PIP3(15%) | 3N0A | 10 × 1500 |
| loopPIP3 | POPC(85%) + PIP3(15%) | 3N0A + loop (resid:76–87) | 5 × 1500 |
| mutPIP3 | POPC(85%) + PIP3(15%) | 3N0A; R301E/R307E/K311E | 10 × 1500 |
| PIP3-AT | POPC(85%) + PIP3(15%) | 3N0A | 2 × 60 |
All simulations were via CG-MD except PIP2-AT and PIP3-AT, which were atomistic.
Recent experimental data (7) suggest that PIP lipids regulate the formation of an auxilin/membrane complex and therefore the main focus of this study was the association of auxilin-1 PTEN-like domain with PIP lipids. In vivo, PIP2 lipids are found in higher concentrations in the plasma membrane than are other PIPs (25) and therefore they were a major focus of the current study. Other types of PIP (i.e., PIP3) were also explored, to examine the possible specificity of association of auxilin/bilayer interactions. We also used a phosphatidyl serine (PS) containing bilayer to study how anionic lipids other than PIPs might influence the association of auxilin with a bilayer. All of the simulation systems are summarized in Table 1. In our simulations PIP2 lipids had a charge of −5 and PIP3 had a charge of −7.
Simulations were performed using GROMACS 4.5.1 (www.gromacs.org) (26). A Berendsen thermostat (27) was used for temperature coupling (coupling constant of 1.0 ps; reference temperature 323 K) and a Berendsen barostat was used for pressure coupling (coupling constant of 1.0 ps, compressibility value of 5.0 × 10-6 bar-1, reference pressure 1 bar). The integration time step was 20 fs for CG simulations. Lennard-Jones interactions were shifted to zero between 9 Å and 12 Å and Coulombic interactions between 0 and 12 Å, respectively. Conversion of resultant CG to atomistic systems was made using a fragment-based approach (21). In this approach the lipids are reconstructed by aligning AT lipid fragments with the corresponding CG templates from an energy-minimized library of atomistic lipid conformations. The protein backbone is reconstructed by the CG backbone trace using PULCHRA (28). Aromatic side chains in the protein are reconstructed by alignment of atomistic ring structure fragments with the CG aromatic chains. All other side chains are reconstructed using Modeler (29,30).
AT-MD simulations
The initial atomistic systems, following conversion from the final state of the corresponding CG systems, were energy minimized and equilibrated for 1 ns with the protein Cα atoms restrained (force constant = 10 kJ/mol/Å2). Unrestrained AT-MD simulations of 60 ns were run, starting from the same initial configuration but with different initial velocities. The simulation temperature was 323 K. The bilayer contained 350 lipids (295 palmitoyloleoyl phosphatidylcholine (POPC) and 55 PIP2 or PIP3; Table 1). At-MD simulations were performed again using GROMACS 4.5.1 (www.gromacs.org) and the GROMOS96 43a1 force field (31). The Parinello-Rahman barostat (32) and the Berendsen thermostat (27) were used for temperature and pressure coupling. The LINCS algorithm was used to constrain bond lengths (33). Long-range electrostatic interactions (i.e., those acting over a distance of >10 Å) were modeled using the particle mesh Ewald method (34). A 10 Å cutoff distance was also used for van der Waals interactions. Analyses used GROMACS (26,35), VMD (36), and locally written codes.
Results and Discussion
Interactions of auxilin-1 PTEN-like domain with membranes that contain PIP2 lipids
Examination of the electrostatic potential mapped onto the surface of the auxilin-1 PTEN-like domain (Fig. 1 B) reveals three highly positively charged regions: residues 240–245 and 299–316 of the C2 domain (region 1), residues 88–122 of the PD domain (PD; region 2), and residues 189–212 of the PD (region 3). The surface that includes C2: region 1 and PD: region 2 is located on one face (the lower) side of the protein. In contrast, PD: region 3 is located on a different (side) face of the protein. Therefore, visualization of surface electrostatics indicates that two possible surfaces might be involved in the membrane binding site. To pursue this further, and to include considerations of local rearrangement of lipids in response to protein binding, we have used simulations to test this initial hypothesis concerning possible binding modes.
