Abstract
The needs of a growing human population require rapid and efficient development of improved cultivars by plant breeders. The doubled haploid (DH) technology enables generating completely homozygous lines in a single step and, thus, is central to modern genetics and breeding approaches. Rapid and reliable identification of seeds with a haploid embryo after in vivo haploid induction is elementary in the method utilized in maize but current systems have severe shortcomings preventing their use in many germplasm types. Here, we describe an alternative method for discrimination of haploid from diploid seeds based on differences in their oil content stemming from pollination with high oil inducers. After presenting some fundamental theory, we provide a proof-of-concept with experimental results, demonstrating acceptable error rates across different germplasm. Our approach represents a breakthrough in DH technology in maize, because it is amenable to automated high-throughput screening and applicable to any maize germplasm worldwide.
The increasing human population in combination with the expected climate change require efficient breeding strategies that especially maximize the selection gain per time unit. Doubled haploids (DH) have replaced the conventional method of producing homozygous lines from heterozygous source germplasm by recurrent selfing or sib-mating in many crop species1. Above all, the DH technology accelerates line development and warrants complete homozygosity already at the start of the selection process (Fig. 1a). In addition, it offers several quantitative-genetic, operational, logistic and economic advantages2,3,4, because the entire genetic variance is available from the very beginning and genetically fixed candidates can be multiplied and tested ad libitum.
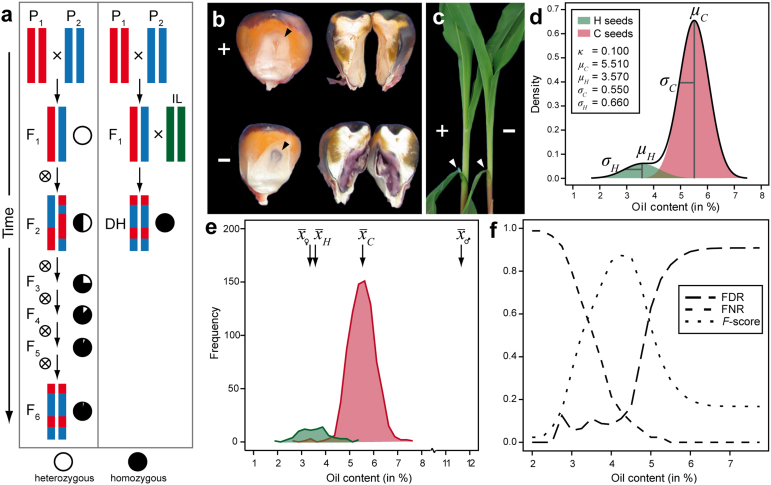
Figure 1. Haploid identification in maize based on oil content.
(a) Schematic representation of obtaining homozygous lines by recurrent selfing as opposed to doubled haploid technology employing an inducer line (IL). (b) R1-nj embryo marker. (c) ligueleless marker. (d) Probability density function for oil content of haploid (H) seeds and diploid crossing (C) seeds and the mixture distribution of all seeds produced by pollinating a source germplasm with pollen from a high oil (HO) inducer. (e) Frequency distribution of oil content of 919 seeds of haploid plants and diploid crossing plants from induction cross (S072 × P213) × UH601, using visual scoring of the phenotype in the field as “gold standard” for classification of H and C seeds. (f) False discovery rate (FDR), false negative rate (FNR), and F-score in classification of seeds from this cross into haploid and diploid crossing seeds based on their oil content as a function of the threshold t (H: OC < t; C: OC ≥ t). Visual scoring of the phenotype in the field was used as a “gold standard” for classification.
In maize, DH lines are commonly produced by in vivo maternal haploid induction5,6. After pollinating the source germplasm with pollen of current inducers, a fraction of 8 to 10% of the developing seeds have a haploid embryo. At present, discrimination of haploid (H) seeds from diploid F1 crossing (C) seeds resulting from normal cross-fertilization relies largely on the dominantly inherited marker gene R1-nj carried by the inducer (Fig. 1b). This gene causes purple coloration of the scutellum and aleurone of C seeds7,8. Thus, it can be used as an embryo– and endosperm-specific marker to distinguish H from C seeds as well as seeds originating from unintended outcrossing6.
Use of the R1-nj marker system for haploid detection in combination with colchicine treatment of haploid seedlings for chromosome duplication have paved the way for routine application of the DH technology in private and public maize breeding programs during the past decade9,10,11. Nevertheless, visual scoring of the R1-nj marker expression is very labor intensive and has so far not been amenable to automation. Moreover, variable color intensity of the embryo marker hampers an unequivocal classification of seeds. The classification system fails entirely if the source germplasm harbors inhibitor alleles such as C1-l12, which suppress expression of the R1-nj marker13. Owing to these shortcomings the DH technology can currently not be applied in many breeding programs that contribute to securing worldwide food production.
These limitations can be overcome by a novel system for discriminating H and C seeds. It is based on their oil content (OC) after pollinating the source germplasm with an inducer distinguished by a high OC in the seeds, subsequently referred to as high oil (HO) inducer. We present the theoretical foundation of this approach and provide a proof-of-concept by numerical examples from four induction crosses with two newly released HO inducers. Further, we discuss the prerequisites and prospects for developing this approach into a high-throughput technique for rapid identification of haploids in maize and its future use in applied maize breeding and basic research.
Results
Theory - errors in classification of seeds after haploid induction
By pollinating a source germplasm as a maternal parent with pollen from an inducer having a haploid induction rate (HIR) κ, one obtains a mixture population of seeds: (i) a fraction κ of H seeds with haploid embryo resulting either from parthenogenesis or chromosome elimination, and (ii) a fraction (1 − κ) of C seeds with a diploid embryo resulting from a normal fertilization event with pollen from the inducer4. For distinguishing between H and C seeds, a classification test T is applied with four possible outcomes (Supplementary Fig. 1), where “T” denotes the test result and “D” the true status of the sample: (i) true positive, where T = H and D = H; (ii) false negative, where T = C and D = H; (iii) false positive, where T = H and D = C; and (iv) true negative, where T = C and D = C.
Adopting standard terminology in the statistical literature, we define two criteria:
False discovery rate (FDR) Q refers to the conditional probability of a truly negative sample when the result of the test T is positive. In our context, Q refers to the proportion of C seeds in the total fraction of seeds classified as H seed, based on the test T. Thus,
where FP and TP denote the number of false positive and true positive test results, respectively.
False negative rate (FNR) β refers to the conditional probability of a negative test result when the true status is positive. In our case, β refers to the proportion of H seeds misclassified as C seeds. Thus,
where FN denotes the number of false negative test results.
Provided a reference method (“gold standard”) for ascertaining the true status of the seeds is available, Eqns. (1) and (2) can be used to determine the FDR and FNR for classification based on the R1-nj marker or any other system.
Theory - application to classification based on seed oil content
Here, we propose an alternative test T based on the oil content (OC) of seeds produced by pollinating the source germplasm with a HO inducer. The seeds harvested from the maternal parent are classified as H or C depending on whether their OC is below or above a predefined threshold t.
Suppose the inducer has a haploid induction rate κ and is used to pollinate plants of the source germplasm. Bulked seeds harvested from the ears of different plants of the cross follow a mixture distribution composed of two fractions (Fig. 1d):
a fraction κ of H seeds displaying a Gaussian normal distribution for OC with
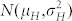 ;
; a fraction (1 − κ) of C seeds displaying a Gaussian normal distribution for OC with
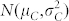 .
.
Thus, the probability density function of all seeds is given by
where 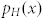 and
and 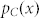 refer to the probability density functions of the Gaussian normal distributions
refer to the probability density functions of the Gaussian normal distributions 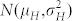 and
and 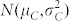 , respectively.
, respectively.
With our alternative test T, seeds from the mixture distribution are classified as H or C seeds, if their OC is below or above the truncation point t. Hence, the proportion of seeds classified as H seeds is given by
where ϕH(t) and ϕC(t) are the cumulative distribution functions of 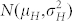 and
and 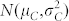 , respectively.
, respectively.
For the FDR Q, we obtain
For the FNR β, we obtain
which does not depend on κ.
Eqns. (5) and (6) show that the FDR and FNR depend on the truncation point t. A low value of t results in a low FDR but a high FNR. On the contrary, a high value of t yields a low FNR but a high FDR. For choosing an appropriate value of t that balances the FDR and FNR, one can use the F-score14 as a measure for the performance of the test T:
Graphs for the FDR, FNR, and F-score are presented for κ = 0.100, 0.050, 0.025 as a function of the threshold t (Supplementary Fig. 2) or the proportion α of selected seeds (Supplementary Fig. 3). The values of 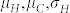 and
and 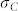 are taken from an experiment with induction cross (S072 × P213) × UH601 described in the next paragraph.
are taken from an experiment with induction cross (S072 × P213) × UH601 described in the next paragraph.