It is thought that the auxilin-1 PTEN-like domain may interact with membranes in a PIP-dependent fashion (7). To this end, CG-MD simulations were performed to examine the interaction of the auxilin-1 PTEN-like domain with PIP2-containing bilayers (Table 1). A PIP2 concentration of 15% was used to mimic that in recent experimental studies of the binding of the auxilin-1 PTEN domain to liposomes (7). The protein was initially positioned ∼120 Å away from the bilayer, and an ensemble of 10 simulations was performed with the protein starting from the same initial position but with different initial velocities. During the simulations the protein initially diffuses in the aqueous environment and subsequently binds to the bilayer (Fig. 2 A). The presence of PIP2 among the bilayer lipids resulted in the association of the PTEN-like domain with the bilayer in 10 out of 10 simulations (each of duration 1.5 μs; Fig. S3 A).
Figure 2.
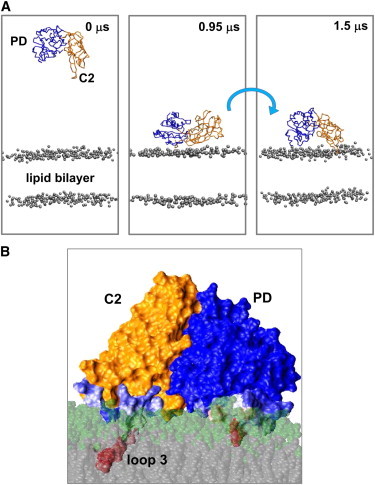
(A) Snapshots from a CG-MD simulation of the auxilin-1 PTEN-like domain with a lipid bilayer (the gray spheres representing the phosphate groups) composed of 15% PIP2 and 85% POPC (i.e., simulation PIP2, see Table 1). (B) Snapshot of the lipid-bound auxilin-1 PTEN-like domain, from end of a simulation in the presence of 15% PIP2. The lipid interacting residues are color coded on a pale blue (infrequent interactions) to red (frequent interactions) scale.
Interestingly, in 5 out of 10 simulations in which the protein was associated with the bilayer it bound initially via its C2 domain loops (res: 240–245 and 301–316) and subsequently the PD (res: 53–60 and 88–99) interacted to form a final stable complex. In the other five simulations, which yielded a protein/bilayer complex, the auxilin-1 PTEN-like domain bound to the bilayer initially in a nonspecific mode. Following the initial encounter the protein reoriented by rotation at the membrane surface (compare the 0.95 and 1.5 μs snapshots in Fig. 2 A) to form more specific interactions with those residues implicated by mutagenesis in binding (i.e., residues 53–60 and 88–99 of the PD domain, and 240–245 and 301–316 of the C2 domain (7)). This orientation and interactions lasted for the remainder of the simulations. This initial encounter followed by rotation can be demonstrated via monitoring the distance of the protein from the bilayer and also the angle between the plane of the protein and the plane of the lipid bilayer as a function of time (see Fig. 3, A and B, Fig. S4, and Fig. S5). The rotation to its final position appears to be driven by the formation of lipid interactions with the C2 domain binding loops (residues 240–245 and 301–316; see Fig. 3 C). A Ca2+- and PIP2-mediated rotation mechanism has previously been suggested for the membrane binding and penetration of the C2 domain of synaptotagmin (37). Because the loops on the C2 domain facilitate interactions with the membrane is also thought to be the case for other Ca2+-independent C2 domains, such as those in PTEN (38) and in Rsp5 (39).
Figure 3.
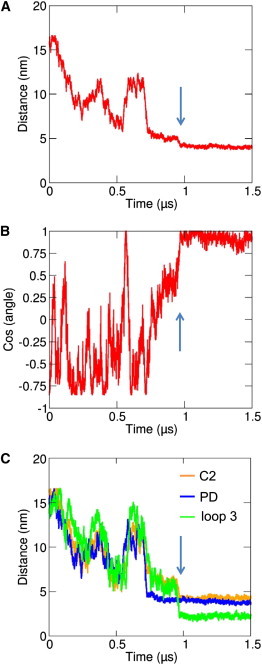
Progress of one of the CG-MD simulations in the presence of 15% PIP2 (PIP2, see Table 1). (A) Shows the distance between the center of mass of the protein and the center of mass of the bilayer as a function of time; (B) shows the cosine of the angle made between the protein plane (defined by residues 214, 216, and 328) and the bilayer plane. This angle is equal to 0° (and hence the cosine is equal to 1) if the protein is in the correct binding orientation of the PTEN-like domain. (C) Shows the distances between the center of mass of the lipid bilayer and centers of mass of loop 3 (green), the C2 domain (orange), and the PD domain (blue) as a function of time. The vertical arrows indicate the approximate time of the reorientation of the protein as illustrated in Fig. 2A and discussed in the text.
During the simulation the auxilin-1 PTEN-like domain interacted preferentially with the negatively charged lipids (i.e., PIP2) and in the final orientation the parts of auxilin that interacted with the lipids were the residues suggested previously to form specific interactions (Fig. S6, A–C). Closer examination of the protein/lipid interactions by calculating the contacts (using a cutoff distance of 7.5 Å) between the lipids and the protein reveals that residues 301–313, 114–115, 243–244, and 267–268 made >50% of the contacts with the lipids during the simulations (Fig. 4 A and Fig. S6 A). In particular, loop 3 of the C2 domain penetrates into the phosphate-containing region of the lipid bilayer (Fig. 4 B). Therefore, these residues may be crucial in determining the orientation of auxilin-1 PTEN-like domain with the membrane.
Figure 4.
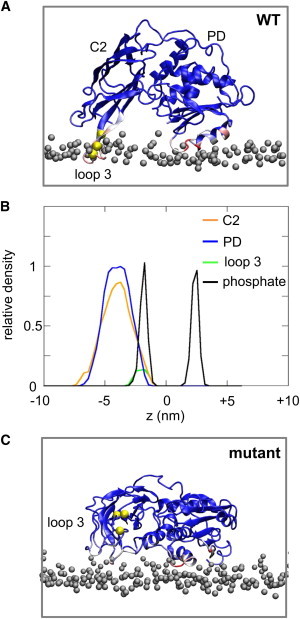
(A) Normalized average number of contacts between the auxilin-1 PTEN-like domain and the lipids mapped onto the final snapshot of the PIP2-AT (see Table 1) simulation. Blue indicates a low number, white indicates a medium number, and red a large number of contacts. The bilayer headgroups are shown as gray spheres and the residues that were mutated in the mutPIP2 (see Table 1) simulations are shown as yellow spheres. (B) Density profiles along the membrane normal for the C2 (orange), PD (blue), and loop 3 (green) regions relative to the positions of the lipid phosphate groups (black), averaged from the four PIP2-AT simulations. (C) Normalized average number of contacts between a mutated (R301E/R307E/K311E) auxilin-1 PTEN-like domain and lipids mapped onto the final snapshot of the mutPIP2 (see Table 1) simulation.
To quantify the behavior of the lipids around auxilin, radial distribution functions of the lipids relative to the protein were calculated for each leaflet separately (Fig. S7, A and B). Interestingly, in the leaflet that is adjacent to the protein a large peak at 0.5 nm for the PIP2 lipids was observed, indicating a clustering of the PIP lipids around the auxilin-1 PTEN-like domain. The same analysis for the POPC lipids revealed a much lower peak in the same region. In the opposite leaflet (i.e., without a protein component) no clustering was observed for the POPC or the PIP2 lipids (Fig. S11 B). Calculation of the number of contacts (using a cutoff distance of 7.5 Å) between the protein and the PIP2 lipids as a function of the angle between the protein and the bilayer suggested that the clustering of PIP2 lipids occurs after auxilin adopts a productive orientation relative to the bilayer (Fig. S8 A). Additionally, the bilayer tension upon protein binding did not change significantly (Fig. S8 B).