Numerical example – selection of haploid seeds based on oil content
We applied our novel classification test to four source germplasm, the mean OC (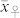 ) of which ranged between 3.35% and 4.15% and was substantially lower (P < 0.001) than that of the HO inducers (
) of which ranged between 3.35% and 4.15% and was substantially lower (P < 0.001) than that of the HO inducers (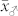 ) (Supplementary Table 1). The mean OC of the H seeds
) (Supplementary Table 1). The mean OC of the H seeds 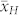 was in relative terms about 10% higher than
was in relative terms about 10% higher than 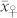 but 25 to 35% lower than the mean OC of the C seeds
but 25 to 35% lower than the mean OC of the C seeds 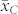 in all four crosses. The H seeds showed consistently larger phenotypic variance for OC than the C seeds.
in all four crosses. The H seeds showed consistently larger phenotypic variance for OC than the C seeds.
The number of seeds classified as H or C based on the two classification systems and the “gold standard” (Supplementary Table 2) were used for calculating the HIR, FDR and FNR. The estimates of HIR based on the “gold standard” ranged between 8.40 and 15.60% and were in close agreement with the HIR determined from the classification based on OC (Supplementary Table 1). By comparison, the estimated HIR in (PDH3 × PDH8) × UH600 based on the R1-nj marker amounted to 5.01% and was much lower than the HIR determined from the “gold standard”. With the exception of a lower value for this cross, the cross-specific thresholds t were chosen such that they exceeded the mean 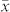 of the respective source germplasm by more than 31 to 38% in relative terms.
of the respective source germplasm by more than 31 to 38% in relative terms.
Classification of H vs. C seeds based on their OC yielded a FDR between 0.08 and 0.18 and a FNR between 0.10 and 0.30 (Supplementary Table 1), By comparison, classification based on the R1-nj marker system in cross (PDH3 × PDH8) × UH600 resulted in a similar FDR but the FNR was more than twice as high.
The OC of H and C seeds from the cross (S072 × P213) × UH601, classified according to the “gold standard” results, displayed distinct distributions with only a minor overlap (Fig. 1e). Investigation of the FDR, FNR, and F-score as a function of the threshold t revealed only a slight increase in FDR with increasing t in the interval between 3.0% and 4.5% OC. A steep rise was observed for higher values of t (Fig. 1f). In contrast, the FNR declined sharply with increasing values of t between 3.0 to 4.5% OC and smoothly leveled off for higher values of t. As a consequence, the F-score reached a maximum for t = 4.35%, close to the threshold t = 4.40% chosen beforehand on prior information on the OC of the parents.
Discussion
A first proposal to discriminate haploid from diploid crossing seeds based on oil content was described by Rotarenco et al.15, yet not based on an inducer with increased OC. The H and C seeds resulting from pollination with an inducer having normal OC can generally not be reliably discriminated due to the small differences in their mean OC relative to the large phenotypic variation within each fraction. Thus, the use of a HO inducer for in vivo induction is compulsory for the success of sorting H and C seeds based on their OC. As illustrated by our theoretical and empirical results, the success of our approach depends on four factors discussed in detail below: (1) the HIR of the inducer; (2) the difference in the mean OC of H and C seeds; (3) the phenotypic variance of OC among seeds within each of these two seed fractions, and (4) the choice of an appropriate threshold t for discriminating putative H and C seeds.
The HIR κ of the inducer reflects the relative weight of the density functions for H and C seeds in the mixture distribution for OC of all seeds (Fig. 1d). For small values of κ, the left tail of the distribution of C seeds outweighs the entire distribution of H seeds unless the means μH and μC differ substantially in comparison to σH and σC. Consequently, even for a rather stringent choice of the threshold t, the FDR remains at a high level. Thus, the maximum F-score achievable by test T is limited by κ. For example, under the assumptions underlying Figure 1d, the maximum F-score is 0.87 for κ = 0.10, but drops to 0.84 and 0.80 for κ = 0.05 and κ = 0.025, respectively (Supplementary Fig. 2). This implies that the HIR of a HO inducer should be at least 5% for a sufficient control of the FDR and for achieving a high F-score.