The previous simulations were performed using a CG model based on the x-ray structure (PDB: 3N0A) of the auxilin-1 PTEN-like domain, from which residues 76–87 of the PD domain are missing. If these residues were modeled back in simulation, similar results were obtained. In contrast, the simulation results were sensitive to mutation of key residues implicated in protein/lipid interactions (7). In particular, we examined a triple mutant (R301E/R307E/K311E) in which the three basic residues of loop 3 were charge reversed and which has been shown to eliminate recruitment of auxilin to coated pits and to remove binding to PIP2-containing liposomes (7). Perhaps surprisingly, simulations of this mutant protein in the presence of 15% PIP2 resulted in 8 out of 10 simulations forming an interaction with the bilayer. This suggests that the second positively charged region identified earlier using the surface electrostatic potential calculation (region 2 in Fig. 1) may localize the protein to the lipid membrane. However, calculation of the angle between the plane of the protein and the plane of the lipid bilayer (Fig. 3 B and Fig. S5) suggested that at the end of all eight simulations, which yielded a mutant auxilin/bilayer complex, the protein was in a perturbed orientation at the bilayer surface relative to the wild-type (WT) protein (Fig. 4 B). Closer examination of the orientation of auxilin-1 PTEN-like domain during the simulation suggested that in the majority of the simulations, the protein interacted with the bilayer in a nonspecific orientation and subsequently rotated to its final perturbed orientation. In the final orientation the protein interacted with the membrane via its second positively charged surface (PD: region 3 in Fig. 1 B; Fig. S6 A). Calculation of the radial distribution function of the lipids relative to the protein suggested a higher density of PIP2 lipids around the protein (Fig. S7, C and D), compared to POPC, however this difference in the radial distribution functions between the anionic and zwitterionic lipids was somewhat lower compared to the simulations with the WT protein (above). Therefore, after mutation of the positively charged surface on C2 region 1, which was suggested previously to drive auxilin to the correct orientation relative to the membrane, the orientation of auxilin-1 PTEN-like domain is driven by its second positively charged patch, resulting in a nonproductive orientation of auxilin-1 PTEN-like domain. This again highlights the crucial role of the C2 loop (i.e., residues 240–245 and 301–316) in orienting auxilin-1 PTEN-like domain relative to the membrane. To further test if the three basic residues in loop 3 specifically drive the productive association of auxilin with the bilayer, simulations with a control triple mutation were performed. This set of mutations was located in region 2 (i.e., away from the WT auxilin/bilayer interface; see simulation mutPIP2-2 in Table 1). In this case auxilin was able to associate with the bilayer with the same mechanism to the one described for the WT auxilin with either a POPC/PIP2 or a POPC/PIP3 bilayer (see Fig. S5 and Fig. S9 A–C). This augments our previous observation that the triple mutation in loop 3 reveals the productive mode of association of auxilin and a model PIP-containing membrane.
To refine our model of auxilin-1 PTEN-like domain/lipid interactions, a final snapshot from the CG-MD simulations in the presence of 15% PIP2 was converted to atomistic representation, and subsequent short AT-MD simulations were performed (duration 2 × 60 ns). In these AT simulations relatively small movements of C2 domain relative to the PD domains were observed, corresponding to rotations of 8.9 ± 1.5° for PIP2-containing bilayers. This suggests that the interface between the PD and C2 domains is largely unchanged when the PTEN-like domain interacts with the membrane. Analysis of the lipid/protein interactions revealed that the interactions with the lipids are dominated by K243 to R245 (C2, loop 1) and by residues R301 to Q316 (C2, loop 3) and also include residues V53 to L61 and V88 to L100 of the PD domain (Fig. 4 A and Fig. S10 A). In particular, loop 3 of the C2 domain penetrates the lipid bilayer reaching into the hydrocarbon core of the bilayer (see Fig. 2 B; Fig. S10 D). The same analysis shows that the PD domain remained on the bilayer surface. Other membrane-binding proteins that contain a C2 domain, e.g., cPLA2 or PKCα, also have binding loops that penetrate to the membrane, although Ca2+-mediated interactions may also play a role in these cases (40–42). Closer examination of the contacts with the bilayer reveals that the same residues described previously formed the largest number of contacts. Calculation of the root mean-square fluctuation (RMSF) and the secondary structure changes during the simulations suggested that no changes in the secondary structure of the protein were observed during the AT-MD simulations and that the RMSF profile was as anticipated with the core secondary structure elements having a lower RMSF than the loops (Fig. S10, B and C). The final snapshots from two additional CG systems, where the auxilin-1 PTEN-like domain was productively associated with the bilayer, were also subjected to AT simulations. The results were identical to the previous simulations (Fig. S10).
Binding of the PTEN-like domain leads to a local clustering of PIP2 within the plane of the bilayer (Fig. 5). This can be seen if one examines the average spatial distribution within the upper (i.e., cytoplasmic) bilayer leaflet, where hotspots of PIP2 distribution (averaged over the duration of the AT simulation) can be seen in the vicinity of both the PD and C2 domains (Fig. 5 A). About 10 of the total ∼30 PIP2 molecules in the upper leaflet formed such interactions. In contrast, there is no evidence for any such clustering of PIP2 in the opposite leaflet, which is not in contact with the protein (Fig. S11, A and B). This clustering of the PIP2 lipids is also evident from the radial distribution functions of the two lipid species around the protein (Fig. 5 C and Fig. S11 C). In particular, it seems that the following interaction with the PTEN-like domain, the PIP2 molecule remains bound for the entire duration of the AT-MD simulation. This local clustering of PIP in response to interaction of binding of a periplasmic protein domain is of interest in relation to, e.g., recent combined experimental and simulation studies of PIP2-clustering (to form ∼75 nm microdomains) in PC12 cell and in liposome membranes in response to the SNARE protein syntaxin-1A (43).
Figure 5.
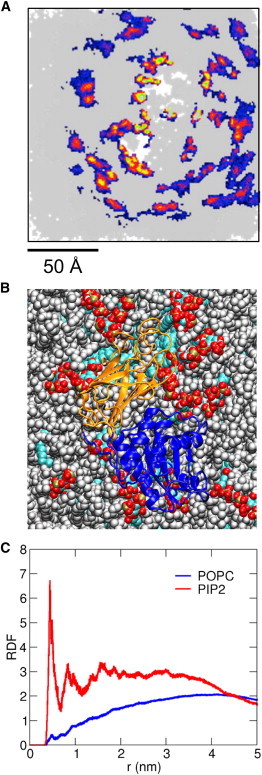
(A) Spatial distribution density in the bilayer plane of the PIP2 headgroups, from the atomistic simulations of the auxilin-1 PTEN-like domain bound to a 15% PIP2/85% POPC bilayer (PIP2-AT; see Table 1). This shows the upper (i.e., cytoplasmic) leaflet to which the auxilin PTEN-like domain was bound. For this analysis all simulation frames were fitted using the protein as a reference structure and the positions of the PIP2 lipid headgroups was calculated for the whole duration (60 ns × 4) of the four simulations. The density of the lipid headgroups is colored from blue (low) through red to green (high). The white regions indicate the footprint of the protein on the lipid bilayer surface. (B) Snapshot from the end of an atomistic simulation in the presence of 15% PIP2 showing the protein (PD domain in blue, C2 domain in orange), PIP2 molecules (red, white, bronze, and cyan) and POPC lipids (gray). (C) The lipid radial distribution functions (for PIP2, red, and POPC, blue) for the same PIP2-AT simulation as in B, calculated around the auxilin-1 PTEN-like domain.