The standardized difference 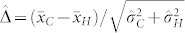 largely determines the amount of overlap between the OC distributions of H and C seeds. In our study,
largely determines the amount of overlap between the OC distributions of H and C seeds. In our study, 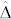 exceeded 1.80 in all induction crosses but (PDH3 × PDH8) × UH600, which had a lower value. However, as illustrated by the relatively high FDR and FNR in the latter cross,
exceeded 1.80 in all induction crosses but (PDH3 × PDH8) × UH600, which had a lower value. However, as illustrated by the relatively high FDR and FNR in the latter cross,  seems necessary and sufficient to reach acceptable levels for both criteria with a HIR of 10%. Obviously, the larger
seems necessary and sufficient to reach acceptable levels for both criteria with a HIR of 10%. Obviously, the larger 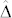 is, the smaller the overlap, and the more successful our approach is likely to be (Supplementary Fig. 2,3). It depends on (i)
is, the smaller the overlap, and the more successful our approach is likely to be (Supplementary Fig. 2,3). It depends on (i) 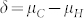 , the difference in the means between both groups, and (ii) the magnitude of variation within each group. The difference δ is mainly determined by the values
, the difference in the means between both groups, and (ii) the magnitude of variation within each group. The difference δ is mainly determined by the values 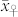 and
and 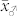 of the parents of the induction cross. In the absence of non-additive gene action and maternal effects, one expects
of the parents of the induction cross. In the absence of non-additive gene action and maternal effects, one expects 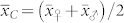 and
and 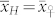 , provided the ploidy level has no influence on the OC of seeds. Hence,
, provided the ploidy level has no influence on the OC of seeds. Hence, 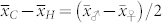 and
and 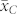 should lie midway between
should lie midway between 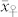 and
and 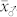 . Most studies in the literature reported predominantly additive gene action for OC in maize16,17,18,19. In all induction crosses with UH600, however,
. Most studies in the literature reported predominantly additive gene action for OC in maize16,17,18,19. In all induction crosses with UH600, however, 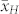 was significantly higher than
was significantly higher than 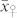 , and
, and 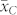 was lower than
was lower than 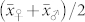 . Poor seed set and negative effects on endosperm development after fertilization with pollen from inducers described by Xu et al.20 might be an explanation for the discrepancies between observed and expected values of
. Poor seed set and negative effects on endosperm development after fertilization with pollen from inducers described by Xu et al.20 might be an explanation for the discrepancies between observed and expected values of 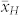 and
and 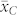 . Evaluation of a large number of induction crosses is warranted to investigate whether the OC of the source germplasm and inducer can provide sufficiently precise approximations of
. Evaluation of a large number of induction crosses is warranted to investigate whether the OC of the source germplasm and inducer can provide sufficiently precise approximations of 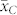 and
and 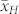 and thus serve as an estimate of
and thus serve as an estimate of 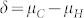 before sorting of H and C seeds.
before sorting of H and C seeds.
Large phenotypic variances 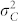 and
and 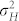 compared with the difference
compared with the difference 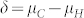 have a negative effect on the FDR and FNR, because this leads to a more pronounced overlap of the density functions of H and C seeds in the mixture distribution of all seeds (Fig. 1d). Both genetic and non-genetic factors contribute to these variances. The genetic component is expected to be higher among H seeds, because they display the full additive genetic variance released from the source germplasm. While in our numerical examples all source germplasm were single crosses and, thus, genetically uniform, information about
have a negative effect on the FDR and FNR, because this leads to a more pronounced overlap of the density functions of H and C seeds in the mixture distribution of all seeds (Fig. 1d). Both genetic and non-genetic factors contribute to these variances. The genetic component is expected to be higher among H seeds, because they display the full additive genetic variance released from the source germplasm. While in our numerical examples all source germplasm were single crosses and, thus, genetically uniform, information about 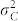 and
and 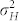 is also required for genetically heterogeneous source germplasm such as landraces or gene bank accessions. This is because the DH method holds great promise for the conservation and exploitation of genetic resources21. Furthermore, the use of HO inducers promises to overcome the previous limitations of the R1-nj marker system for such materials. Actually, we even envision to employ the OC marker for inducing DH lines in teosinte (Zea mays mexicana L.) and other species crossable with maize, because it relies exclusively on the OC of seeds and not on any other properties of the embryo or endosperm.
is also required for genetically heterogeneous source germplasm such as landraces or gene bank accessions. This is because the DH method holds great promise for the conservation and exploitation of genetic resources21. Furthermore, the use of HO inducers promises to overcome the previous limitations of the R1-nj marker system for such materials. Actually, we even envision to employ the OC marker for inducing DH lines in teosinte (Zea mays mexicana L.) and other species crossable with maize, because it relies exclusively on the OC of seeds and not on any other properties of the embryo or endosperm.
Choice of the threshold t for distinguishing H from C seeds should balance the FDR and FNR. To achieve this goal, one can use our theoretical results in Eq. (5) to (6) together with the empirical estimates of 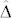 to determine the value t* that maximizes the F-score numerically. Since the latter has a flat curve as a function of t in the vicinity of t*, the experimenter might prefer to choose t somewhat more stringent than t* so that the FDR is reduced at the expense of a higher value of FNR, because carrying a false positive to the next step of the DH process is more expensive than discarding a false negative at this early stage. This compromise could be offset by using a greater number of plants of the source germplasm in the isolation plot for producing the induction crosses so that the desired number of putative H seeds can still be achieved. An alternative criterion for choosing the threshold t could be to minimize the costs of producing a desired number of H plants.
to determine the value t* that maximizes the F-score numerically. Since the latter has a flat curve as a function of t in the vicinity of t*, the experimenter might prefer to choose t somewhat more stringent than t* so that the FDR is reduced at the expense of a higher value of FNR, because carrying a false positive to the next step of the DH process is more expensive than discarding a false negative at this early stage. This compromise could be offset by using a greater number of plants of the source germplasm in the isolation plot for producing the induction crosses so that the desired number of putative H seeds can still be achieved. An alternative criterion for choosing the threshold t could be to minimize the costs of producing a desired number of H plants.
As pointed out before, the size of 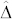 is of crucial importance to distinguish H from C seeds by their OC. While the reduction in
is of crucial importance to distinguish H from C seeds by their OC. While the reduction in 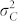 and
and 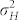 is limited by their corresponding genetic components, the size of
is limited by their corresponding genetic components, the size of 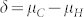 depends to a large extent on the OC
depends to a large extent on the OC 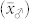 of the inducer. For maize germplasm with normal OC, the HO inducers UH600 and UH601 with an OC of 9.9 and 11.6%, respectively, warrant
of the inducer. For maize germplasm with normal OC, the HO inducers UH600 and UH601 with an OC of 9.9 and 11.6%, respectively, warrant 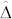 values exceeding 1.8. This allows a suitable choice of t to discriminate H and C seeds with acceptable levels of FDR and FNR. Nevertheless, since specially bred maize germplasm such as the Illinois High Oil populations22 have OC above 20%, a further substantial increase in OC of inducers without compromising the HIR seems realistic. If HO inducers with HIR of 10% and OC of 15% are available, reliable sorting of H and C seeds should be possible even for source germplasm with an OC as high as 7%, which is the upper limit of OC found in normal maize. In addition to a breakthrough in sorting seeds produced by in vivo haploid induction, the OC marker system also offers substantial advantages for maintenance and de novo breeding of inducers.
values exceeding 1.8. This allows a suitable choice of t to discriminate H and C seeds with acceptable levels of FDR and FNR. Nevertheless, since specially bred maize germplasm such as the Illinois High Oil populations22 have OC above 20%, a further substantial increase in OC of inducers without compromising the HIR seems realistic. If HO inducers with HIR of 10% and OC of 15% are available, reliable sorting of H and C seeds should be possible even for source germplasm with an OC as high as 7%, which is the upper limit of OC found in normal maize. In addition to a breakthrough in sorting seeds produced by in vivo haploid induction, the OC marker system also offers substantial advantages for maintenance and de novo breeding of inducers.
To gain wide acceptance in practice, our approach will depend heavily on high-throughput platforms for determining OC of individual seeds. With an OC between 10 and 12% in the newly released HO inducers, the degree of precision required for discriminating between H and C seeds can at present only be achieved with highly repeatable NMR measurements in combination with a high precision balance. Using these components, we are currently developing a high throughput system for OC measurements with a rate of one seed every three seconds.