Interactions of auxilin-1 PTEN-like domain with membranes that contain PIP3 lipids
Our results highlight the crucial role of the PIP2 in regulating the auxilin-1 PTEN-like domain orientation relative to the membrane. To examine the role of PIPs further, a bilayer with 15% PIP3 lipids were constructed. Although PIP3 occurs at a very low frequency within cell membranes (the overall concentration of PIPs in the plasma membrane is ∼5%, comprised mainly of PIP2 and PIP3 (25)), from a more methodological perspective it allows us to examine the effects on auxilin-1 PTEN-like domain interactions with the bilayer in an environment with increased specificity in the bilayer. The presence of PIP3 among the lipids again resulted in binding of protein to the bilayer (albeit to a lesser extent compared to PIP2). Thus, in the presence of 15% PIP3 6 out of 10 simulations (each of duration 1.5 μs) resulted in the PTEN-like domain binding to the membrane suggesting a somewhat lower relative affinity to the lipid bilayer (Fig. S12 A). The final orientation of the protein was identical to the orientation of the protein proposed by the simulations with the PIP2 lipids (Fig. S12 E and Fig. S13 A). Calculation of the angle between the plane of the protein and the bilayer again revealed a rotation of the protein in 4 out of the 6 simulations in which an auxilin/bilayer complex was formed (Fig. S5 and Fig. S12 B). Similar to the simulations with PIP2 lipids, calculation of the contacts between the protein and the bilayer revealed a preference of auxilin-1 PTEN-like domain to interact with the negatively charged PIP3 lipids (Fig. S13, B and C). The same areas of the protein also made the largest number of contacts with the lipids in the bilayer (Fig. S13 A). The AT simulations with the PIP3 lipids revealed similar auxilin/lipids interactions surface and clustering around the C2 and the PD domain was also observed (see above and Fig. S14).
As for our simulations with PIP2 lipids in the bilayer, for PIP3-containing bilayers we examined the effect of the C2 loop 3 triple mutant R301E/R307E/K311E (Fig. S2 B; Table 1, and mutPIP3). In the mutPIP3 simulations, auxilin associated with the bilayer in only 3 out of 10 simulations (Fig. S12 C). Calculation of the orientation of the protein relative to the bilayer suggested that the mutant auxilin was associated with the bilayer in a perturbed orientation, which was the same as that seen in the mutPIP2 simulations (Fig. S5 and Fig. S12 D). Thus, auxilin interacted with the bilayer via its other positively charged surface (PD: region 3 in Fig. 1 B; Fig. S12 F and Fig. S13 A). This further supports our previous observation that the C2 loop (i.e., and in particular the loop 3 residues 301–311) are crucial in orienting auxilin-1 PTEN-like domain relative to a PIP-containing membrane.
Interactions of auxilin-1 PTEN-like domain with zwitterionic membranes
We further explored the specificity of the auixilin-1 PTEN-like domain interaction with bilayers by varying the lipid composition of the bilayer. If only a zwitterionic lipid (i.e., POPC) was present, the PTEN-like domain remained in the aqueous region throughout each of 10 simulations without binding to the bilayer (Fig. S15 A). If an anionic lipid was present (e.g., 15% 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS)) again no binding interactions between the protein and the bilayer were observed. If the fraction of POPS was increased to 30% or 60% (which is above levels likely to be present in cell membranes; see Fig. S15) only 3 or 4 out of 10 simulations yielded an auxilin/bilayer complex, indicative of a weakened auxilin/bilayer interaction than is the case for bilayers containing PIP2 (Fig. 6). It must be noted that in the PS simulations, which yielded an auxilin/bilayer complex, the auxilin interacted with the bilayer with mainly the same regions of the C2 domain as seen for the PIP2 simulations (see Fig. S15, E and F, and Fig. S16, E and F). The PD domain made some transient interactions with the lipids. In this case, however, calculation of the radial distribution functions of the different lipids did not reveal any significant clustering of the POPS lipids around the protein (Fig. S16, A–D). Significantly, PS does not lead to significant auxilin-1 PTEN-like domain binding to membranes in experimental studies (7). This suggests that PIPs are needed for strong interactions of auxilin-1 PTEN-like domain with a membrane, and that clustering of PIP-lipids plays a key part in these interactions. Other proteins with C2 domains have also been shown to interact specifically with PIP2 (44).