In conclusion, this study provides a convincing proof-of-concept and demonstrates the tremendous potential of our approach for haploid seed identification based on HO inducers. This approach can serve as a more widely applicable, reliable high-throughput system for DH production that promises to accelerate genomic approaches and to revolutionize maize breeding in germplasm pools thus far not amenable to DH technology.
Methods
Two new HO inducer lines, UH600 and UH601 developed by the maize breeding program of the University of Hohenheim (https://plant-breeding.uni-hohenheim.de), were used for this proof-of-concept. In addition to a high OC of seeds, both inducers express the R1-nj marker in the embryo and endosperm. Inducer UH600 with 9.91% OC in the seeds was used as a pollen parent in the nursery 2009 in Eckartsweier, Germany, to pollinate three source germplasm: the single (2 W) cross PDH3 × PDH8 between two dent lines carrying the recessive liguleless gene (lg), the 2 W cross F103 × F087 between two European flint lines, and the 2 W cross P204 × P211 between two European Iodent lines. Inducer UH601 with 11.6% OC served in the winter nursery 2011/2012 in Chile as a pollinator for the 2 W cross S072 × P213 between a European Stiff Stalk and an Iodent line. After harvest and discarding embryoless seeds based on visual scoring, individual seeds from all four induction crosses, ranging in total between 1003 and 8709 seeds, as well as 100 seeds from each inducer and each parent line of the four 2 W crosses were measured for OC by nuclear magnetic resonance (NMR) according to manufacturer's instructions (Bruker BioSpin GmbH, Rheinstetten, Germany).
Classification into putative H vs. C seeds based on their OC was performed by choosing a cross-specific threshold t between 4.40% and 5.50% OC using prior information about the OC of the source germplasm and the inducer as well as our theoretical results. In the cross (PDH3 × PDH8) × UH600, seeds were additionally classified as white (W) or purple (P) based on expression of the R1-nj embryo marker.
For determination of the true status (H vs. C) of the seeds, we used two test systems as a “gold standard”. In the induction cross (PDH3 × PDH8) × UH600, seeds were grown in the greenhouse and each plantlet was scored in the 3rd leaf stage for absence or presence of the liguleless (lg) phenotype to infer whether it had originated from an H or C seed, respectively. For all other induction crosses, seeds were sown in the nursery and the corresponding plants were visually scored at flowering time for the H or C phenotype: compared to the C phenotype, the H phenotype is characterized by shorter stature, erect and narrow leaves, as well as reduced growth and fertility, as reflected particularly by the number of anthers produced per tassel. For the induction cross (S072 × P213) × UH601, individually tagged seeds were planted for associating their OC directly with the phenotype of the respective plant. For the remaining two crosses, all seeds with an OC below the cross-specific threshold t and a random subset of about 200 seeds with OC above the threshold t were grown as separate bulks.
Based on the results for the “gold standard”, the FDR and FNR for classification based on (i) the OC of the seed or (ii) the R1-nj marker expression in the embryo were calculated as described in the theory section. The number of germinated plants in each category was used as a reference basis. In addition to the HIR determined from the “gold standard” results, the estimates of HIR were also calculated for each classification method. The number of putative H seeds classified on the basis of their OC or R1-nj marker expression was divided by the total number of H plus C seeds with necessary adjustments, if only a random subset of the putative C seeds was tested for false negatives.
Supplementary Material
Melchinger et al Supplementary Information
Acknowledgments
We are grateful to the technical staff of the maize breeding group at the University of Hohenheim for their dedicated work in the field trials. We thank Dr. Vanessa Prigge for valuable suggestions in preparing the manuscript. Funding in support of this research came from the foundation “Fiat panis”, Ulm, Germany, and from the BILL & MELINDA GATES Foundation under the grant ”A Double Haploid Facility for Strengthening Maize Breeding Programs in Africa”.
Footnotes
Author Contributions A.E.M. developed the theory, W.S. designed and supervised the experiments, S.C. provided ideas and materials, F.T. performed the statistical analysis, and A.E.M., F.T., T.W. wrote the manuscript.
References
- Wedzony M. et al. Progress in doubled haploid technology in higher plants. In: Touraev, A., Forster, B. P., Jain, S. M. (eds) Advances in haploid production in higher plants. Springer Science + Business Media BV, Heidelberg, pp 1–33 (2009).