Figure 6.
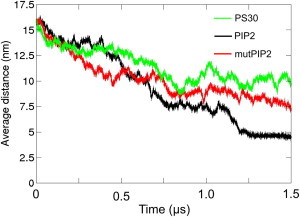
Average distances of the systems with the WT form of the auxilin-1 PTEN-like domain with 15% PIP2 (PIP2, black), the mutant form of the auxilin-1 PTEN-like domain with 15% PIP2 (mutPIP2, red), and the WT form of the auxilin-1 PTEN-like domain with 30% POPS lipids (PS30, green). The average distances were calculated as the ensemble average of the separation between the centers of mass of the protein and the bilayer for all 10 repeats for each system. All distances for the 10 repeats of each system can be found in the Supporting Material Fig. S3 and Fig. S15.
Methodological considerations
It is useful to compare the methodology in the current study with that in a recent simulation study of PTEN/bilayer interactions (12). In the PTEN study, a PIP3 molecule was docked at the active site of the enzyme (based on coordinates for tartrate in the crystal structure) and CG-MD simulations were used to self-assemble a bilayer around the protein-PIP3 complex. In contrast, in the current study, we started with a preformed bilayer (which included either PS or PIP2 or PIP3 as the anionic lipid component). The protein was not docked onto this bilayer but rather CG-MD simulations were used to explore the encounter with and subsequent binding to the membrane of the protein. Subsequent AT-MD simulations are used to help define the nature of interactions with and clustering of PIP2 around the bound protein. Thus, the current approach both relaxes the starting assumptions as to the mode of interaction with the membrane, and also can provide mechanistic details on the encounter process.
Proposed model for auxilin-1 PTEN-like domain interaction with the membrane
Overall, our study provides direct computational support for a model of binding of the auxilin-1 PTEN-like domain to PIP-containing membranes. In this model the auxilin-1 PTEN-like domain binds initially to the bilayer in a nonspecific manner, and subsequently rotates to form a more preferred interaction. This reorientation is regulated by the interactions of the C2 loop 1 with anionic lipids. The presence of PIP2 significantly enhances this process. After the reorientation of the protein, interactions of the binding loops on the PTEN-like domain with negatively charged PIP headgroups result in the formation of nanoclusters of PIP molecules in the adjacent bilayer leaflet. This extends previous observations of such nanoclustering of lipids induced by simple transmembrane proteins (43); and is of interest in the context of suggested roles of membrane localization in regulation of PTEN activity (45,46).
Acknowledgments
This research was funded by the Wellcome Trust. G.M. acknowledges support from the Nuffield foundation.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References
- 1.Kirchhausen T. Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 2009;19:596–605. doi: 10.1016/j.tcb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 3.Massol R.H., Boll W., Kirchhausen T. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl. Acad. Sci. USA. 2006;103:10265–10270. doi: 10.1073/pnas.0603369103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee D.W., Wu X., Greene L.E. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell Sci. 2006;119:3502–3512. doi: 10.1242/jcs.03092. [DOI] [PubMed] [Google Scholar]
- 5.Schlossman D.M., Schmid S.L., Rothman J.E. An enzyme that removes clathrin coats: purification of an uncoating ATPase. J. Cell Biol. 1984;99:723–733. doi: 10.1083/jcb.99.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ungewickell E., Ungewickell H., Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 7.Guan R., Dai H., Kirchhausen T. Structure of the PTEN-like region of auxilin, a detector of clathrin-coated vesicle budding. Structure. 2010;18:1191–1198. doi: 10.1016/j.str.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J.O., Yang H., Pavletich N.P. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 9.Nalefski E.A., Falke J.J. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizo J., Südhof T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 11.Murray D., Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol. Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 12.Lumb C.N., Sansom M.S.P. Defining the membrane-associated state of the PTEN tumor suppressor protein. Biophys. J. 2013;104:613–621. doi: 10.1016/j.bpj.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenoy S., Shekhar P., Lösche M. Membrane association of the PTEN tumor suppressor: molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS ONE. 2012;7:e32591. doi: 10.1371/journal.pone.0032591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stansfeld P.J., Sansom M.S.P. Molecular simulation approaches to membrane proteins. Structure. 2011;19:1562–1572. doi: 10.1016/j.str.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Jaud S., Tobias D.J., White S.H. Self-induced docking site of a deeply embedded peripheral membrane protein. Biophys. J. 2007;92:517–524. doi: 10.1529/biophysj.106.090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Psachoulia E., Sansom M.S.P. Interactions of the pleckstrin homology domain with phosphatidylinositol phosphate and membranes: characterization via molecular dynamics simulations. Biochemistry. 2008;47:4211–4220. doi: 10.1021/bi702319k. [DOI] [PubMed] [Google Scholar]
- 17.Psachoulia E., Sansom M.S.P. PX- and FYVE-mediated interactions with membranes: simulation studies. Biochemistry. 2009;48:5090–5095. doi: 10.1021/bi900435m. [DOI] [PubMed] [Google Scholar]
- 18.Arcario M.J., Ohkubo Y.Z., Tajkhorshid E. Capturing spontaneous partitioning of peripheral proteins using a biphasic membrane-mimetic model. J. Phys. Chem. B. 2011;115:7029–7037. doi: 10.1021/jp109631y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lumb C.N., He J., Sansom M.S.P. Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain. Structure. 2011;19:1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalli A.C., Wegener K.L., Sansom M.S.P. The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure. 2010;18:1280–1288. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stansfeld P.J., Sansom M.S.P. From coarse-grained to atomistic: a serial multi-scale approach to membrane protein simulations. J. Chem. Theory Comput. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 22.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 23.Monticelli L., Kandasamy S.K., Marrink S.J. The MARTINI coarse grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 24.Fiser A., Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 25.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 27.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 28.Rotkiewicz P., Skolnick J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J. Comput. Chem. 2008;29:1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Eswar N., Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Prot. Protein Sci. 2007 doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 31.Scott W.R.P., Hunenberger P.H., van Gunsteren W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A. 1999;103:3596–3607. [Google Scholar]
- 32.Parrinello M., Rahman A. Polymorphic transitions in single-crystals - a new molecular-dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 33.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 34.Darden T., York D., Pedersen L. Particle mesh Ewald - an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 35.Van Der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 37.Bai J., Tucker W.C., Chapman E.R. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 38.Das S., Dixon J.E., Cho W.W. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. USA. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn R., Klos D.A., Hicke L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 2004;165:135–144. doi: 10.1083/jcb.200309026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frazier A.A., Wisner M.A., Cafiso D.S. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 41.Kohout S.C., Corbalán-García S., Falke J.J. C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho W., Stahelin R.V. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 43.van den Bogaart G., Meyenberg K., Jahn R. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montaville P., Coudevylle N., Becker S. The PIP2 binding mode of the C2 domains of rabphilin-3A. Protein Sci. 2008;17:1025–1034. doi: 10.1110/ps.073326608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koldsö H., Sansom M.S.P. Local lipid reorganization by a transmembrane protein domain. J. Phys. Chem. Lett. 2012;3:3498–3502. doi: 10.1021/jz301570w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao X., Lowry P.R., Zhang J. PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proc. Natl. Acad. Sci. USA. 2011;108:14509–14514. doi: 10.1073/pnas.1019386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLano, W. L. 2002. The PyMOL Molecular Graphics System. http://www.pymol.org. Accessed April 30, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