- Nei M. The efficiency of haploid methods of plant breeding. Heredity 18, 95–100 (1963). [Google Scholar]
- Melchinger A. E., Longin C. F. H., Utz H. F. & Reif J. C. Hybrid maize breeding with doubled haploid lines: quantitative genetic and selection theory for optimum allocation of resources. In: Proc. 41st Annual Illinois Corn Breeders' School 2005. Urbana-Champaign, Illinois, USA. pp 8–21 (2005). [Google Scholar]
- Geiger H. H. Doubled haploids. In: Bennetzen, J. L., Hake, S. (Eds.), Maize Handbook. Vol. II: Genetics and Genomics. Springer Verlag, Heidelberg, New York, pp. 641–659 (2009).
- Bordes J., Dumas de Vaulx R., Lapierre A. & Pollacsek M. Haplodiploidization of maize (Zea mays L.) through induced gynogenesis assisted by glossy markers and its use in breeding. Agronomie 17, 291–297 (1997). [Google Scholar]
- Prigge V. & Melchinger A. E. Production of haploids and doubled haploids in maize. In: Loyola-Vargas, V. M., Ochoa-Alejo, N. (eds.) Plant Cell Culture Protocols. 3rd edition. Humana Press, Totowa, pp 161–172 (2012). [Google Scholar]
- Nanda D. K. & Chase S. S. An embryo marker for detecting monoploids of maize (Zea mays L.). Crop Sci. 6, 213–215 (1966). [Google Scholar]
- Neuffer M. G., Coe E. H. & Wessler S. R. Mutants of maize. CSHL Press, New York (1997). [Google Scholar]
- Schmidt W. Hybridmaizüchtung bei der KWS SAAT AG. In: Bericht über die Arbeitstagung der Vereinigung der Pflanzenzüchter und Saatgutkaufleute Österreichs, Gumpenstein, Österreich, 25–27, Nov. 2003, pp 1–6 (2003).
- Seitz G. The use of doubled haploids in corn breeding. In: Proc. 41st Annual Illinois Corn Breeders' School 2005. Urbana-Champaign, Illinois, USA. pp 1–7 (2005). [Google Scholar]
- Smith J. S. C. et al. Use of doubled haploids in maize breeding: implications for intellectual property protection and genetic diversity in hybrid crops. Mol. Breed. 22, 51–59 (2008). [Google Scholar]
- Paz-Ares J., Ghosal D. & Saedler H. Molecular analysis of the C1-I allele from Zea mays:a dominant mutant of the regulatory C 1 locus. EMBO J. 9, 315–321 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E. H. Spontaneous mutation of the aleurone color inhibitor in maize. Genetics 47, 779–783 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijsbergen C. J. Information retrieval. 2nd ed. London. Butterworths Press, (1979).
- Rotarenco V. A., Kirtoca I. H. & Jacota A. G. Possibility to identify kernels with haploid embryo by oil content. Maize Genet. Newsl. 81, 11 (2007). [Google Scholar]
- Moreno-Gonzalez J., Dudley J. W. & Lambert R. J. A. Design III Study of Linkage Disequilibrium for Percent Oil in Maize. Contribution from the Agronomy Dep., Univ. of Illinois, Urbana, and the Illinois Agric. Exp. Stn. (1975).
- Berke T. G. & Rocheford T. R. Quantitative trait loci for flowering, plant and ear height, and kernel traits in maize. Crop Sci. 35, 1542–1549 (1995). [Google Scholar]
- Môro G. V., Santos M. F., Bento D. A. V., Aguiar A. M. & Lopes de Souza Jr C. Genetic analysis of kernel oil content in tropical maize with design III and QTL mapping. Euphytica 185, 419–428 (2012). [Google Scholar]
- Laurie C. C. et al. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168, 2141–2155 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. et al. Gametophytic and zygotic selection leads to segregation distortion associated with in vivo maternal haploid in maize. J. Exp. Botany 64, 1083–1096 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif J. C. et al. Genetic structure and diversity of European flint maize population determined with SSR analyses of individuals and bulks. Theor. Appl. Genet. 111, 906–913 (2005). [DOI] [PubMed] [Google Scholar]
- Dudley J. W. & Lambert R. J. 100 Generations of selection for oil and protein in corn. Plant Breeding Reviews. Vol. 24, Long-term Selection: Maize, John Wiley & Sons, Inc. (2004).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Melchinger et al Supplementary Information